Key Points
-
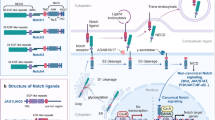
A causative role for Notch signalling is well established in T cell acute lymphoblastic leukaemias (T-ALLs), which have activating mutations in the Notch genes resulting in a constitutively active pathway. By contrast, solid tumours, which have ample opportunity to activate the pathway, exhibit inappropriate activation by multiple mechanisms, such as overexpression of ligand or loss of negative regulators of the pathway.
-
The role of Notch signalling in solid tumours is highly dependent on the spatial and temporal context of Notch activation, as well as the status of other signalling pathways in the cells.
-
Notch signalling has opposing roles in tumorigenesis depending on the cell type. Opposite interactions of the Notch pathway have been documented with the WNT and p53 pathways. Although synergy with WNT and antagonism of the p53 pathway directs the oncogenic role of Notch, the opposite is seen in the tumour suppressor context.
-
Notch signalling has a major role in the maintenance and progression of tumours by promoting epithelial to mesenchymal transition (EMT) and angiogenesis. It also confers resistance to radiation and chemotherapeutic agents.
-
The knowledge of the extensive crosstalk of the Notch pathway with other pathways such as the epidermal growth factor receptor (EGFR) pathway could prove useful in developing combinatorial cancer therapies.
Abstract
The discovery of Notch in Drosophila melanogaster nearly a century ago opened the door to an ever-widening understanding of cellular processes that are controlled or influenced by Notch signalling. As would be expected with such a pleiotropic pathway, the deregulation of Notch signalling leads to several pathological conditions, including cancer. A role for Notch is well established in haematological malignancies, and more recent studies have provided evidence for the importance of Notch activity in solid tumours. As it is thought to act as an oncogene in some cancers but as a tumour suppressor in others, the role of Notch in solid tumours seems to be highly context dependent.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
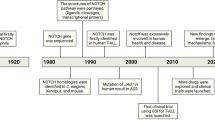
Ellisen, L. W. et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661 (1991). NOTCH1 was found to be affected by the chromosomal translocation t(7;9)(q34;q34.3), which was found in several patients with T-ALL. This was the first instance in which NOTCH1 was implicated in human cancer.
Weng, A. P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004). The authors found that more than 50% of human T-ALLs have activating mutations in NOTCH1.
Malecki, M. J. et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell. Biol. 26, 4642–4651 (2006).
Koch, U. & Radtke, F. Notch and cancer: a double-edged sword. Cell. Mol. Life Sci. 64, 2746–2762 (2007).
Hanlon, L. et al. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 70, 4280–4286 (2010).
Plentz, R. et al. Inhibition of γ-secretase activity inhibits tumor progression in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology 136, 1741–1749 e1746 (2009).
Santagata, S. et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 64, 6854–6857 (2004).
Miyamoto, Y. et al. Notch mediates TGF α-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 3, 565–576 (2003).
Brennan, C. et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS ONE 4, e7752 (2009).
Reedijk, M. et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 65, 8530–8537 (2005).
Aster, J. C., Pear, W. S. & Blacklow, S. C. Notch signaling in leukemia. Annu. Rev. Pathol. 3, 587–613 (2008).
Gordon, W. R., Arnett, K. L. & Blacklow, S. C. The molecular logic of Notch signaling-a structural and biochemical perspective. J. Cell Sci. 121, 3109–3119 (2008).
Bray, S. J. Notch signalling: a simple pathway becomes complex. Nature Rev. Mol. Cell Biol. 7, 678–689 (2006).
Kopan, R. & Ilagan, M. X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 (2009). A comprehensive review on Notch signalling.
Kovall, R. A. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene 27, 5099–5109 (2008).
Fortini, M. E. Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633–647 (2009).
LaVoie, M. J. & Selkoe, D. J. The Notch ligands, Jagged and Delta, are sequentially processed by α-secretase and presenilin/γ-secretase and release signaling fragments. J. Biol. Chem. 278, 34427–34437 (2003).
Six, E. et al. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and γ-secretase. Proc. Natl Acad. Sci. USA 100, 7638–7643 (2003).
Bland, C. E., Kimberly, P. & Rand, M. D. Notch-induced proteolysis and nuclear localization of the Delta ligand. J. Biol. Chem. 278, 13607–13610 (2003).
Klueg, K. M., Parody, T. R. & Muskavitch, M. A. Complex proteolytic processing acts on Delta, a transmembrane ligand for Notch, during Drosophila development. Mol. Biol. Cell 9, 1709–1723 (1998).
D'Souza, B., Meloty-Kapella, L. & Weinmaster, G. Canonical and non-canonical Notch ligands. Curr. Top. Dev. Biol. 92, 73–129 (2010).
Ascano, J. M., Beverly, L. J. & Capobianco, A. J. The C-terminal PDZ-ligand of JAGGED1 is essential for cellular transformation. J. Biol. Chem. 278, 8771–8779 (2003). The first evidence suggesting a bidirectional signalling by Notch ligands independently of Notch. The authors demonstrate that JAG1 can transform cells and this requires the PDZ ligand motif of the ICD.
Kolev, V. et al. The intracellular domain of Notch ligand Delta1 induces cell growth arrest. FEBS Lett. 579, 5798–5802 (2005).
Gallahan, D., Kozak, C. & Callahan, R. A new common integration region (int-3) for mouse mammary tumor virus on mouse chromosome 17. J. Virol. 61, 218–220 (1987).
Gallahan, D. & Callahan, R. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4). Oncogene 14, 1883–1890 (1997). This study, together with reference 24, identified the Notch locus as a common integration site in MMTV-induced tumours. Subsequently identified as NOTCH4, it was suggested that this has a role in mammary tumorigenesis.
Gallahan, D. et al. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 56, 1775–1785 (1996). Expression of a truncated form of the Int3 (Notch4 ) gene in the mammary epithelium of mice resulted in highly malignant mammary tumours and lung metastases.
Capobianco, A. J., Zagouras, P., Blaumueller, C. M., Artavanis-Tsakonas, S. & Bishop, J. M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 17, 6265–6273 (1997). A constitutively active truncated form of NOTCH1 and NOTCH2, in cooperation with the E1A adenovirus oncogene, can transform RKE cells.
Demarest, R. M., Ratti, F. & Capobianco, A. J. It's T-ALL about Notch. Oncogene 27, 5082–5091 (2008).
Ronchini, C. & Capobianco, A. J. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic). Mol. Cell. Biol. 21, 5925–5934 (2001).
Ling, H., Sylvestre, J. R. & Jolicoeur, P. Notch1-induced mammary tumor development is cyclin D1-dependent and correlates with expansion of pre-malignant multipotent duct-limited progenitors. Oncogene 29, 4543–4554 (2010).
Cohen, B. et al. Cyclin D1 is a direct target of JAG1-mediated Notch signaling in breast cancer. Breast Cancer Res. Treat. 123, 113–124 (2010).
Sicinska, E. et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell 4, 451–461 (2003).
Sharma, V. M. et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol. Cell. Biol. 26, 8022–8031 (2006).
Weng, A. P. et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096–2109 (2006).
Klinakis, A. et al. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl Acad. Sci. USA 103, 9262–9267 (2006).
Palomero, T. et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl Acad. Sci. USA 103, 18261–18266 (2006).
Murata, J. et al. Notch-Hes1 pathway contributes to the cochlear prosensory formation potentially through the transcriptional down-regulation of p27Kip1. J. Neurosci. Res. 87, 3521–3534 (2009).
Monahan, P., Rybak, S. & Raetzman, L. T. The notch target gene HES1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology 150, 4386–4394 (2009).
Riccio, O. et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 9, 377–383 (2008).
Dohda, T. et al. Notch signaling induces SKP2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp. Cell Res. 313, 3141–3152 (2007).
Mittal, S., Subramanyam, D., Dey, D., Kumar, R. & Rangarajan, A. Cooperation of Notch and Ras/MAPK signaling pathways in human breast carcinogenesis. Mol. Cancer 8, 128 (2009).
Xu, P. et al. The oncogenic roles of Notch1 in astrocytic gliomas in vitro and in vivo. J. Neurooncol. 97, 41–51 (2010).
Liu, W., Singh, S. R. & Hou, S. X. JAK-STAT is restrained by Notch to control cell proliferation of the Drosophila intestinal stem cells. J. Cell. Biochem. 109, 992–999 (2010).
Li, T. et al. Epidermal growth factor receptor and notch pathways participate in the tumor suppressor function of γ-secretase. J. Biol. Chem. 282, 32264–32273 (2007).
Lefort, K. et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKα kinases. Genes Dev. 21, 562–577 (2007).
Wang, C. et al. Notch1 signaling sensitizes tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human hepatocellular carcinoma cells by inhibiting Akt/Hdm2-mediated p53 degradation and up-regulating p53-dependent DR5 expression. J. Biol. Chem. 284, 16183–16190 (2009). This study suggests a mechanism by which Notch signalling induces apoptosis by activating p53.
Wang, L. et al. Overexpressed active Notch1 induces cell growth arrest of HeLa cervical carcinoma cells. Int. J. Gynecol. Cancer 17, 1283–1292 (2007).
Talora, C. et al. Constitutively active Notch1 induces growth arrest of HPV-positive cervical cancer cells via separate signaling pathways. Exp. Cell Res. 305, 343–354 (2005).
Dotto, G. P. Crosstalk of Notch with p53 and p63 in cancer growth control. Nature Rev. Cancer 9, 587–595 (2009). A comprehensive review on the crosstalk between Notch and p53, which probably determines whether Notch is oncogenic or tumour suppressive.
Shou, J., Ross, S., Koeppen, H., de Sauvage, F. J. & Gao, W. Q. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 61, 7291–7297 (2001).
Whelan, J. T., Kellogg, A., Shewchuk, B. M., Hewan-Lowe, K. & Bertrand, F. E. Notch-1 signaling is lost in prostate adenocarcinoma and promotes PTEN gene expression. J. Cell. Biochem. 107, 992–1001 (2009).
Bin Hafeez, B. et al. Targeted knockdown of Notch1 inhibits invasion of human prostate cancer cells concomitant with inhibition of matrix metalloproteinase-9 and urokinase plasminogen activator. Clin. Cancer Res. 15, 452–459 (2009).
Niimi, H., Pardali, K., Vanlandewijck, M., Heldin, C. H. & Moustakas, A. Notch signaling is necessary for epithelial growth arrest by TGF-β. J. Cell Biol. 176, 695–707 (2007).
Sun, Y. et al. Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-β signaling. Oncogene 24, 5365–5374 (2005).
Masuda, S. et al. Notch1 oncoprotein antagonizes TGF-β/Smad-mediated cell growth suppression via sequestration of coactivator p300. Cancer Sci. 96, 274–282 (2005).
Demehri, S., Turkoz, A. & Kopan, R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell 16, 55–66 (2009). This study demonstrates the role of the microenvironment in determining the role of Notch signalling in the skin, where Notch is known to have a tumour suppressor effect. The study shows that intact Notch signalling is not sufficient to prevent tumorigenesis when the microenvironment is tumour promoting.
Dumortier, A. et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS ONE 5, e9258 (2010).
Demehri, S. et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 6, e123 (2008).
Palomero, T., Dominguez, M. & Ferrando, A. A. The role of the PTEN/AKT Pathway in NOTCH1-induced leukemia. Cell Cycle 7, 965–970 (2008).
Nair, P., Somasundaram, K. & Krishna, S. Activated Notch1 inhibits p53-induced apoptosis and sustains transformation by human papillomavirus type 16 E6 and E7 oncogenes through a PI3K-PKB/Akt-dependent pathway. J. Virol. 77, 7106–7112 (2003).
Meurette, O. et al. Notch activation induces Akt signaling via an autocrine loop to prevent apoptosis in breast epithelial cells. Cancer Res. 69, 5015–5022 (2009).
Wendorff, A. A. et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity 33, 671–684 (2010).
Mungamuri, S., Yang, X., Thor, A. & Somasundaram, K. Survival signaling by Notch1: mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res. 66, 4715–4724 (2006).
Beverly, L., Felsher, D. & Capobianco, A. Suppression of p53 by Notch in lymphomagenesis: implications for initiation and regression. Cancer Res. 65, 7159–7168 (2005). A very elegant tetracycline-inducible mouse model, Top-Notchic, was used to show that Notch suppresses p53 through repression of the ARF–MDM2–p53 tumour surveillance pathway in lymphomagenesis.
Song, L. L. et al. Notch-1 associates with IKKα and regulates IKK activity in cervical cancer cells. Oncogene 27, 5833–5844 (2008).
Ramdass, B. et al. Coexpression of Notch1 and NF-κB signaling pathway components in human cervical cancer progression. Gynecol. Oncol. 104, 352–361 (2007).
Osipo, C., Golde, T. E., Osborne, B. A. & Miele, L. A. Off the beaten pathway: the complex cross talk between Notch and NF-κB. Lab. Invest. 88, 11–17 (2008).
Nakamura, T., Tsuchiya, K. & Watanabe, M. Crosstalk between Wnt and Notch signaling in intestinal epithelial cell fate decision. J. Gastroenterol. 42, 705–710 (2007).
Bienz, M. & Clevers, H. Linking colorectal cancer to Wnt signaling. Cell 103, 311–320 (2000).
Inomata, M., Ochiai, A., Akimoto, S., Kitano, S. & Hirohashi, S. Alteration of β-catenin expression in colonic epithelial cells of familial adenomatous polyposis patients. Cancer Res. 56, 2213–2217 (1996).
Satoh, S. et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nature Genet. 24, 245–250 (2000).
Korinek, V. et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC-/- colon carcinoma. Science 275, 1784–1787 (1997).
Caldwell, G. et al. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 64, 883–888 (2004).
Suzuki, H. et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nature Genet. 36, 417–422 (2004).
Katoh, M., Kirikoshi, H., Terasaki, H. & Shiokawa, K. WNT2B2 mRNA, up-regulated in primary gastric cancer, is a positive regulator of the WNT- β-catenin-TCF signaling pathway. Biochem. Biophys. Res. Commun. 289, 1093–1098 (2001).
Kirikoshi, H., Sekihara, H. & Katoh, M. Expression of WNT14 and WNT14B mRNAs in human cancer, up-regulation of WNT14 by IFNγ and up-regulation of WNT14B by β-estradiol. Int. J. Oncol. 19, 1221–1225 (2001).
Okino, K. et al. Up-regulation and overproduction of DVL-1, the human counterpart of the Drosophila dishevelled gene, in cervical squamous cell carcinoma. Oncol. Rep. 10, 1219–1223 (2003).
Terasaki, H., Saitoh, T., Shiokawa, K. & Katoh, M. Frizzled-10, up-regulated in primary colorectal cancer, is a positive regulator of the WNT - β-catenin - TCF signaling pathway. Int. J. Mol. Med. 9, 107–112 (2002).
Bjorklund, P., Akerstrom, G. & Westin, G. An LRP5 receptor with internal deletion in hyperparathyroid tumors with implications for deregulated WNT/β-catenin signaling. PLoS Med. 4, e328 (2007).
van Es, J. et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005).
Axelrod, J., Matsuno, K., Artavanis-Tsakonas, S. & Perrimon, N. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science 271, 1826–1832 (1996). D. melanogaster DSH, which is a transducer of WNT signalling, antagonizes the Notch pathway through a direct physical interaction with the C terminus of Notch.
Couso, J. & Martinez Arias, A. Notch is required for wingless signaling in the epidermis of Drosophila. Cell 79, 259–272 (1994).
Giraldez, A. & Cohen, S. Wingless and Notch signaling provide cell survival cues and control cell proliferation during wing development. Development 130, 6533–6543 (2003).
Jin, Y. et al. β-catenin modulates the level and transcriptional activity of Notch1/NICD through its direct interaction. Biochim. Biophys. Acta 1793, 290–299 (2009).
Alves-Guerra, M., Ronchini, C. & Capobianco, A. Mastermind-like 1 is a specific coactivator of β-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 67, 8690–8698 (2007). This paper is the first report of a role for MAML1outside of the Notch pathway. MAML1is recruited to the CCND1 promoter by β-catenin, and knock down of MAML1in colonic carcinoma cells results in cell death.
Nicolas, M. et al. Notch1 functions as a tumor suppressor in mouse skin. Nature Genet. 33, 416–421 (2003). A key paper demonstrating a tumour suppressor role for NOTCH1 and also the interactions with the WNT pathway.
Devgan, V., Mammucari, C., Millar, S. E., Brisken, C. & Dotto, G. P. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 19, 1485–1495 (2005).
De La, O. J. et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc. Natl Acad. Sci. USA 105, 18907–18912 (2008).
De La, O. J. & Murtaugh, L. C. Notch and Kras in pancreatic cancer: at the crossroads of mutation, differentiation and signaling. Cell Cycle 8, 1860–1864 (2009).
Weijzen, S. et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nature Med. 8, 979–986 (2002). A study that gives an insight into the role of Notch signalling in RAS-driven cancers.
Claxton, S. & Fruttiger, M. Periodic Delta-like 4 expression in developing retinal arteries. Gene Expr. Patterns 5, 123–127 (2004).
Hellstrom, M. et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 445, 776–780 (2007).
Suchting, S. et al. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc. Natl Acad. Sci. USA 104, 3225–3230 (2007).
Mailhos, C. et al. Delta4, an endothelial specific notch ligand expressed at sites of physiological and tumor angiogenesis. Differentiation 69, 135–144 (2001).
Patel, N. et al. Up-regulation of endothelial delta-like 4 expression correlates with vessel maturation in bladder cancer. Clin. Cancer Res. 12, 4836–4844 (2006).
Thurston, G., Noguera-Troise, I. & Yancopoulos, G. The Delta paradox: DLL4 blockade leads to more tumour vessels but less tumour growth. Nature Rev. Cancer 7, 327–331 (2007).
Patel, N. et al. Up-regulation of delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function. Cancer Res. 65, 8690–8697 (2005).
Noguera-Troise, I. et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature 444, 1032–1037 (2006).
Benedito, R. et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell 137, 1124–1135 (2009). Differential effects of Notch ligands on angiogenesis. The study suggests that modification by Fringe probably modulates Notch activity during angiogenesis.
Panin, V. M., Papayannopoulos, V., Wilson, R. & Irvine, K. D. Fringe modulates Notch-ligand interactions. Nature 387, 908–912 (1997).
Fu, Y. et al. Differential regulation of transforming growth factor β signaling pathways by Notch in human endothelial cells. J. Biol. Chem. 284, 19452–19462 (2009).
Itoh, F. et al. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 23, 541–551 (2004).
Thiery, J. P. Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15, 740–746 (2003). A comprehensive review on EMT.
Leong, K. et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J. Exp. Med. 204, 2935–2948 (2007).
Chen, J., Imanaka, N. & Griffin, J. D. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br. J. Cancer 102, 351–360 (2010).
Sahlgren, C., Gustafsson, M. V., Jin, S., Poellinger, L. & Lendahl, U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl Acad. Sci. USA 105, 6392–6397 (2008).
Zavadil, J., Cermak, L., Soto-Nieves, N. & Bottinger, E. P. Integration of TGF-β/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 23, 1155–1165 (2004).
Mani, S. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). This paper demonstrates that the induction of an EMT in immortalized mammary epithelial cells results in the expression of stem cell markers and an increased ability to self-renew. The paper suggests EMT as a mechanism of conferring stem-like properties to cells.
Lapidot, T. et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648 (1994).
Bonnet, D. & Dick, J. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature Med. 3, 730–737 (1997).
Cicalese, A. et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138, 1083–1095 (2009).
Al-Hajj, M., Wicha, M., Benito-Hernandez, A., Morrison, S. & Clarke, M. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA 100, 3983–3988 (2003).
Boiko, A. D. et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature 466, 133–137 (2010).
Lee, C., Dosch, J. & Simeone, D. Pancreatic cancer stem cells. J. Clin. Oncol. 26, 2806–2812 (2008).
Maitland, N. & Collins, A. Prostate cancer stem cells: a new target for therapy. J. Clin. Oncol. 26, 2862–2870 (2008).
O'Brien, C., Pollett, A., Gallinger, S. & Dick, J. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110 (2007).
Peacock, C. & Watkins, D. Cancer stem cells and the ontogeny of lung cancer. J. Clin. Oncol. 26, 2883–2889 (2008).
Prince, M. & Ailles, L. Cancer stem cells in head and neck squamous cell cancer. J. Clin. Oncol. 26, 2871–2875 (2008).
Sell, S. & Leffert, H. Liver cancer stem cells. J. Clin. Oncol. 26, 2800–2805 (2008).
Singh, S. et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 63, 5821–5828 (2003).
Bao, S. et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 (2006).
Dean, M., Fojo, T. & Bates, S. Tumour stem cells and drug resistance. Nature Rev. Cancer 5, 275–284 (2005).
Bao, S. et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 66, 7843–7848 (2006).
Pistollato, F. et al. Interaction of hypoxia-inducible factor-1α and Notch signaling regulates medulloblastoma precursor proliferation and fate. Stem Cells 28, 1918–1929 (2010).
Varnum-Finney, B. et al. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nature Med. 6, 1278–1281 (2000).
Dontu, G. et al. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 6, R605–R615 (2004).
Farnie, G. & Clarke, R. Mammary stem cells and breast cancer-role of Notch signalling. Stem Cell Rev. 3, 169–175 (2007).
Farnie, G. et al. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J. Natl Cancer Inst. 99, 616–627 (2007).
Zhang, X. et al. Notch activation promotes cell proliferation and the formation of neural stem cell-like colonies in human glioma cells. Mol. Cell. Biochem. 307, 101–108 (2008).
Hitoshi, S. et al. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 16, 846–858 (2002).
Hitoshi, S. et al. Primitive neural stem cells from the mammalian epiblast differentiate to definitive neural stem cells under the control of Notch signaling. Genes Dev. 18, 1806–1811 (2004).
Nakamura, Y. et al. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J. Neurosci. 20, 283–293 (2000).
Fan, X. et al. Notch pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells 28, 5–16 (2009).
Meng, R. D. et al. γ-secretase inhibitors abrogate oxaliplatin-induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer Res. 69, 573–582 (2009).
Doucas, H. et al. Expression of nuclear Notch3 in pancreatic adenocarcinomas is associated with adverse clinical features, and correlates with the expression of STAT3 and phosphorylated Akt. J. Surg. Oncol. 97, 63–68 (2008).
Akiyoshi, T. et al. γ-secretase inhibitors enhance taxane-induced mitotic arrest and apoptosis in colon cancer cells. Gastroenterology 134, 131–144 (2008).
Ristorcelli, E., Beraud, E., Mathieu, S., Lombardo, D. & Verine, A. Essential role of Notch signaling in apoptosis of human pancreatic tumoral cells mediated by exosomal nanoparticles. Int. J. Cancer 125, 1016–1026 (2009).
Wang, Z. et al. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-κB signaling pathways. J. Cell. Biochem. 109, 726–736 (2010).
Osipo, C. et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a γ-secretase inhibitor. Oncogene 27, 5019–5032 (2008).
Hao, L. et al. Notch-1 activates estrogen receptor-α-dependent transcription via IKKα in breast cancer cells. Oncogene 29, 201–213 (2010).
Lee, C. W. et al. A functional Notch-survivin gene signature in basal breast cancer. Breast Cancer Res. 10, R97 (2008).
Lee, C. W., Raskett, C. M., Prudovsky, I. & Altieri, D. C. Molecular dependence of estrogen receptor-negative breast cancer on a notch-survivin signaling axis. Cancer Res. 68, 5273–5281 (2008).
Wang, J. et al. Notch promotes radioresistance of glioma stem cells. Stem Cells 28, 17–28 (2010). Glioma stem cells are resistant to radiation and this paper shows that blocking Notch signalling with GSIs sensitizes these cells to radiation and impairs xenograft formation. The mechanism is thought to be through the reduction of AKT activity.
Gilbert, C. A., Daou, M. C., Moser, R. P. & Ross, A. H. γ-secretase inhibitors enhance temozolomide treatment of human gliomas by inhibiting neurosphere repopulation and xenograft recurrence. Cancer Res. 70, 6870–6879 (2010). This study, together with references 139 and 142, demonstrates that Notch signalling is responsible for resistance to chemotherapeutic agents and shows the importance of using other therapeutic agents in combination with Notch inhibitors (GSI) for effective therapy.
Dikic, I. & Schmidt, M. H. Notch: implications of endogenous inhibitors for therapy. Bioessays 32, 481–487 (2010).
Wu, Y. et al. Therapeutic antibody targeting of individual Notch receptors. Nature 464, 1052–1057 (2010).
Yin, L., Velazquez, O. C. & Liu, Z. J. Notch signaling: emerging molecular targets for cancer therapy. Biochem. Pharmacol. 80, 690–701 (2010).
Harrison, H. et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 70, 709–718 (2010).
Mazur, P. K. et al. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc. Natl Acad. Sci. USA 107, 13438–13443 (2010).
Tonon, G. et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nature Genet. 33, 208–213 (2003).
Enlund, F. et al. Altered Notch signaling resulting from expression of a WAMTP1-MAML2 gene fusion in mucoepidermoid carcinomas and benign Warthin's tumors. Exp. Cell Res. 292, 21–28 (2004).
Lennerz, J. K., Perry, A., Mills, J. C., Huettner, P. C. & Pfeifer, J. D. Mucoepidermoid carcinoma of the cervix: another tumor with the t(11;19)-associated CRTC1-MAML2 gene fusion. Am. J. Surg. Pathol. 33, 835–843 (2009).
Behboudi, A. et al. Clear cell hidradenoma of the skin-a third tumor type with a t(11;19)-associated TORC1-MAML2 gene fusion. Genes Chromosomes Cancer 43, 202–205 (2005).
Martinez, A. M. et al. Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nature Genet. 41, 1076–1082 (2009).
Ferres-Marco, D. et al. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature 439, 430–436 (2006).
Moshkin, Y. M. et al. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol. Cell 35, 782–793 (2009).
Goodfellow, H. et al. Gene-specific targeting of the histone chaperone Asf1 to mediate silencing. Dev. Cell 13, 593–600 (2007).
Takeuchi, T., Adachi, Y. & Ohtsuki, Y. Skeletrophin, a novel ubiquitin ligase to the intracellular region of Jagged-2, is aberrantly expressed in multiple myeloma. Am. J. Pathol. 166, 1817–1826 (2005).
Takeuchi, T., Adachi, Y., Sonobe, H., Furihata, M. & Ohtsuki, Y. A ubiquitin ligase, skeletrophin, is a negative regulator of melanoma invasion. Oncogene 25, 7059–7069 (2006).
Wu, G. et al. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 21, 7403–7415 (2001).
Akhoondi, S. et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 67, 9006–9012 (2007).
Miyaki, M. et al. Somatic mutations of the CDC4 (FBXW7) gene in hereditary colorectal tumors. Oncology 76, 430–434 (2009).
Lan, K. et al. Kaposi's sarcoma herpesvirus-encoded latency-associated nuclear antigen stabilizes intracellular activated Notch by targeting the Sel10 protein. Proc. Natl Acad. Sci. USA 104, 16287–16292 (2007).
McGill, M. A. & McGlade, C. J. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278, 23196–23203 (2003).
Pece, S. et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167, 215–221 (2004).
Colaluca, I. N. et al. NUMB controls p53 tumour suppressor activity. Nature 451, 76–80 (2008).
Matsuno, K., Diederich, R. J., Go, M. J., Blaumueller, C. M. & Artavanis-Tsakonas, S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121, 2633–2644 (1995).
Matsuno, K. et al. Human deltex is a conserved regulator of Notch signalling. Nature Genet. 19, 74–78 (1998).
Izon, D. J. et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16, 231–243 (2002).
Yamamoto, N. et al. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J. Biol. Chem. 276, 45031–45040 (2001).
Acknowledgements
The authors would like to thank members of the Capobianco laboratory for support and D. Robbins and J. Diez for critical reading of the manuscript and useful suggestions. Work in the authors' laboratory is partly supported by grants from the US National Cancer Institute (ROI CA 83736) and the Samuel Waxman Foundation for Cancer Research, USA. The authors sincerely apologize to all their colleagues whose work has not been referenced in this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Role of Notch in solid tumors (PDF 265 kb)
Related links
Glossary
- Negative selection
-
The intrathymic elimination of CD4+CD8+ thymocytes that express T cell receptors with high affinity for self antigens.
- Type I transmembrane receptors
-
Proteins that span the plasma membrane once, with the carboxy-terminal end extending into the cytoplasm.
- Exocrine pancreas
-
The portion of the pancreas that secretes digestive enzymes that are then passed on to the small intestine.
Rights and permissions
About this article
Cite this article
Ranganathan, P., Weaver, K. & Capobianco, A. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer 11, 338–351 (2011). https://doi.org/10.1038/nrc3035
Published:
Issue date:
DOI: https://doi.org/10.1038/nrc3035
This article is cited by
-
Inhibition of NOTCH4 sensitizes FLT3/ITD acute myeloid leukemia cells to FLT3 tyrosine kinase inhibition
Leukemia (2024)
-
Desmoid Tumors: Current Perspective and Treatment
Current Treatment Options in Oncology (2024)
-
Angiogenic signaling pathways and anti-angiogenic therapy for cancer
Signal Transduction and Targeted Therapy (2023)
-
Enhancer remodeling activates NOTCH3 signaling to confer chemoresistance in advanced nasopharyngeal carcinoma
Cell Death & Disease (2023)
-
Notch signaling pathway: a comprehensive prognostic and gene expression profile analysis in breast cancer
BMC Cancer (2022)