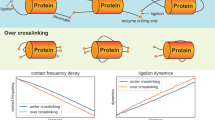
Abstract
Chromosome conformation capture (3C) technology is a pioneering methodology that allows in vivo genomic organization to be explored at a scale encompassing a few tens to a few hundred kilobase-pairs. Understanding the folding of the genome at this scale is particularly important in mammals where dispersed regulatory elements, like enhancers or insulators, are involved in gene regulation. 3C technology involves formaldehyde fixation of cells, followed by a polymerase chain reaction (PCR)-based analysis of the frequency with which pairs of selected DNA fragments are crosslinked in the population of cells. Accurate measurements of crosslinking frequencies require the best quantification techniques. We recently adapted the real-time TaqMan PCR technology to the analysis of 3C assays, resulting in a method that more accurately determines crosslinking frequencies than current semiquantitative 3C strategies that rely on measuring the intensity of ethidium bromide-stained PCR products separated by gel electrophoresis. Here, we provide a detailed protocol for this method, which we have named 3C-qPCR. Once preliminary controls and optimizations have been performed, the whole procedure (3C assays and quantitative analyses) can be completed in 7–9 days.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Carter, D., Chakalova, L., Osborne, C.S., Dai, Y.F. & Fraser, P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32, 623–626 (2002).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Dekker, J. A closer look at long-range chromosomal interactions. Trends Biochem. Sci. 28, 277–280 (2003).
Splinter, E., Grosveld, F. & de Laat, W. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 375, 493–507 (2004).
Tolhuis, B., Palstra, R.J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465 (2002).
Dostie, J. & Dekker, J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat. Protoc. 2, 988–1002 (2007).
Dostie, J. et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006).
Lomvardas, S. et al. Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413 (2006).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 (2006).
Wurtele, H. & Chartrand, P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 14, 477–495 (2006).
Zhao, Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 (2006).
Heid, C.A., Stevens, J., Livak, K.J. & Williams, P.M. Real time quantitative PCR. Genome Res. 6, 986–994 (1996).
Livak, K.J., Flood, S.J., Marmaro, J., Giusti, W. & Deetz, K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4, 357–362 (1995).
Lutfalla, G. & Uze, G. Performing quantitative reverse-transcribed polymerase chain reaction experiments. Methods Enzymol. 410, 386–400 (2006).
Vernimmen, D., De Gobbi, M., Sloane-Stanley, J.A., Wood, W.G. & Higgs, D.R. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 26, 2041–2051 (2007).
Splinter, E. et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 20, 2349–2354 (2006).
Holland, P.M., Abramson, R.D., Watson, R. & Gelfand, D.H. Detection of specific polymerase chain reaction product by utilizing the 5′— —3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88, 7276–7280 (1991).
Dekker, J. The three “C”s of chromosome conformation capture: controls, controls, controls. Nat. Methods 3, 17–21 (2006).
Palstra, R.J. et al. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35, 190–194 (2003).
Weber, M. et al. A real-time polymerase chain reaction assay for quantification of allele ratios and correction of amplification bias. Anal. Biochem. 320, 252–258 (2003).
Kurukuti, S. et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA 103, 10684–10689 (2006).
Murrell, A., Heeson, S. & Reik, W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36, 889–893 (2004).
Milligan, L. et al. H19 gene expression is up-regulated exclusively by stabilization of the RNA during muscle cell differentiation. Oncogene 19, 5810–5816 (2000).
Leighton, P.A., Saam, J.R., Ingram, R.S., Stewart, C.L. & Tilghman, S.M. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 9, 2079–2089 (1995).
Acknowledgements
We thank Robert Feil and Franck Court for discussion and comments on the manuscript. This work was supported by grants from the Association pour la Recherche contre le Cancer (ARC contract no. 3279), the “GIS Longévité” (contract no. GISLO401), the “Fond National de la Science” (ACI jeune chercheur) given to T. Forné and by funds from the “Centre National de la Recherche Scientifique” (CNRS). C.B. was supported by an ARC fellowship (JR/MLD/MDV—P05/2 and P06/2). W.L. was supported by grants from the Dutch Scientific Organization (NWO) (016-006-026) and (912-04-082). J.D. was supported by a grant from the NIH (HG003143).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Hagège, H., Klous, P., Braem, C. et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2, 1722–1733 (2007). https://doi.org/10.1038/nprot.2007.243
Published:
Issue date:
DOI: https://doi.org/10.1038/nprot.2007.243
This article is cited by
-
Prioritization of risk genes in colorectal cancer by integrative analysis of multi-omics data and gene networks
Science China Life Sciences (2024)
-
A chromatin-regulated biphasic circuit coordinates IL-1β-mediated inflammation
Nature Genetics (2024)
-
GATA2 co-opts TGFβ1/SMAD4 oncogenic signaling and inherited variants at 6q22 to modulate prostate cancer progression
Journal of Experimental & Clinical Cancer Research (2023)
-
Epigenetic reprogramming-induced guanidinoacetic acid synthesis promotes pancreatic cancer metastasis and transcription-activating histone modifications
Journal of Experimental & Clinical Cancer Research (2023)
-
Lack of GPR180 ameliorates hepatic lipid depot via downregulation of mTORC1 signaling
Scientific Reports (2023)