Abstract
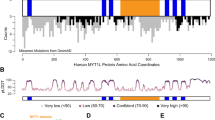
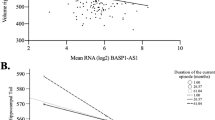
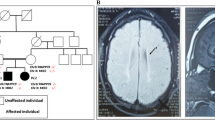
The fundamental role of the brain-specific myelin transcription factor 1-like (MYT1L) gene in cases of intellectual disability and in the etiology of neurodevelopmental disorders is increasingly recognized. Yet, its function remains under-investigated. Here, we identify a network of helix–loop–helix (HLH) transcriptional regulators controlled by MYT1L, as indicated by our analyses in human neural stem cells and in the human brain. Using cell-based knockdown approaches and microarray analyses we found that (1) MYT1L is required for neuronal differentiation and identified ID1, a HLH inhibitor of premature neurogenesis, as a target. (2) Although MYT1L prevented expression of ID1, it induced expression of a large number of terminal differentiation genes. (3) Consistently, expression of MYT1L in the human brain coincided with neuronal maturation and inversely correlated with that of ID1 and ID3 throughout the lifespan. (4) Genetic polymorphisms that reduced expression of MYT1L in the hippocampus resulted in increased expression of ID1 and ID3, decreased levels of the proneural basic HLH (bHLH) transcriptional regulators TCF4 and NEUROD6 and decreased expression of genes involved in long-term potentiation and synaptic transmission, cancer and neurodegeneration. Furthermore, our neuroimaging analyses indicated that MYT1L expression associated with hippocampal volume and activation during episodic memory recall, as measured by blood-oxygen-level-dependent (BOLD) signals. Overall, our findings suggest that MYT1L influences memory-related processes by controlling a neuronal proliferation/differentiation switch of ID-bHLH factors.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bliss TV, Collingridge GL (1993). A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361: 31–39.
Bonaglia MC, Giorda R, Zanini S (2014). A new patient with a terminal de novo 2p25.3 deletion of 1.9 Mb associated with early-onset of obesity, intellectual disabilities and hyperkinetic disorder. Mol Cytogenet 7: 53.
Chaudhary J, Johnson J, Kim G, Skinner MK (2001). Hormonal regulation and differential actions of the helix-loop-helix transcriptional inhibitors of differentiation (Id1, Id2, Id3, and Id4) in Sertoli cells. Endocrinology 142: 1727–1736.
de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T et al (2012). Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 367: 1921–1929.
De Rocker N, Vergult S, Koolen D, Jacobs E, Hoischen A, Zeesman S et al (2015). Refinement of the critical 2p25.3 deletion region: the role of MYT1L in intellectual disability and obesity. Genet Med 17: 460–466.
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE et al (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515: 209–215.
Deary IJ, Penke L, Johnson W (2010). The neuroscience of human intelligence differences. Nat Rev Neurosci 11: 201–211.
Desrivières S, Lourdusamy A, Tao C, Toro R, Jia T, Loth E et al (2015). Single nucleotide polymorphism in the neuroplastin locus associates with cortical thickness and intellectual ability in adolescents. Mol Psychiatry 20: 263–274.
Doco-Fenzy M, Leroy C, Schneider A, Petit F, Delrue MA, Andrieux J et al (2014). Early-onset obesity and paternal 2pter deletion encompassing the ACP1, TMEM18, and MYT1L genes. Eur J Hum Genet 22: 471–479.
Erk S, Meyer-Lindenberg A, Schnell K, Opitz von Boberfeld C, Esslinger C, Kirsch P et al (2010). Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch Gen Psychiatry 67: 803–811.
Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E, Blake DJ (2014). The emerging roles of TCF4 in disease and development. Trends Mol Med 20: 322–331.
Gao H, Bu Y, Wu Q, Wang X, Chang N, Lei L et al (2015). Mecp2 regulates neural cell differentiation by suppressing the Id1 to Her2 axis in zebrafish. J Cell Sci 128: 2340–2350.
Haworth CM, Wright MJ, Luciano M, Martin NG, de Geus EJ, van Beijsterveldt CE et al (2010). The heritability of general cognitive ability increases linearly from childhood to young adulthood. Mol Psychiatry 15: 1112–1120.
Hu J, Ho AL, Yuan L, Hu B, Hua S, Hwang SS et al (2013). From the Cover: neutralization of terminal differentiation in gliomagenesis. Proc Natl Acad Sci USA 110: 14520–14527.
Huang DW, Sherman BT, Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57.
Jankiewicz M, Groner B, Desrivières S (2006). Mammalian target of rapamycin regulates the growth of mammary epithelial cells through the inhibitor of deoxyribonucleic acid binding Id1 and their functional differentiation through Id2. Mol Endocrinol 20: 2369–2381.
Lee Y, Mattai A, Long R, Rapoport JL, Gogtay N, Addington AM (2012). Microduplications disrupting the MYT1L gene (2p25.3) are associated with schizophrenia. Psychiatr Genet 22: 206–209.
Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R et al (1999). Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401: 670–677.
Mayo S, Rosello M, Monfort S, Oltra S, Orellana C, Martinez F (2015). Haploinsufficiency of the MYT1L gene causes intellectual disability frequently associated with behavioral disorder. Genet Med 17: 683–684.
Meyer KJ, Axelsen MS, Sheffield VC, Patil SR, Wassink TH (2012). Germline mosaic transmission of a novel duplication of PXDN and MYT1L to two male half-siblings with autism. Psychiatr Genet 22: 137–140.
Nadel L (2003). Down's syndrome: a genetic disorder in biobehavioral perspective. Genes Brain Behav 2: 156–166.
Niola F, Zhao X, Singh D, Castano A, Sullivan R, Lauria M et al (2012). Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat Cell Biol 14: 477–487.
Niola F, Zhao X, Singh D, Sullivan R, Castano A, Verrico A et al (2013). Mesenchymal high-grade glioma is maintained by the ID-RAP1 axis. J Clin Invest 123: 405–417.
Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ et al (2011). Induction of human neuronal cells by defined transcription factors. Nature 476: 220–223.
Peddada S, Yasui DH, LaSalle JM (2006). Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome. Hum Mol Genet 15: 2003–2014.
Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M et al (2006). In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502.
Ren H, Yang BF, Rainov NG (2007). Receptor tyrosine kinases as therapeutic targets in malignant glioma. Rev Recent Clin Trials 2: 87–101.
Rio M, Royer G, Gobin S, de Blois MC, Ozilou C, Bernheim A et al (2013). Monozygotic twins discordant for submicroscopic chromosomal anomalies in 2p25.3 region detected by array CGH. Clin Genet 84: 31–36.
Ruzinova MB, Benezra R (2003). Id proteins in development, cell cycle and cancer. Trends Cell Biol 13: 410–418.
Schumann G, Loth E, Banaschewski T, Barbot A, Barker G, Buchel C et al (2010). The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry 15: 1128–1139.
Schwarz TJ, Ebert B, Lie DC (2012). Stem cell maintenance in the adult mammalian hippocampus: a matter of signal integration? Dev Neurobiol 72: 1006–1015.
Soroceanu L, Murase R, Limbad C, Singer E, Allison J, Adrados I et al (2013). Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res 73: 1559–1569.
Stevens SJ, van Ravenswaaij-Arts CM, Janssen JW, Klein Wassink-Ruiter JS, van Essen AJ, Dijkhuizen T et al (2011). MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions. Am J Med Genet A 155A: 2739–2745.
Sugimori M, Nagao M, Bertrand N, Parras CM, Guillemot F, Nakafuku M (2007). Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development 134: 1617–1629.
Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C et al (2012). MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet 21: 4094–4103.
Vaillend C, Poirier R, Laroche S (2008). Genes, plasticity and mental retardation. Behav Brain Res 192: 88–105.
Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041.
Visel A, Minovitsky S, Dubchak I, Pennacchio LA (2007). VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res 35 (Database issue): D88–D92.
Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, Geurts van Kessel A et al (2008). Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet 83: 504–510.
Wang LH, Baker NE (2015). E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev Cell 35: 269–280.
Wang T, Zeng Z, Li T, Liu J, Li J, Li Y et al (2010). Common SNPs in myelin transcription factor 1-like (MYT1L): association with major depressive disorder in the Chinese Han population. PLoS ONE 5: e13662.
Whalen S, Heron D, Gaillon T, Moldovan O, Rossi M, Devillard F et al (2012). Novel comprehensive diagnostic strategy in Pitt-Hopkins syndrome: clinical score and further delineation of the TCF4 mutational spectrum. Hum Mutat 33: 64–72.
Ying QL, Nichols J, Chambers I, Smith A (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115: 281–292.
Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S et al (1999). Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397: 702–706.
Acknowledgements
We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics for the generation of the Gene Expression data. A.K. and L.M.M were recipients of a studentship form the Medical Research Council, UK and Consejo Nacional de Ciencia y Tecnología (CONACyT; México), respectively.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Neuropsychopharmacology website
Rights and permissions
About this article
Cite this article
Kepa, A., Martinez Medina, L., Erk, S. et al. Associations of the Intellectual Disability Gene MYT1L with Helix–Loop–Helix Gene Expression, Hippocampus Volume and Hippocampus Activation During Memory Retrieval. Neuropsychopharmacol. 42, 2516–2526 (2017). https://doi.org/10.1038/npp.2017.91
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/npp.2017.91
This article is cited by
-
MYT1L deficiency impairs excitatory neuron trajectory during cortical development
Nature Communications (2024)
-
St18 specifies globus pallidus projection neuron identity in MGE lineage
Nature Communications (2022)
-
MYT1L in the making: emerging insights on functions of a neurodevelopmental disorder gene
Translational Psychiatry (2022)
-
A causal association of ANKRD37 with human hippocampal volume
Molecular Psychiatry (2022)