Key Points
-
Two highly pathogenic human coronaviruses, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), have emerged in the past decade. The lack of any clinically approved antiviral treatments or vaccines for either virus emphasizes the importance of the design of effective therapeutics and preventives.
-
Bats have been implicated as reservoirs of both SARS-CoV and MERS-CoV as well as related viruses and other human coronaviruses (HCoVs), such as HCoV-229E and HCoV-NL63. The dispersion of bat species over much of the globe probably enhances their potential to act as reservoirs for pathogens, some of which are extremely virulent and potentially lethal to other animals and humans.
-
Multiple animal models for SARS-CoV infection exist, although mouse models have been the most thoroughly characterized. Mouse-adapted SARS-CoV is capable of causing pathology that is representative of human infections in both young and aged animals.
-
Small animal models for MERS-CoV infection have not yet been reported, although the possibility of further ongoing selection in the receptor-binding sequence in the spike protein or other sequences that are important for host specificity might contribute to this limitation. A mild disease phenotype that can include either localized or widespread pneumonia is observed in inoculated macaques.
-
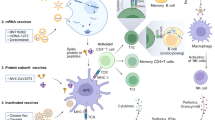
Multiple vaccine strategies have been attempted with coronaviruses, mostly (but not exclusively) targeting the spike glycoprotein. Successful live-attenuated vaccines have utilized reverse genetic strategies to delete the envelope protein or inactivate the exonuclease activity of non-structural protein 14 (nsp14) .
-
MERS-CoV, similarly to SARS-CoV in 2003, has the potential to have a profound impact on the human population; however, its low penetrance thus far suggests that the virus might either ultimately fail to develop a niche in humans or it might still be adapting to human hosts and that the worst of its effects are yet to come.
-
Coronavirus phylogeny shows an incredible diversity in antigenic variants, which leads to limited cross-protection against infection with different strains, even within a phylogenetic subcluster. Consequently, the risk of introducing novel coronaviruses into naive human and animal populations remains high.
Abstract
Two novel coronaviruses have emerged in humans in the twenty-first century: severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), both of which cause acute respiratory distress syndrome (ARDS) and are associated with high mortality rates. There are no clinically approved vaccines or antiviral drugs available for either of these infections; thus, the development of effective therapeutic and preventive strategies that can be readily applied to new emergent strains is a research priority. In this Review, we describe the emergence and identification of novel human coronaviruses over the past 10 years, discuss their key biological features, including tropism and receptor use, and summarize approaches for developing broadly effective vaccines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Zhong, N. Management and prevention of SARS in China. Phil. Trans. R. Soc. Lond. B Biol. Sci. 359, 1115–1116 (2004).
Peiris, J. S., Guan, Y. & Yuen, K. Y. Severe acute respiratory syndrome. Nature Med. 10, S88–S97 (2004).
Cherry, J. D. The chronology of the 2002–2003 SARS mini pandemic. Paediatr. Respir. Rev. 5, 262–269 (2004).
Anthony, S. J. et al. Coronaviruses in bats from Mexico. J. Gen. Virol. 94, 1028–1038 (2013).
Woo, P. C. et al. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 86, 3995–4008 (2012). This paper describes phylogenetic and molecular clock analyses that implicate bat coronaviruses as the forebearers of alphacoronaviruses and betacoronaviruses, and thus all currently known human coronaviruses.
Lau, S. K. et al. Recent transmission of a novel Alphacoronavirus, bat coronavirus HKU10, from Leschenault's rousettes to pomona leaf-nosed bats: first evidence of interspecies transmission of coronavirus between bats of different suborders. J. Virol. 86, 11906–11918 (2012).
Pfefferle, S. et al. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 15, 1377–1384 (2009).
Chu, D. K., Peiris, J. S., Chen, H., Guan, Y. & Poon, L. L. Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J. Gen. Virol. 89, 1282–1287 (2008).
Woo, P. C. et al. Molecular diversity of coronaviruses in bats. Virology 351, 180–187 (2006).
Tang, X. C. et al. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80, 7481–7490 (2006).
Lau, S. K. et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 44, 2063–2071 (2006).
Li, W. et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679 (2005).
Lau, S. K. et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA 102, 14040–14045 (2005). This is the first report of a SARS-related coronavirus in bats, and suggests that bats were a primary reservoir of the precursor to human SARS-CoV.
Lu, L., Liu, Q., Du, L. & Jiang, S. Middle East respiratory syndrome coronavirus (MERS-CoV): challenges in identifying its source and controlling its spread. Microbes Infect. 15, 625–629 (2013).
Annan, A. et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 19, 456–459 (2013).
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). This is the first description of the novel coronavirus that is responsible for severe respiratory illness in the Middle East. The virus was later named MERS-CoV.
van Boheemen, S. et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3 e00473-12 (2012).
Chan, J. F. et al. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J. Infect. 65, 477–489 (2012).
Bolles, M. et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 85, 12201–12215 (2011).
Pyrc, K., Berkhout, B. & van der Hoek, L. Identification of new human coronaviruses. Expert Rev. Anti Infect. Ther. 5, 245–253 (2007).
Reed, S. E. The behaviour of recent isolates of human respiratory coronavirus in vitro and in volunteers: evidence of heterogeneity among 229E-related strains. J. Med. Virol. 13, 179–192 (1984).
Peiris, J. S. et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361, 1319–1325 (2003). This is one of three papers to describe the novel coronavirus that is responsible for SARS.
Shi, Z. & Hu, Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 133, 74–87 (2008).
Li, W. et al. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2–S-protein interactions. J. Virol. 80, 4211–4219 (2006).
Guan, Y. et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302, 276–278 (2003).
Woo, P. C. et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79, 884–895 (2005).
van der Hoek, L. et al. Identification of a new human coronavirus. Nature Med. 10, 368–373 (2004).
Fouchier, R. A. et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl Acad. Sci. USA 101, 6212–6216 (2004).
Talbot, H. K. et al. Coronavirus infection and hospitalizations for acute respiratory illness in young children. J. Med. Virol. 81, 853–856 (2009).
Wertheim, J. O., Chu, D. K., Peiris, J. S., Kosakovsky Pond, S. L. & Poon, L. L. A case for the ancient origin of coronaviruses. J. Virol. 87, 7039–7045 (2013).
Huynh, J. et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 86, 12816–12825 (2012).
Memish, Z. A., Zumla, A. I., Al-Hakeem, R. F., Al-Rabeeah, A. A. & Stephens, G. M. Family cluster of Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 368, 2487–2494 (2013).
Assiri, A. et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 369, 407–416 (2013).
Assiri, A. et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 13, 752–761 (2013).
Reusken, C. B. et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 13, 859–866 (2013).
Wang, N. et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 23, 986–993 (2013). This paper reports the crystal structure of the MERS-CoV spike RBD in complex with the human DPP4 receptor molecule and identifies the key spike–DPP4 interaction residues.
Raj, V. S. et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254 (2013). This paper identifies DPP4 as the receptor for MERS-CoV.
Li, W. et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). This paper identifies ACE2 as the receptor for SARS-CoV.
Kindler, E. et al. Efficient replication of the novel human Betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio 4, e00611–00612 (2013).
Haagmans, B. L. et al. Pegylated interferon-α protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nature Med. 10, 290–293 (2004).
Chow, K. C., Hsiao, C. H., Lin, T. Y., Chen, C. L. & Chiou, S. H. Detection of severe acute respiratory syndrome-associated coronavirus in pneumocytes of the lung. Am. J. Clin. Pathol. 121, 574–580 (2004).
Guery, B. et al. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet 381, 2265–2272 (2013).
Cotten, M. et al. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet http://dx.doi.org/10.1016/S0140-6736(13)61887-5 (2013).
Kan, B. et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 79, 11892–11900 (2005).
Song, H. D. et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl Acad. Sci. USA 102, 2430–2435 (2005).
Yu, S. et al. Retrospective serological investigation of severe acute respiratory syndrome coronavirus antibodies in recruits from mainland China. Clin. Diagn. Lab. Immunol. 12, 552–554 (2005).
Zheng, B. J. et al. SARS-related virus predating SARS outbreak, Hong Kong. Emerg. Infect. Dis. 10, 176–178 (2004).
Smith, I. & Wang, L. F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 3, 84–91 (2013).
Wang, L. F. et al. Review of bats and SARS. Emerg. Infect. Dis. 12, 1834–1840 (2006).
Plowright, R. K. et al. Urban habituation, ecological connectivity and epidemic dampening: the emergence of Hendra virus from flying foxes (Pteropus spp.). Proc. Biol. Sci. 278, 3703–3712 (2011).
Becker, M. M. et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl Acad. Sci. USA 105, 19944–19949 (2008). This paper describes the reverse genetic design and recovery of bat SARS-related CoV with the epidemic and mouse-adapted SARS-CoV RBDs, including its capacity to infect mice and stimulate the production of cross-neutralizing antibodies.
Carrington, C. V. et al. Detection and phylogenetic analysis of group 1 coronaviruses in South American bats. Emerg. Infect. Dis. 14, 1890–1893 (2008).
Vijgen, L. et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 79, 1595–1604 (2005).
Lau, S. K. et al. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of Pipistrellus bat coronavirus HKU5 in Japanese Pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J. Virol. 87, 8638–8650 (2013).
Muller, M. A. et al. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. MBio 3, e00515-12 (2012).
Munster, V. J., de Wit, E. & Feldmann, H. Pneumonia from human coronavirus in a macaque model. N. Engl. J. Med. 368, 1560–1562 (2013).
Frieman, M. et al. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 86, 884–897 (2012).
Day, C. W. et al. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology 395, 210–222 (2009).
Roberts, A. et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3, e5 (2007). This is the first description of an in vivo mouse-adapted variant of SARS-CoV that is capable of recapitulating the clinical features of severe human disease in aged BALB/c mice.
Zhao, J., Zhao, J., Legge, K. & Perlman, S. Age-related increases in PGD2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J. Clin. Invest. 121, 4921–4930 (2011).
Zhao, J., Zhao, J. & Perlman, S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 84, 9318–9325 (2010).
Zhao, J., Zhao, J., Van Rooijen, N. & Perlman, S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 5, e1000636 (2009).
Sheahan, T. et al. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 4, e1000240 (2008).
Chen, W. et al. Antibody response and viraemia during the course of severe acute respiratory syndrome (SARS)-associated coronavirus infection. J. Med. Microbiol. 53, 435–438 (2004).
Li, C. K. et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 181, 5490–5500 (2008).
Roberts, A. et al. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 79, 5833–5838 (2005).
Subbarao, K. et al. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 78, 3572–3577 (2004).
Hogan, R. J. et al. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 78, 11416–11421 (2004).
Glass, W. G., Subbarao, K., Murphy, B. & Murphy, P. M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 173, 4030–4039 (2004).
Frieman, M. B. et al. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 6, e1000849 (2010).
Graham, R. L. et al. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nature Med. 18, 1820–1826 (2012).
Schaecher, S. R. et al. An immunosuppressed Syrian golden hamster model for SARS-CoV infection. Virology 380, 312–321 (2008).
Luo, D. et al. Protection from infection with severe acute respiratory syndrome coronavirus in a Chinese hamster model by equine neutralizing Fab′2. Viral Immunol. 20, 495–502 (2007).
Subbarao, K. & Roberts, A. Is there an ideal animal model for SARS? Trends Microbiol. 14, 299–303 (2006).
Roberts, A. et al. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 79, 503–511 (2005).
ter Meulen, J. et al. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet 363, 2139–2141 (2004).
Martina, B. E. et al. Virology: SARS virus infection of cats and ferrets. Nature 425, 915 (2003).
Roper, R. L. & Rehm, K. E. SARS vaccines: where are we? Expert Rev. Vaccines 8, 887–898 (2009).
See, R. H. et al. Severe acute respiratory syndrome vaccine efficacy in ferrets: whole killed virus and adenovirus-vectored vaccines. J. Gen. Virol. 89, 2136–2146 (2008).
Nagata, N. et al. Participation of both host and virus factors in induction of severe acute respiratory syndrome (SARS) in F344 rats infected with SARS coronavirus. J. Virol. 81, 1848–1857 (2007).
Fukushi, S. et al. Amino acid substitutions in the s2 region enhance severe acute respiratory syndrome coronavirus infectivity in rat angiotensin-converting enzyme 2-expressing cells. J. Virol. 81, 10831–10834 (2007).
Rockx, B. et al. Comparative pathogenesis of three human and zoonotic SARS-CoV strains in cynomolgus macaques. PLoS ONE 6, e18558 (2011).
Greenough, T. C. et al. Pneumonitis and multi-organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome-associated coronavirus. Am. J. Pathol. 167, 455–463 (2005).
McAuliffe, J. et al. Replication of SARS coronavirus administered into the respiratory tract of African green, rhesus and cynomolgus monkeys. Virology 330, 8–15 (2004).
Kuiken, T. et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362, 263–270 (2003). This is one of three papers to describe the novel coronavirus that is responsible for SARS.
Smits, S. L. et al. Distinct severe acute respiratory syndrome coronavirus-induced acute lung injury pathways in two different nonhuman primate species. J. Virol. 85, 4234–4245 (2011).
Falzarano, D. et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nature Med. 19, 1313–1317 (2013).
Adedeji, A. O. et al. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 87, 8017–8028 (2013).
Breban, R., Riou, J. & Fontanet, A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet 382, 694–699 (2013).
Qu, D. et al. Intranasal immunization with inactivated SARS-CoV (SARS-associated coronavirus) induced local and serum antibodies in mice. Vaccine 23, 924–931 (2005).
Takasuka, N. et al. A subcutaneously injected UV-inactivated SARS coronavirus vaccine elicits systemic humoral immunity in mice. Int. Immunol. 16, 1423–1430 (2004).
He, Y., Zhou, Y., Siddiqui, P. & Jiang, S. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 325, 445–452 (2004).
Enjuanes, L. et al. Vaccines to prevent severe acute respiratory syndrome coronavirus-induced disease. Virus Res. 133, 45–62 (2008).
See, R. H. et al. Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. J. Gen. Virol. 87, 641–650 (2006).
Tseng, C. T. et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE 7, e35421 (2012).
Ishioka, T. et al. Effects of respiratory syncytial virus infection and major basic protein derived from eosinophils in pulmonary alveolar epithelial cells (A549). Cell Biol. Int. 35, 467–474 (2011).
Lin, J. T. et al. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir Ther. 12, 1107–1113 (2007).
Vignuzzi, M., Wendt, E. & Andino, R. Engineering attenuated virus vaccines by controlling replication fidelity. Nature Med. 14, 154–161 (2008).
DeDiego, M. L. et al. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 81, 1701–1713 (2007).
Netland, J. et al. Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology 399, 120–128 (2010).
Fett, C., DeDiego, M. L., Regla-Nava, J. A., Enjuanes, L. & Perlman, S. Complete protection against severe acute respiratory syndrome coronavirus-mediated lethal respiratory disease in aged mice by immunization with a mouse-adapted virus lacking E protein. J. Virol. 87, 6551–6559 (2013).
Minskaia, E. et al. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl Acad. Sci. USA 103, 5108–5113 (2006).
Eckerle, L. D. et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6, e1000896 (2010).
Yount, B., Roberts, R. S., Lindesmith, L. & Baric, R. S. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc. Natl Acad. Sci. USA 103, 12546–12551 (2006).
Scobey, T. et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl Acad. Sci. USA (2013). This paper describes the design and implementation of an infectious cDNA clone of MERS-CoV.
Almazan, F. et al. Engineering a replication-competent, propagation-defective middle East respiratory syndrome coronavirus as a vaccine candidate. mBio 4, e00650-13 (2013).
Deming, D. et al. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 3, e525 (2006).
Baric, R. S. et al. SARS coronavirus vaccine development. Adv. Exp. Med. Biol. 581, 553–560 (2006).
Chen, Z. et al. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 79, 2678–2688 (2005).
Bisht, H. et al. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl Acad. Sci. USA 101, 6641–6646 (2004).
Bukreyev, A. et al. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet 363, 2122–2127 (2004).
Buchholz, U. J. et al. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl Acad. Sci. USA 101, 9804–9809 (2004).
Faber, M. et al. A single immunization with a rhabdovirus-based vector expressing severe acute respiratory syndrome coronavirus (SARS-CoV) S protein results in the production of high levels of SARS-CoV-neutralizing antibodies. J. Gen. Virol. 86, 1435–1440 (2005).
Kapadia, S. U. et al. Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology 340, 174–182 (2005).
Du, L. et al. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 87, 9939–9942 (2013).
Li, F., Li, W., Farzan, M. & Harrison, S. C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868 (2005). This paper reports the crystal structure of the SARS-CoV spike RBD in complex with the human ACE2 receptor molecule and identifies the key spike–ACE interaction residues.
Bisht, H., Roberts, A., Vogel, L., Subbarao, K. & Moss, B. Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology 334, 160–165 (2005).
Liu, S. J. et al. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 24, 3100–3108 (2006).
Gupta, V. et al. SARS coronavirus nucleocapsid immunodominant T-cell epitope cluster is common to both exogenous recombinant and endogenous DNA-encoded immunogens. Virology 347, 127–139 (2006).
Woo, P. C. et al. SARS coronavirus spike polypeptide DNA vaccine priming with recombinant spike polypeptide from Escherichia coli as booster induces high titer of neutralizing antibody against SARS coronavirus. Vaccine 23, 4959–4968 (2005).
Yang, Z. Y. et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428, 561–564 (2004).
Zakhartchouk, A. N., Liu, Q., Petric, M. & Babiuk, L. A. Augmentation of immune responses to SARS coronavirus by a combination of DNA and whole killed virus vaccines. Vaccine 23, 4385–4391 (2005).
Zhao, P. et al. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology 331, 128–135 (2005).
Rockx, B. et al. Escape from human monoclonal antibody neutralization affects in vitro and in vivo fitness of severe acute respiratory syndrome coronavirus. J. Infect. Dis. 201, 946–955 (2010).
Anthony, S. J. et al. A strategy to estimate unknown viral diversity in mammals. mBio 4, e00598-13 (2013).
Pensaert, M. B. & de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 58, 243–247 (1978).
Snodgrass, D. R. et al. Aetiology of diarrhoea in young calves. Vet. Rec. 119, 31–34 (1986).
Ksiazek, T. G. et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966 (2003). This is one of three papers to describe the novel coronavirus that is responsible for SARS.
Drosten, C. et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003).
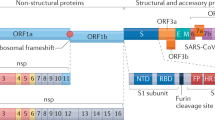
Narayanan, K., Huang, C. & Makino, S. SARS coronavirus accessory proteins. Virus Res. 133, 113–121 (2008).
Snijder, E. J. et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331, 991–1004 (2003).
Hofmann, H. et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl Acad. Sci. USA 102, 7988–7993 (2005).
Perlman, S. & Netland, J. Coronaviruses post-SARS: update on replication and pathogenesis. Nature Rev. Microbiol. 7, 439–450 (2009).
Pacciarini, F. et al. Persistent replication of severe acute respiratory syndrome coronavirus in human tubular kidney cells selects for adaptive mutations in the membrane protein. J. Virol. 82, 5137–5144 (2008).
Vijgen, L. et al. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 80, 7270–7274 (2006).
Louz, D., Bergmans, H. E., Loos, B. P. & Hoeben, R. C. Cross-species transfer of viruses: implications for the use of viral vectors in biomedical research, gene therapy and as live-virus vaccines. J. Gene Med. 7, 1263–1274 (2005).
Baric, R. S., Sullivan, E., Hensley, L., Yount, B. & Chen, W. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J. Virol. 73, 638–649 (1999).
Glowacka, I. et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85, 4122–4134 (2011).
Bertram, S. et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 85, 13363–13372 (2011).
Drosten, C. et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 13, 745–751 (2013).
Luyt, C. E., Combes, A., Trouillet, J. L., Nieszkowska, A. & Chastre, J. Virus-induced acute respiratory distress syndrome: epidemiology, management and outcome. Presse Med. 40, e561–568 (2011).
Imai, Y. et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 133, 235–249 (2008).
Ware, L. B. & Matthay, M. A. The acute respiratory distress syndrome. N. Engl. J. Med. 342, 1334–1349 (2000).
Cameron, M. J. et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 81, 8692–8706 (2007).
Tang, N. L. et al. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin. Chem. 51, 2333–2340 (2005).
Reghunathan, R. et al. Expression profile of immune response genes in patients with Severe Acute Respiratory syndrome. BMC Immunol. 6, 2 (2005).
Jiang, Y. et al. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am. J. Respir. Crit. Care Med. 171, 850–857 (2005).
Tsushima, K. et al. Acute lung injury review. Intern. Med. 48, 621–630 (2009).
Li, T. S. et al. Long-term outcome of acute respiratory distress syndrome caused by severe acute respiratory syndrome (SARS): an observational study. Crit. Care Resusc. 8, 302–308 (2006).
Gralinski, L. E. et al. Mechanisms of severe acute respiratory syndrome coronavirus-induced acute lung injury. mBio 4, e00271-13 (2013).
Graham, R. L. & Baric, R. S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 84, 3134–3146 (2010).
Ge, X.-Y. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature http://dx.doi.org/10.1038/nature12711 (2013).
Acknowledgements
This work was funded by US National Institutes of Health grants U19-AI100625 and U54-AI057157 (Southeast Regional Center of Excellence for Emerging Infections and Biodefense; SERCEB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Purifying selection
-
(Also called negative selection). A type of natural selection that removes deleterious alleles.
- Seropositivity
-
The positive reaction of a serum sample tested for a pathogen: for example, detection of antibodies against a virus.
- Type I pneumocytes
-
Epithelial cells that line the lung alveoli; type I cells are flat and thin to enable efficient gas exchange.
- Type II pneumocytes
-
Epithelial cells that line the lung alveoli; type II cells are round and produce surfactants.
- Synanthropy
-
The process of an organism, particularly a wild animal, becoming ecologically associated with humans.
- TALEN-mediated mutagenesis
-
Mutagenesis mediated by transcription activator-like effector nucleases (TALENs); involves the use of a series of TAL effector (DNA-binding) repeats fused to a Fok I-specific cleavage domain, which enables site-specific DNA double-strand breaks, non-homologous end joining and repair, which result in hybrid gene construction.
- CRISPR–Cas-mediated mutagenesis
-
Clustered regularly interspaced short palindromic repeats (CRISPRs), which consist of multiple short, conserved nucleotide repeats and function as a bacterial immune system that resists the incorporation of exogenous genetic elements. For mutagenesis, the CRISPR–Cas (CRISPR-associated) system targets foreign DNA with short, complementary single-stranded RNA that directs the Cas9 nuclease to the target DNA, causing double-strand breaks and resulting in silencing of that DNA sequence.
- Super-spreader events
-
Pathogen transmission events that are characterized by the identification of a host (human or animal) that transmits and spreads infection to a significantly greater number of susceptible organisms than the statistical average for that pathogen–host combination.
- Adjuvants
-
Pharmacological or immunological substances that modify and enhance the effect of an agent, such as a vaccine.
- Viral replicon particles
-
(VRPs). Virus-like particles that encode the components required to mediate replication of the genome but that do not encode genes necessary for the production of new virions. Viral replicon particles are typically generated by co-transfecting the replicon genome with helper cassettes that encode structural components, which enables a single round of replication and particle production to occur.
- Delayed-type hypersensitivity
-
(Also called type IV hypersensitivity). A CD4+ T cell-mediated immunological response rather than an antibody-mediated response. The immune reaction develops 24–72 hours after exposure to an immunogen.
Rights and permissions
About this article
Cite this article
Graham, R., Donaldson, E. & Baric, R. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol 11, 836–848 (2013). https://doi.org/10.1038/nrmicro3143
Published:
Issue date:
DOI: https://doi.org/10.1038/nrmicro3143
This article is cited by
-
The incidence of psychosocial disturbances during the coronavirus disease-19 pandemic in an Iranian sample
Current Psychology (2023)
-
In Silico Studies of Phytoconstituents from Piper longum and Ocimum sanctum as ACE2 and TMRSS2 Inhibitors: Strategies to Combat COVID-19
Applied Biochemistry and Biotechnology (2023)
-
A virus–target host proteins recognition method based on integrated complexes data and seed extension
BMC Bioinformatics (2022)
-
The recombinant pseudorabies virus expressing porcine deltacoronavirus spike protein is safe and effective for mice
BMC Veterinary Research (2022)