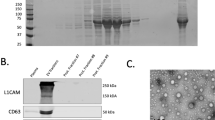
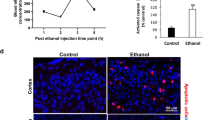
Abstract
Local translation of mRNAs in the synapse has a major role in synaptic structure and function. Chronic alcohol use causes persistent changes in synaptic mRNA expression, possibly mediated by microRNAs localized in the synapse. We profiled the transcriptome of synaptoneurosomes (SN) obtained from the amygdala of mice that consumed 20% ethanol (alcohol) in a 30-day continuous two-bottle choice test to identify the microRNAs that target alcohol-induced mRNAs. SN are membrane vesicles containing pre- and post-synaptic compartments of neurons and astroglia and are a unique model for studying the synaptic transcriptome. We previously showed that chronic alcohol regulates mRNA expression in a coordinated manner. Here, we examine microRNAs and mRNAs from the same samples to define alcohol-responsive synaptic microRNAs and their predicted interactions with targeted mRNAs. The aim of the study was to identify the microRNA–mRNA synaptic interactions that are altered by alcohol. This was accomplished by comparing the effect of alcohol in SN and total homogenate preparations from the same samples. We used a combination of unbiased bioinformatic methods (differential expression, correlation, co-expression, microRNA-mRNA target prediction, co-targeting, and cell type-specific analyses) to identify key alcohol-sensitive microRNAs. Prediction analysis showed that a subset of alcohol-responsive microRNAs was predicted to target many alcohol-responsive mRNAs, providing a bidirectional analysis for identifying microRNA–mRNA interactions. We found microRNAs and mRNAs with overlapping patterns of expression that correlated with alcohol consumption. Cell type-specific analysis revealed that a significant number of alcohol-responsive mRNAs and microRNAs were unique to glutamate neurons and were predicted to target each other. Chronic alcohol consumption appears to perturb the coordinated microRNA regulation of mRNAs in SN, a mechanism that may explain the aberrations in synaptic plasticity affecting the alcoholic brain.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Betel D, Koppal A, Agius P, Sander C, Leslie C (2010). Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites’. Genome Biol 11: R90.
Blednov YA, Mayfield RD, Belknap J, Harris RA (2012). Behavioral actions of alcohol: phenotypic relations from multivariate analysis of mutant mouse data. Genes Brain Behav 11: 424–435.
Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS et al (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28: 264–278.
Chandrasekar V, Dreyer JL (2011). Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 36: 1149–1164.
Edgar R, Domrachev M, Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210.
Eipper-Mains JE, Kiraly DD, Palakodeti D, Mains RE, Eipper BA, Graveley BR (2011). MicroRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA 17: 1529–1543.
Escudeiro SS, Soares PM, Almeida AB, de Freitas Guimaraes Lobato R, de Araujo DP, Macedo DS et al (2013). Antidepressant effect of aminophylline after ethanol exposure. Sci Pharm 81: 211–222.
Gatch MB, Selvig M (2002). Theophylline blocks ethanol withdrawal-induced hyperalgesia. Alcohol Alcohol 37: 313–317.
Gorini G, Nunez YO, Mayfield RD (2013). Integration of miRNA and protein profiling reveals coordinated neuroadaptations in the alcohol-dependent mouse brain. PLoS One 8: e82565.
He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ (2012). Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 73: 35–48.
He Y, Yang C, Kirkmire CM, Wang ZJ (2010). Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J Neurosci 30: 10251–10258.
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19.
Hollingsworth EB, McNeal ET, Burton JL, Williams RJ, Daly JW, Creveling CR (1985). Biochemical characterization of a filtered synaptoneurosome preparation from guinea pig cerebral cortex: cyclic adenosine 3':5'-monophosphate-generating systems, receptors, and enzymes. J Neurosci 5: 2240–2253.
John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004). Human MicroRNA targets. PLoS Biol 2: e363.
Kauffmann A, Gentleman R, Huber W (2009). arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25: 415–416.
Kauffmann A, Huber W (2010). Microarray data quality control improves the detection of differentially expressed genes. Genomics 95: 138–142.
Kolbert CP, Feddersen RM, Rakhshan F, Grill DE, Simon G, Middha S et al (2013). Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. PLoS ONE 8: e52517.
Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M et al (2010). MicroRNA loss enhances learning and memory in mice’. J Neurosci 30: 14835–14842.
Langfelder P, Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559.
Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD (2011). Up-regulation of microRNAs in brain of human alcoholics. Alcohol Clin Exp Res 35: 1928–1937.
Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR (2005). Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem 94: 896–905.
Lugli G, Torvik VI, Larson J, Smalheiser NR (2008). Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem 106: 650–661.
Maciotta S, Meregalli M, Torrente Y (2013). The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci 7: 265.
Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA (2002). Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem 81: 802–813.
Mayfield RD, Nunez YO (2012). Understanding alcoholism through microRNA signatures in brains of human alcoholics. Front Genetics 3: 43.
Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C et al (2014). Evaluation of quantitative miRNA expression platforms in the microRNA quality control(miRQC) study. Nat Methods 11: 809–815.
Most D, Ferguson L, Blednov Y, Mayfield RD, Harris RA (2014). The synaptoneurosome transcriptome: a model for profiling the synaptic molecular effects of alcohol. J Pharmacogenomics 15: 177–188.
Novak M, Halbout B, O'Connor EC, Rodriguez Parkitna J, Su T, Chai M et al (2010). Incentive learning underlying cocaine-seeking requires mGluR5 receptors located on dopamine D1 receptor-expressing neurons. J Neurosci 30: 11973–11982.
Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA et al (2013). Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genomics 14: 725.
Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S et al (2008). ‘Functional organization of the transcriptome in human brain.’. Nat Neurosci 11: 1271–1282.
Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM et al (2008). Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron 59: 274–287.
Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD (2012). Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci 32: 1884–1897.
Quinlan EM, Philpot BD, Huganir RL, Bear MF (1999). Rapid, experience-dependent expression of synaptic NMDA receptors in visual cortex in vivo. Nat Neurosci 2: 352–357.
Raab-Graham KF, Haddick PC, Jan YN, Jan LY (2006). Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science 314: 144–148.
Smalheiser NR, Lugli G (2009). microRNA regulation of synaptic plasticity. Neuromol Med 11: 133–140.
Smyth GK (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 Article3.
Soares PM, Patrocinio MC, Assreuy AM, Siqueira RC, Lima NM, Arruda Mde O et al (2009). Aminophylline (a theophylline-ethylenediamine complex) blocks ethanol behavioral effects in mice. Behav Pharmacol 20: 297–302.
Sosanya NM, Huang PP, Cacheaux LP, Chen CJ, Nguyen K, Perrone-Bizzozero NI et al (2013). Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Biol 202: 53–69.
Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C et al (2006). Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci 9: 99–107.
Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A et al (2014). microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J Neurosci 34: 4581–4588.
Tapocik JD, Solomon M, Flanigan M, Meinhardt M, Barbier E, Schank JR et al (2013). Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenomics J 13: 286–296.
Tsai G, Coyle JT (1998). The role of glutamatergic neurotransmission in the pathophysiology of alcoholism. Annu Rev Med 49: 173–184.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A et al (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 (7).
Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC (2009). Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 324: 1536–1540.
Wang DO, Martin KC, Zukin RS (2010). Spatially restricting gene expression by local translation at synapses. Trends Neurosci 33: 173–182.
Acknowledgements
We thank Jody Mayfield for many valuable comments and scientific editing.
Author contributions
Dana Most designed, performed experiments, and wrote the paper; Courtney Leiter performed the PCR experiments; Yuri A. Blednov designed the behavioral experiments; R. Adron Harris designed the experiments and edited the paper; R Dayne Mayfield designed experiments and edited the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Neuropsychopharmacology website
Rights and permissions
About this article
Cite this article
Most, D., Leiter, C., Blednov, Y. et al. Synaptic microRNAs Coordinately Regulate Synaptic mRNAs: Perturbation by Chronic Alcohol Consumption. Neuropsychopharmacol 41, 538–548 (2016). https://doi.org/10.1038/npp.2015.179
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/npp.2015.179
This article is cited by
-
Differential regulation of alcohol consumption and reward by the transcriptional cofactor LMO4
Molecular Psychiatry (2021)
-
The Regulatory Role of miRNAs in Ethanol-induced TLR4 Activation and Neuroinflammation
Current Pathobiology Reports (2020)
-
Transcriptome analysis of alcohol-treated microglia reveals downregulation of beta amyloid phagocytosis
Journal of Neuroinflammation (2018)
-
Regulatory mechanisms of microRNA expression
Journal of Translational Medicine (2016)
-
Molecular mechanisms underlying alcohol-drinking behaviours
Nature Reviews Neuroscience (2016)