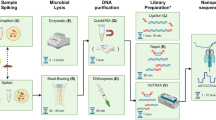
Abstract
Metagenomic sequencing has been widely used for the study of microbial communities from various environments such as soil, ocean, sediment and fresh water. Nonetheless, metagenomic sequencing of microbial communities in the air remains technically challenging, partly owing to the limited mass of collectable atmospheric particulate matter and the low biological content it contains. Here we present an optimized protocol for extracting up to tens of nanograms of airborne microbial genomic DNA from collected particulate matter. With an improved sequencing library preparation protocol, this quantity is sufficient for downstream applications, such as metagenomic sequencing for sampling various genes from the airborne microbial community. The described protocol takes ∼12 h of bench time over 2–3 d, and it can be performed with standard molecular biology equipment in the laboratory. A modified version of this protocol may also be used for genomic DNA extraction from other environmental samples of limited mass or low biological content.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Eisen, J.A. Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol. 5, e82 (2007).
Yooseph, S. et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468, 60–66 (2010).
Christner, B.C. et al. A microbial ecosystem beneath the West Antarctic ice sheet. Nature 512, 310–313 (2014).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Fierer, N. et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 109, 21390–21395 (2012).
Fierer, N. et al. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017 (2012).
Foster, J.S. et al. Molecular and morphological characterization of cyanobacterial diversity in the stromatolites of Highborne Cay, Bahamas. ISME J. 3, 573–587 (2009).
Cao, C. et al. Inhalable microorganisms in Beijing′s PM2.5 and PM10 pollutants during a severe smog event. Environ. Sci. Technol. 48, 1499–1507 (2014).
Bertolini, V. et al. Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Appl. Microbiol. Biotechnol. 97, 6561–6570 (2013).
Bowers, R.M., McLetchie, S., Knight, R. & Fierer, N. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 5, 601–612 (2011).
Brodie, E.L. et al. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. USA 104, 299–304 (2007).
Deleon-Rodriguez, N. et al. Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc. Natl. Acad. Sci. USA 110, 2575–2580 (2013).
Robertson, C.E. et al. Culture-independent analysis of aerosol microbiology in a metropolitan subway system. Appl. Environ. Microbiol. 79, 3485–3493 (2013).
Bowers, R.M. et al. Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environ. Sci. Technol. 47, 12097–12106 (2013).
Steffen, M.M. et al. Comparative metagenomics of toxic freshwater cyanobacteria bloom communities on two continents. PloS ONE 7, e44002 (2012).
Rinke, C. et al. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat. Protoc. 9, 1038–1048 (2014).
Leung, M. & Chan, A.H. Control and management of hospital indoor air quality. Med. Sci. Monit. 12, SR17–23 (2006).
Peiris, J.S. et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361, 1319–1325 (2003).
Uyeki, T.M. & Cox, N.J. Global concerns regarding novel influenza A (H7N9) virus infections. N. Engl. J. Med. 11, 11 (2013).
He, K. et al. The characteristics of PM2.5 in Beijing, China. Atmos. Environ. 35, 4959–4970 (2001).
Wang, S. et al. Quantifying the air pollutants emission reduction during the 2008 Olympic games in Beijing. Environ. Sci. Technol. 44, 2490–2496 (2010).
Yang, F. et al. Characteristics of PM2.5 speciation in representative megacities and across China. Atmos. Chem. Phys. 11, 5207–5219 (2011).
Tolocka, M.P., Peters, T.M., Vanderpool, R.W., Chen, F.L. & Wiener, R.W. On the modification of the low-flow-rate PM10 dichotomous sampler inlet. Aerosol. Sci. Technol. 34, 407–415 (2001).
Metzker, M.L. Sequencing technologies - the next generation. Nat. Rev. Genet. 11, 31–46 (2010).
Metzker, M.L. Emerging technologies in DNA sequencing. Genome Res. 15, 1767–1776 (2005).
Segata, N. et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814 (2012).
Segata, N. et al. Computational meta′omics for microbial community studies. Mol. Syst. Biol. 9, 666 (2013).
Meyer, F. et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 1–8 (2008).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Caporaso, J.G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 108, 4516–4522 (2011).
Acknowledgements
We thank M. Kayani, C. Liu and C. Cao for helpful discussions and for help taking the experimental setup photos. This work was supported in part by funding from the Ministry of Science and Technology of China (973 grant no. 2015CB553402), the National Natural Science Foundation of China (grant nos. 31470532, 21190054, 21221004), the Tsinghua University-Peking University Center for Life Sciences (CLS), a Tsinghua-Janssen Scholarship and research grant and the Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases.
Author information
Authors and Affiliations
Contributions
W.J., P.L., B.W., J.F. and J.L., with inputs from T.F.Z., J.J. and G.T., performed the experiments and analyzed the data. T.F.Z. conceived and organized the study. W.J. and T.F.Z., with inputs from the other coauthors, wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Jiang, W., Liang, P., Wang, B. et al. Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat Protoc 10, 768–779 (2015). https://doi.org/10.1038/nprot.2015.046
Published:
Issue date:
DOI: https://doi.org/10.1038/nprot.2015.046
This article is cited by
-
Characteristics, diffusion, and exposure risk of bioaerosol pollution during haze period on a campus of central China
Air Quality, Atmosphere & Health (2023)
-
Release of airborne antibiotic resistance genes from municipal solid waste transfer stations
Sustainable Environment Research (2022)
-
Inhalable antibiotic resistomes emitted from hospitals: metagenomic insights into bacterial hosts, clinical relevance, and environmental risks
Microbiome (2022)
-
Particulate matter emission sources and meteorological parameters combine to shape the airborne bacteria communities in the Ligurian coast, Italy
Scientific Reports (2021)
-
Efficiency of bioaerosol samplers: a comparison study
Aerobiologia (2021)