Abstract
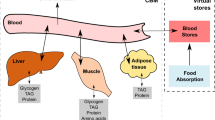
Multiple models of human metabolism have been reconstructed, but each represents only a subset of our knowledge. Here we describe Recon 2, a community-driven, consensus 'metabolic reconstruction', which is the most comprehensive representation of human metabolism that is applicable to computational modeling. Compared with its predecessors, the reconstruction has improved topological and functional features, including ∼2× more reactions and ∼1.7× more unique metabolites. Using Recon 2 we predicted changes in metabolite biomarkers for 49 inborn errors of metabolism with 77% accuracy when compared to experimental data. Mapping metabolomic data and drug information onto Recon 2 demonstrates its potential for integrating and analyzing diverse data types. Using protein expression data, we automatically generated a compendium of 65 cell type–specific models, providing a basis for manual curation or investigation of cell-specific metabolic properties. Recon 2 will facilitate many future biomedical studies and is freely available at http://humanmetabolism.org/.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Palsson, B. Systems biology: properties of reconstructed networks. (Cambridge University Press, 2006).
Thiele, I. & Palsson, B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5, 93–121 (2010).
Oberhardt, M.A., Palsson, B.O. & Papin, J.A. Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol. 5, 320 (2009).
Orth, J.D., Thiele, I. & Palsson, B.O. What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010).
Schellenberger, J. et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 6, 1290–1307 (2011).
Bordbar, A. & Palsson, B.O. Using the reconstructed genome-scale human metabolic network to study physiology and pathology. J. Intern. Med. 271, 131–141 (2012).
Duarte, N.C. et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. USA 104, 1777–1782 (2007).
Sahoo, S., Franzson, L., Jonsson, J.J. & Thiele, I. A compendium of inborn errors of metabolism mapped onto the human metabolic network. Mol. Biosyst. 8, 2545–2558 (2012).
Folger, O. et al. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 7, 501 (2011).
Frezza, C. et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature 477, 225–228 (2011).
Chang, R.L., Xie, L., Bourne, P.E. & Palsson, B.O. Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput. Biol. 6, e1000938 (2010).
Rolfsson, O., Palsson, B.O. & Thiele, I. The human metabolic reconstruction Recon 1 directs hypotheses of novel human metabolic functions. BMC Syst. Biol. 5, 155 (2011).
Rolfsson, O., Paglia, G., Magnusdottir, M., Palsson, B.O. & Thiele, I. Inferring the metabolism of human orphan metabolites from their metabolic network context affirms human gluconokinase activity. Biochem. J. 449, 427–435 (2013).
Bordbar, A., Lewis, N.E., Schellenberger, J., Palsson, B.O. & Jamshidi, N. Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol. Syst. Biol. 6, 422 (2010).
Heinken, A., Sahoo, S., Fleming, R.M. & Thiele, I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes 4, 28–40 (2013).
Stobbe, M.D., Houten, S.M., Jansen, G.A., van Kampen, A.H. & Moerland, P.D. Critical assessment of human metabolic pathway databases: a stepping stone for future integration. BMC Syst. Biol. 5, 165 (2011).
Hao, T., Ma, H.W., Zhao, X.M. & Goryanin, I. Compartmentalization of the Edinburgh Human Metabolic Network. BMC Bioinformatics 11, 393 (2010).
Gille, C. et al. HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol. Syst. Biol. 6, 411 (2010).
Sahoo, S. & Thiele, I. Predicting the impact of diet and enzymopathies on human small intestinal epithelial cells. Human Mol. Genet. (in the press).
Jerby, L., Shlomi, T. & Ruppin, E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol. Syst. Biol. 6, 401 (2010).
McHugh, D.M. et al. Clinical validation of cutoff target ranges in newborn screening of metabolic disorders by tandem mass spectrometry: a worldwide collaborative project. Genet. Med. 13, 230–254 (2011).
Blazier, A.S. & Papin, J.A. Integration of expression data in genome-scale metabolic network reconstructions. Front Physiol. 3, 299 (2012).
Agren, R. et al. Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput. Biol. 8, e1002518 (2012).
Sigurdsson, M.I., Jamshidi, N., Steingrimsson, E., Thiele, I. & Palsson, B.O. A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst. Biol. 4, 140 (2010).
Thiele, I. & Palsson, B.O. Reconstruction annotation jamborees: a community approach to systems biology. Mol. Syst. Biol. 6, 361 (2010).
Herrgard, M.J. et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 26, 1155–1160 (2008).
Heavner, B.D., Smallbone, K., Barker, B., Mendes, P. & Walker, L.P. Yeast 5—an expanded reconstruction of the Saccharomyces cerevisiae metabolic network. BMC Syst. Biol. 6, 55 (2012).
Thiele, I. et al. A community effort towards a knowledge-base and mathematical model of the human pathogen Salmonella Typhimurium LT2. BMC Syst. Biol. 5, 8 (2011).
Kell, D.B. et al. Metabolic footprinting and systems biology: the medium is the message. Nat. Rev. Microbiol. 3, 557–565 (2005).
Wishart, D.S. et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–D906 (2008).
Swainston, N., Smallbone, K., Mendes, P., Kell, D. & Paton, N. The SuBliMinaL Toolbox: automating steps in the reconstruction of metabolic networks. J. Integr. Bioinform. 8, 186 (2011).
Hoppe, A., Hoffmann, S., Gerasch, A., Gille, C. & Holzhutter, H.G. FASIMU: flexible software for flux-balance computation series in large metabolic networks. BMC Bioinformatics 12, 28 (2011).
Jain, M. et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 (2012).
Gudmundsson, S. & Thiele, I. Computationally efficient flux variability analysis. BMC Bioinformatics 11, 489 (2010).
Zelena, E. et al. Development of a robust and repeatable UPLC-MS method for the long-term metabolomic study of human serum. Anal. Chem. 81, 1357–1364 (2009).
Wishart, D.S. et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 37, D603–D610 (2009).
Uhlen, M. et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 (2010).
Shlomi, T., Cabili, M.N., Herrgard, M.J., Palsson, B.O. & Ruppin, E. Network-based prediction of human tissue-specific metabolism. Nat. Biotechnol. 26, 1003–1010 (2008).
Smallbone, K., Simeonidis, E., Broomhead, D.S. & Kell, D.B. Something from nothing: bridging the gap between constraint-based and kinetic modelling. FEBS J. 274, 5576–5585 (2007).
Smallbone, K., Simeonidis, E., Swainston, N. & Mendes, P. Towards a genome-scale kinetic model of cellular metabolism. BMC Syst. Biol. 4, 6 (2010).
Le Novère, N. et al. Minimum information requested in the annotation of biochemical models (MIRIAM). Nat. Biotechnol. 23, 1509–1515 (2005).
Paglia, G. et al. Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS). Anal. Bioanal. Chem. 402, 1183–1198 (2012).
Suhre, K. et al. A genome-wide association study of metabolic traits in human urine. Nat. Genet. 43, 565–569 (2011).
Reed, J.L. et al. Systems approach to refining genome annotation. Proc. Natl. Acad. Sci. USA 103, 17480–17484 (2006).
Wikoff, W.R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 106, 3698–3703 (2009).
Claus, S.P. et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 4, 219 (2008).
Haraldsdottir, H.S., Thiele, I. & Fleming, R.M. Quantitative assignment of reaction directionality in a multicompartmental human metabolic reconstruction. Biophys. J. 102, 1703–1711 (2012).
Thorleifsson, S.G. & Thiele, I. rBioNet: A COBRA toolbox extension for reconstructing high-quality biochemical networks. Bioinformatics 27, 2009–2010 (2011).
Shlomi, T., Cabili, M.N. & Ruppin, E. Predicting metabolic biomarkers of human inborn errors of metabolism. Mol. Syst. Biol. 5, 263 (2009).
Hucka, M. et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19, 524–531 (2003).
Swainston, N. & Mendes, P. libAnnotationSBML: a library for exploiting SBML annotations. Bioinformatics 25, 2292–2293 (2009).
Acknowledgements
I.T. was supported, in part, by a Marie Curie International Reintegration Grant (249261) within the 7th European Community Framework Program. I.T., O.R., S.G.T. and M.A. were supported by the European Research Council grant proposal number 232816. S.S. and H.H. were supported by a Rannis research grant (100406022). Authors from Manchester thank the Biotechnology and Biological Sciences Research Council (BBSRC), and Engineering and Physical Sciences Research Council for their funding of the Manchester Centre for Integrative Systems Biology (grant BB/C008219/1), H.W. for Bioprocessing Research Industry Club grants, and P.M. and N. Swainston for support from the European Union FP7 project UNICELLSYS (grant agreement 201142). The Knut and Alice Wallenberg Foundation supported R.A., S.B. and I.N.; D.J. thanks the BBSRC for funding of Systems Approaches to Biological Research grants BB/F005938 and BB/F00561X. A.H. and C.B. were supported by the German Federal Ministry for Education and Research within the Virtual Liver Network (grant numbers 0315756 and 0315741). A.K.C. and J.A.P. acknowledge funding from the US National Institutes of Health (grant GM088244), National Science Foundation (grant 0643548) and Cystic Fibrosis Research Foundation (grant 1060). N.D.P. was supported by a National Cancer Institute to Independence Award in Cancer Research. I.G. thanks the Science and Technology Facilities Council for Scottish Bioinformatics Research Network funding. M.H. and P.M. thank the US National Institute of General Medical Sciences for support under grants R01GM070923 and R01GM080219. M.D.S. thanks the BioRange programme (project SP1.2.4) of The Netherlands Bioinformatics Centre for support under a Besluit Subsidies Investeringen Kennisinfrastructuur grant through The Netherlands Genomics Initiative.
Author information
Authors and Affiliations
Contributions
I.T. and N. Swainston led the project and developed the reconstruction. I.T., N. Swainston, B.Ø.P., P.M. and D.K. wrote the manuscript. B.Ø.P., D.B.K. and P.M. conceived the project. R.M.T.F. and A.H. performed validation of the reconstruction. S.S. and M.L.M. performed substantial manual curation of the reconstruction. M.K.A., H.H., O.R., M.D.S. and S.G.T. contributed to the analysis of the reconstruction and its models. I.T., N. Swainston, R.M.T.F., A.H., S.S., M.K.A., H.H., M.L.M., O.R., M.D.S., S.G.T., R.A., C.B., S.B., A.K.C., P.D., W.B.D., L.E., D. Hala, M.H., D. Hull, D.J., N. Jamshidi, J.J.J., N. Juty, S.K., I.N., N.L.N., N.M., A.M., J.A.P., N.D.P., E. Selkov, M.I., E. Simeonidis, N. Sonnenschein, K.S., A.S., J.H.G.M.v.B., D.W., I.G., J.N., H.V.W., D.B.K., P.M. and B.Ø.P. attended one or more jamboree meetings and provided manual curation of the reconstruction.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Notes 1–4, Supplementary Figures 1–6 (PDF 2031 kb)
Supplementary Data
Recon 2 and cell type–specific models in SBML format. (ZIP 31200 kb)
Supplementary Table 1
Unbalanced reactions and missing chemical formulae. (XLS 116 kb)
Supplementary Table 2
Metabolic task results. (XLS 380 kb)
Supplementary Table 3
Cancer exometabolome results. (XLS 357 kb)
Supplementary Table 4
Cell-type reactions. (XLS 338 kb)
Supplementary Table 5
IEM information (XLS 22 kb)
Supplementary Table 6
Evidence code (ECO) terms associated with Recon 2 reactions. (XLS 21 kb)
Supplementary Table 7
Modeling constraints. (XLS 793 kb)
Rights and permissions
About this article
Cite this article
Thiele, I., Swainston, N., Fleming, R. et al. A community-driven global reconstruction of human metabolism. Nat Biotechnol 31, 419–425 (2013). https://doi.org/10.1038/nbt.2488
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/nbt.2488
This article is cited by
-
A new dawn beyond lysine ubiquitination
Nature Chemical Biology (2022)
-
Dynamic partitioning of branched-chain amino acids-derived nitrogen supports renal cancer progression
Nature Communications (2022)
-
Argininosuccinate lyase is a metabolic vulnerability in breast development and cancer
npj Systems Biology and Applications (2021)
-
A hexokinase isoenzyme switch in human liver cancer cells promotes lipogenesis and enhances innate immunity
Communications Biology (2021)
-
A dynamic multi-tissue model to study human metabolism
npj Systems Biology and Applications (2021)