Abstract
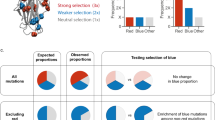
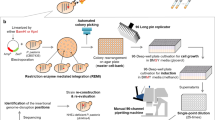
We describe a new method to mutate target genes through somatic hypermutation (SHM) and to evolve proteins directly in living mammalian cells. Target genes are expressed under the control of an inducible promoter in a B-cell line that hypermutates its immunoglobulin (Ig) V genes constitutively. Mutations can be introduced into the target gene through SHM upon transcription. Mutant genes are then expressed and selected or screened for desired properties in cells. Identified cells are subjected to another round of mutation and selection or screening. This process can be iterated easily for numerous rounds, and multiple reinforcing mutations can be accumulated to produce desirable phenotypes. This approach bypasses labor-intensive in vitro mutagenesis and samples a large protein sequence space. In this protocol a monomeric red fluorescent protein (mRFP1.2) was evolved in Ramos cells to afford a mutant (mPlum) with far-red emission. This method can be adapted to evolve other eukaryotic proteins and to be used in other cells able to perform SHM. For each round of evolution, it takes ∼1 d to mutate the target gene, ∼0.5–1 d to select or screen, and 2–4 d to propagate the cells for the next round depending on how many cells are collected.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lin, H. & Cornish, V.W. Screening and selection methods for large-scale analysis of protein function. Angew. Chem. Int. Edn. Engl. 41, 4402–4425 (2002).
Bloom, J.D. et al. Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 15, 447–452 (2005).
Minshull, J. & Stemmer, W.P. Protein evolution by molecular breeding. Curr. Opin. Chem. Biol. 3, 284–290 (1999).
Papavasiliou, F.N. & Schatz, D.G. Somatic hypermutation of immunoglobulin genes: merging mechanisms for genetic diversity. Cell 109 (Suppl.): S35–S44 (2002).
Martin, A. & Scharff, M.D. AID and mismatch repair in antibody diversification. Nat. Rev. Immunol. 2, 605–614 (2002).
Maizels, N. Immunoglobulin gene diversification. Annu. Rev. Genet. 39, 23–46 (2005).
Neuberger, M.S. et al. Somatic hypermutation at A.T pairs: polymerase error versus dUTP incorporation. Nat. Rev. Immunol. 5, 171–178 (2005).
Rajewsky, K., Forster, I. & Cumano, A. Evolutionary and somatic selection of the antibody repertoire in the mouse. Science 238, 1088–1094 (1987).
Cumbers, S.J. et al. Generation and iterative affinity maturation of antibodies in vitro using hypermutating B-cell lines. Nat. Biotechnol. 20, 1129–1134 (2002).
Bachl, J., Carlson, C., Gray-Schopfer, V., Dessing, M. & Olsson, C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 166, 5051–5057 (2001).
Wang, C.L., Harper, R.A. & Wabl, M. Genome-wide somatic hypermutation. Proc. Natl. Acad. Sci. USA 101, 7352–7356 (2004).
Wang, L., Jackson, W.C., Steinbach, P.A. & Tsien, R.Y. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc. Natl. Acad. Sci. USA 101, 16745–16749 (2004).
Sale, J.E. & Neuberger, M.S. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity 9, 859–869 (1998).
Campbell, R.E. et al. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882 (2002).
Camps, M., Naukkarinen, J., Johnson, B.P. & Loeb, L.A. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc. Natl. Acad. Sci. USA 100, 9727–9732 (2003).
Larijani, M., Frieder, D., Basit, W. & Martin, A. The mutation spectrum of purified AID is similar to the mutability index in Ramos cells and in ung−/−msh2−/− mice. Immunogenetics 56, 840–845 (2005).
Naviaux, R.K., Costanzi, E., Haas, M. & Verma, I.M. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70, 5701–5705 (1996).
Go, W.Y. & Ho, S.N. Optimization and direct comparison of the dimerizer and reverse tet transcriptional control systems. J. Gene Med. 4, 258–270 (2002).
Baird, G.S., Zacharias, D.A. & Tsien, R.Y. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97, 11984–11989 (2000).
Li, J. et al. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat. Genet. 23, 348–353 (1999).
Sale, J.E., Calandrini, D.M., Takata, M., Takeda, S. & Neuberger, M.S. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature 412, 921–926 (2001).
Kardinal, C., Selmayr, M. & Mocikat, R. Genetic stability of gene targeted immunoglobulin loci. I. Heavy chain isotype exchange induced by a universal gene replacement vector. Immunology 89, 309–315 (1996).
Acknowledgements
This work was supported by the Howard Hughes Medical Institute, National Institutes of Health Grant NS27177, and US Department of Energy Grant DE-FG03-01ER63276 to R.Y.T. L.W. was supported by the Damon Runyon Cancer Research Foundation (Merck Fellow, Grant DRG-1767-03), the Searle Scholars Program, and the Beckman Young Investigators Program.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Wang, L., Tsien, R. Evolving proteins in mammalian cells using somatic hypermutation. Nat Protoc 1, 1346–1350 (2006). https://doi.org/10.1038/nprot.2006.243
Published:
Issue date:
DOI: https://doi.org/10.1038/nprot.2006.243
This article is cited by
-
Use of red, far-red, and near-infrared light in imaging of yeasts and filamentous fungi
Applied Microbiology and Biotechnology (2022)
-
Affinity maturation of anti-TNF-alpha scFv with somatic hypermutation in non-B cells
Protein & Cell (2012)
-
Practical and reliable FRET/FLIM pair of fluorescent proteins
BMC Biotechnology (2009)
-
Protein and Genome Evolution in Mammalian Cells for Biotechnology Applications
Molecular Biotechnology (2009)