Key Points
-
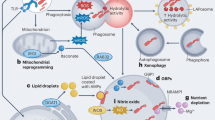
Autophagy is used by the cell to degrade various substrates; this is achieved either through the canonical, non-selective autophagy pathway or through selective autophagy. Both pathways proceed via distinct key steps and use specific molecular mechanisms.
-
The canonical autophagy pathway has been studied in detail in mammalian cells and in model organisms, such as yeast. The molecular mechanisms underlying non-canonical autophagy, in addition to alternative pathways that are independent of some of the key autophagy machinery, are beginning to become clear.
-
Besides degradation of cellular proteins, autophagy proteins are also involved in many other functions, some of which are important during bacterial infections.
-
Autophagy functions as an antibacterial mechanism. The induction and recognition mechanisms for several bacterial species have been elucidated.
-
Bacteria can escape killing by autophagy and some can even use autophagy to promote infection of host cells, through the interaction between bacterial effector proteins and autophagy components.
-
The knowledge about bacteria–autophagy interactions will inform the design of new drugs and treatments against bacterial infections.
Abstract
Autophagy is a cellular process that targets proteins, lipids and organelles to lysosomes for degradation, but it has also been shown to combat infection with various pathogenic bacteria. In turn, bacteria have developed diverse strategies to avoid autophagy by interfering with autophagy signalling or the autophagy machinery and, in some cases, they even exploit autophagy for their growth. In this Review, we discuss canonical and non-canonical autophagy pathways and our current knowledge of antibacterial autophagy, with a focus on the interplay between bacterial factors and autophagy components.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Klionsky, D. J. et al. How shall I eat thee? Autophagy 3, 413–416 (2007).
Shahnazari, S. & Brumell, J. H. Mechanisms and consequences of bacterial targeting by the autophagy pathway. Curr. Opin. Microbiol. 14, 68–75 (2011).
Baxt, L. A., Garza-Mayers, A. C. & Goldberg, M. B. Bacterial subversion of host innate immune pathways. Science 340, 697–701 (2013).
Shahnazari, S., Namolovan, A., Mogridge, J., Kim, P. K. & Brumell, J. H. Bacterial toxins can inhibit host cell autophagy through cAMP generation. Autophagy 7, 957–965 (2011).
Tattoli, I. et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563–575 (2012). This study shows that bacteria inhibit autophagy induction signalling by affecting mTOR localization.
Ogawa, M. et al. Escape of intracellular Shigella from autophagy. Science 307, 727–731 (2005). This work shows that some bacterial pathogens can avoid autophagy recognition by camouflaging themselves with effector proteins.
Dortet, L. et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 7, e1002168 (2011).
Yoshikawa, Y. et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nature Cell Biol. 11, 1233–1240 (2009).
Dong, N. et al. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150, 1029–1041 (2012). This study shows that a bacterial effector can turn off an autophagy-regulating RAB GTPase to escape killing of the bacterium by autophagy.
Choy, A. et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338, 1072–1076 (2012). This study shows that a bacterial effector can directly uncouple the LC3–PE conjugation to inhibit autophagy.
Chargui, A. et al. Subversion of autophagy in adherent invasive Escherichia coli-infected neutrophils induces inflammation and cell death. PLoS ONE 7, e51727 (2012).
Niu, H., Yamaguchi, M. & Rikihisa, Y. Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell. Microbiol. 10, 593–605 (2008).
Schnaith, A. et al. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J. Biol. Chem. 282, 2695–2706 (2007).
Parzych, K. R. & Klionsky, D. J. An overview of autophagy: morphology, mechanism, and regulation. Antioxid. Redox Signal http://dx.doi.org/10.1089/ars.2013.5371 (2013).
Axe, E. L. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 (2008).
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009).
Mizushima, N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139 (2010).
Jung, C. H. et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009).
Ganley, I. G. et al. ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 (2009).
Itakura, E. & Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776 (2010).
Komduur, J. A., Veenhuis, M. & Kiel, J. A. The Hansenula polymorpha PDD7 gene is essential for macropexophagy and microautophagy. FEMS Yeast Res. 3, 27–34 (2003).
Kanki, T. et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell 20, 4730–4738 (2009).
Kim, J. & Klionsky, D. J. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 69, 303–342 (2000).
Kageyama, S. et al. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol. Biol. Cell 22, 2290–2300 (2011).
Nair, U., Cao, Y., Xie, Z. & Klionsky, D. J. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J. Biol. Chem. 285, 11476–11488 (2010).
Polson, H. E. et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6, 506–522 (2010).
Mari, M. et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190, 1005–1022 (2010).
Yamamoto, H. et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 198, 219–233 (2012).
Orsi, A. et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 23, 1860–1873 (2012).
Young, A. R. et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 (2006).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Mizushima, N. et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–668 (2001).
Mizushima, N. et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116, 1679–1688 (2003).
Moreau, K., Ravikumar, B., Renna, M., Puri, C. & Rubinsztein, D. C. Autophagosome precursor maturation requires homotypic fusion. Cell 146, 303–317 (2012).
Fujita, N. et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 (2008).
Hanada, T. et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 (2007).
Tanida, I., Ueno, T. & Kominami, E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 36, 2503–2518 (2004).
Nair, U. et al. A role for Atg8–PE deconjugation in autophagosome biogenesis. Autophagy 8, 780–793 (2012).
Yu, Z. Q. et al. Dual roles of Atg8–PE deconjugation by Atg4 in autophagy. Autophagy 8, 883–892 (2012).
Nakatogawa, H., Ichimura, Y. & Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 (2007).
Xie, Z., Nair, U. & Klionsky, D. J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298 (2008).
Jager, S. et al. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117, 4837–4848 (2004).
Itakura, E., Kishi-Itakura, C. & Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012).
Tanaka, Y. et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406, 902–906 (2000).
Eskelinen, E. L. et al. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol. Biol. Cell 13, 3355–3368 (2002).
Reggiori, F. & Klionsky, D. J. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194, 341–361 (2013).
Hanna, R. A. et al. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 287, 19094–19104 (2012).
Liu, L. et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nature Cell Biol. 14, 177–185 (2012).
Novak, I. et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 (2010).
Boyle, K. B. & Randow, F. The role of 'eat-me' signals and autophagy cargo receptors in innate immunity. Curr. Opin. Microbiol. 16, 339–348 (2013).
Dupont, N. et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 6, 137–149 (2009).
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007).
Kim, P. K., Hailey, D. W., Mullen, R. T. & Lippincott-Schwartz, J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl Acad. Sci. USA 105, 20567–20574 (2008).
Mostowy, S. et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J. Biol. Chem. 286, 26987–26995 (2011).
Zheng, Y. T. et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 183, 5909–5916 (2009).
Kirkin, V. et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 (2009).
Thurston, T. L., Ryzhakov, G., Bloor, S., von Muhlinen, N. & Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nature Immunol. 10, 1215–1221 (2009).
Wild, P. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 (2011).
Deosaran, E. et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126, 939–952 (2013).
Chong, A. et al. Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy 8, 1342–1356 (2012).
Cheong, H., Lindsten, T., Wu, J., Lu, C. & Thompson, C. B. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl Acad. Sci. USA 108, 11121–11126 (2011).
Nishida, Y. et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658 (2009). This study shows that autophagy can occur in the absence of the two ubiquitin-like conjugation systems that are essential for autophagosome formation.
Grishchuk, Y., Ginet, V., Truttmann, A. C., Clarke, P. G. & Puyal, J. Beclin 1-independent autophagy contributes to apoptosis in cortical neurons. Autophagy 7, 1115–1131 (2011).
Scarlatti, F., Maffei, R., Beau, I., Codogno, P. & Ghidoni, R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 15, 1318–1329 (2008).
Zhu, J. H. et al. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol. 170, 75–86 (2007).
Liao, C. C., Ho, M. Y., Liang, S. M. & Liang, C. M. Recombinant protein rVP1 upregulates BECN1-independent autophagy, MAPK1/3 phosphorylation and MMP9 activity via WIPI1/WIPI2 to promote macrophage migration. Autophagy 9, 5–19 (2012).
Wong, C. H. et al. Simultaneous induction of non-canonical autophagy and apoptosis in cancer cells by ROS-dependent ERK and JNK activation. PLoS ONE 5, e9996 (2010).
Kondylis, V. et al. Endosome-mediated autophagy: An unconventional MIIC-driven autophagic pathway operational in dendritic cells. Autophagy 9 861–880 (2013).
Sanjuan, M. A. et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 (2007). This study reports the discovery of the LAP pathway and shows that recruiting autophagy components to the phagosome promotes phagosome fusion with the lysosome.
Huang, J. et al. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl Acad. Sci. USA 106, 6226–6231 (2009).
Martinez, J. et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl Acad. Sci. USA 108, 17396–17401 (2011).
Gong, L. et al. The Burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS ONE 6, e17852 (2011).
Jia, K. et al. Autophagy genes protect against Salmonella Typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc. Natl Acad. Sci. USA 106, 14564–14569 (2009).
Brumell, J. H., Steele-Mortimer, O. & Finlay, B. B. Bacterial invasion: force feeding by Salmonella. Curr. Biol. 9, R277–R280 (1999).
Perrin, A. J., Jiang, X., Birmingham, C. L., So, N. S. & Brumell, J. H. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr. Biol. 14, 806–811 (2004).
Birmingham, C. L., Smith, A. C., Bakowski, M. A., Yoshimori, T. & Brumell, J. H. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 281, 11374–11383 (2006). This study shows that S . Typhimurium is targeted by autophagy in a damaged vacuole; on the basis of this work, extensive research has been done on the mechanisms of antibacterial autophagy, using S . Typhimurium as a model.
Huett, A. et al. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe 12, 778–790 (2012).
Thurston, T. L., Wandel, M. P., von Muhlinen, N., Foeglein, A. & Randow, F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012). This work reveals that sugar molecules on the plasma membrane can serve as signals to trigger antibacterial autophagy.
Shahnazari, S. et al. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 8, 137–146 (2010).
Fontayne, A., Dang, P. M., Gougerot-Pocidalo, M. A. & El-Benna, J. Phosphorylation of p47phox sites by PKC α, βII, δ, and ζ: effect on binding to p22phox and on NADPH oxidase activation. Biochemistry 41, 7743–7750 (2002).
Lam, G. Y. et al. Listeriolysin O suppresses phospholipase C-mediated activation of the microbicidal NADPH oxidase to promote Listeria monocytogenes infection. Cell Host Microbe 10, 627–634 (2011).
Watson, R. O., Manzanillo, P. S. & Cox, J. S. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815 (2012).
Wang, W., Sacher, M. & Ferro-Novick, S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J. Cell Biol. 151, 289–296 (2000).
Manzanillo, P. S. et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501, 512–516 (2013).
Nakagawa, I. et al. Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040 (2004). This is the first study to show that autophagy is an innate immune defence mechanism against an intracellular bacterial pathogen during natural infection.
Sakurai, A. et al. Specific behavior of intracellular Streptococcus pyogenes that has undergone autophagic degradation is associated with bacterial streptolysin O and host small G proteins Rab5 and Rab7. J. Biol. Chem. 285, 22666–22675 (2010).
Joubert, P. E. et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 6, 354–366 (2009).
Nozawa, T. et al. The small GTPases Rab9A and Rab23 function at distinct steps in autophagy during Group A Streptococcus infection. Cell. Microbiol. 14, 1149–1165 (2012).
O'Seaghdha, M. & Wessels, M. R. Streptolysin O and its co-toxin NAD-glycohydrolase protect group A Streptococcus from xenophagic killing. PLoS Pathog. 9, e1003394 (2013).
Checroun, C., Wehrly, T. D., Fischer, E. R., Hayes, S. F. & Celli, J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl Acad. Sci. USA 103, 14578–14583 (2006).
Deuretzbacher, A. et al. β1 integrin-dependent engulfment of Yersinia enterocolitica by macrophages is coupled to the activation of autophagy and suppressed by type III protein secretion. J. Immunol. 183, 5847–5860 (2009).
Choi, J. H. et al. Orientia tsutsugamushi subverts dendritic cell functions by escaping from autophagy and impairing their migration. PLoS Negl. Trop. Dis. 7, e1981 (2013).
Ko, Y. et al. Active escape of Orientia tsutsugamushi from cellular autophagy. Infect. Immun. 81, 552–559 (2012).
Eriksson, S., Lucchini, S., Thompson, A., Rhen, M. & Hinton, J. C. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47, 103–118 (2003).
Shin, D. M. et al. Mycobacterium tuberculosis Eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 6, e1001230 (2010).
Kim, K. H. et al. Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc. Natl Acad. Sci. USA 109, 7729–7734 (2012).
Beebe, S. J. The cAMP-dependent protein kinases and cAMP signal transduction. Semin. Cancer Biol. 5, 285–294 (1994).
Serezani, C. H., Ballinger, M. N., Aronoff, D. M. & Peters-Golden, M. Cyclic AMP: master regulator of innate immune cell function. Am. J. Respir. Cell. Mol. Biol. 39, 127–132 (2008).
Holen, I., Gordon, P. B., Stromhaug, P. E. & Seglen, P. O. Role of cAMP in the regulation of hepatocytic autophagy. Eur. J. Biochem. 236, 163–170 (1996).
Leppla, S. H. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc. Natl Acad. Sci. USA 79, 3162–3166 (1982).
Cassel, D. & Pfeuffer, T. Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc. Natl Acad. Sci. USA 75, 2669–2673 (1978).
Amer, A. O. & Swanson, M. S. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell. Microbiol. 7, 765–778 (2005).
Huang, J. et al. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy 7, 17–26 (2010).
Zoppino, F. C., Militello, R. D., Slavin, I., Alvarez, C. & Colombo, M. I. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11, 1246–1261 (2010).
Mochizuki, Y. et al. Phosphatidylinositol 3-phosphatase myotubularin-related protein 6 (MTMR6) is regulated by small GTPase Rab1B in the early secretory and autophagic pathways. J. Biol. Chem. 288, 1009–1021 (2012).
Allaoui, A., Mounier, J., Prevost, M. C. & Sansonetti, P. J. & Parsot, C. icsB: a Shigella flexneri virulence gene necessary for the lysis of protrusions during intercellular spread. Mol. Microbiol. 6, 1605–1616 (1992).
Ogawa, M., Suzuki, T., Tatsuno, I., Abe, H. & Sasakawa, C. IcsB, secreted via the type III secretion system, is chaperoned by IpgA and required at the post-invasion stage of Shigella pathogenicity. Mol. Microbiol. 48, 913–931 (2003).
Mostowy, S. et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe 8, 433–444 (2010).
Dussurget, O., Pizarro-Cerda, J. & Cossart, P. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58, 587–610 (2004).
Gouin, E., Welch, M. D. & Cossart, P. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 8, 35–45 (2005).
Birmingham, C. L. et al. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy 3, 442–451 (2007).
Lam, G. Y., Cemma, M., Muise, A. M., Higgins, D. E. & Brumell, J. H. Host and bacterial factors that regulate LC3 recruitment to Listeria monocytogenes during the early stages of macrophage infection. Autophagy 9 985–995 (2013).
Rich, K. A., Burkett, C. & Webster, P. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 5, 455–468 (2003).
D'Cruze, T. et al. Role for the Burkholderia pseudomallei type three secretion system cluster 1 bpscN gene in virulence. Infect. Immun. 79, 3659–3664 (2011).
Li, X., Prescott, M., Adler, B., Boyce, J. D. & Devenish, R. J. Beclin 1 is required for starvation-enhanced, but not rapamycin-enhanced, LC3-associated phagocytosis of Burkholderia pseudomallei in RAW 264.7 cells. Infect. Immun. 81, 271–277 (2012).
Gutierrez, M. G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004).
Seto, S., Tsujimura, K. & Koide, Y. Coronin-1a inhibits autophagosome formation around Mycobacterium tuberculosis-containing phagosomes and assists mycobacterial survival in macrophages. Cell. Microbiol. 14, 710–727 (2012).
Lapaquette, P., Bringer, M. A. & Darfeuille-Michaud, A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell. Microbiol. 14, 791–807 (2012).
Lerena, M. C. & Colombo, M. I. Mycobacterium marinum induces a marked LC3 recruitment to its containing phagosome that depends on a functional ESX-1 secretion system. Cell. Microbiol. 13, 814–835 (2011).
Romagnoli, A. et al. ESX-1 dependent impairment of autophagic flux by Mycobacterium tuberculosis in human dendritic cells. Autophagy 8, 1357–1370 (2012).
Al-Younes, H. M., Brinkmann, V. & Meyer, T. F. Interaction of Chlamydia trachomatis serovar L2 with the host autophagic pathway. Infect. Immun. 72, 4751–4762 (2004).
Yasir, M., Pachikara, N. D., Bao, X., Pan, Z. & Fan, H. Regulation of chlamydial infection by host autophagy and vacuolar ATPase-bearing organelles. Infect. Immun. 79, 4019–4028 (2011).
Pujol, C. et al. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect. Immun. 77, 2251–2261 (2009).
Raju, D. et al. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote Helicobacter pylori infection in humans. Gastroenterology 142, 1160–1171 (2012).
Wang, Y. H., Wu, J. J. & Lei, H. Y. The autophagic induction in Helicobacter pylori-infected macrophage. Exp. Biol. Med. (Maywood) 234, 171–180 (2009).
Niu, H., Xiong, Q., Yamamoto, A., Hayashi-Nishino, M. & Rikihisa, Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc. Natl Acad. Sci. USA 109, 20800–20807 (2012). This study shows that a bacterial effector directly binds autophagy components to actively recruit the autophagosome to the bacteria-containing inclusions to promote bacterial replication.
Gutierrez, M. G. et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell. Microbiol. 7, 981–993 (2005).
Beron, W., Gutierrez, M. G., Rabinovitch, M. & Colombo, M. I. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect. Immun. 70, 5816–5821 (2002).
Romano, P. S., Gutierrez, M. G., Beron, W., Rabinovitch, M. & Colombo, M. I. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell. Microbiol. 9, 891–909 (2007).
Vazquez, C. L. & Colombo, M. I. Coxiella burnetii modulates Beclin 1 and Bcl-2, preventing host cell apoptosis to generate a persistent bacterial infection. Cell Death Differ. 17, 421–438 (2009).
Starr, T. et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 11, 33–45 (2012).
Fedrigo, G. V., Campoy, E. M., Di Venanzio, G., Colombo, M. I. & Garcia Vescovi, E. Serratia marcescens is able to survive and proliferate in autophagic-like vacuoles inside non-phagocytic cells. PLoS ONE 6, e24054 (2011).
Fine, K. L. et al. Involvement of the autophagy pathway in trafficking of Mycobacterium tuberculosis bacilli through cultured human type II epithelial cells. Cell. Microbiol. 14, 1402–1414 (2012).
Amer, A. O., Byrne, B. G. & Swanson, M. S. Macrophages rapidly transfer pathogens from lipid raft vacuoles to autophagosomes. Autophagy 1, 53–58 (2005).
Guo, F. et al. Autophagy favors Brucella melitensis survival in infected macrophages. Cell. Mol. Biol. Lett. 17, 249–257 (2012).
Dorn, B. R., Dunn, W. A. Jr & Progulske-Fox, A. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect. Immun. 69, 5698–5708 (2001).
Wang, C. et al. Atg16L1 deficiency confers protection from uropathogenic Escherichia coli infection in vivo. Proc. Natl Acad. Sci. USA 109, 11008–11013 (2012).
Moreau, K. et al. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell. Microbiol. 12, 1108–1123 (2010).
Mestre, M. B., Fader, C. M., Sola, C. & Colombo, M. I. α-hemolysin is required for the activation of the autophagic pathway in Staphylococcus aureus-infected cells. Autophagy 6, 110–125 (2010).
Mestre, M. B. & Colombo, M. I. cAMP and EPAC are key players in the regulation of the signal transduction pathway involved in the α-hemolysin autophagic response. PLoS Pathog. 8, e1002664 (2012).
Travassos, L. H. et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature Immunol. 11, 55–62 (2010).
Ogawa, M. et al. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe 9, 376–389 (2011).
Boada-Romero, E. et al. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 32, 566–582 (2013).
Florey, O., Kim, S. E., Sandoval, C. P., Haynes, C. M. & Overholtzer, M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nature Cell Biol. 13, 1335–1343 (2011).
Liang, C. et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nature Cell Biol. 10, 776–787 (2008).
Kim, H. J. et al. Beclin-1-interacting autophagy protein Atg14L targets the SNARE-associated protein Snapin to coordinate endocytic trafficking. J. Cell Sci. 125, 4740–4750 (2012).
Yousefi, S. et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nature Cell Biol. 8, 1124–1132 (2006).
Dupont, N. et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 30, 4701–4711 (2011).
Jounai, N. et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl Acad. Sci. USA 104, 14050–14055 (2007).
Zhao, Z. et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469 (2008).
Lee, H. K. et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32, 227–239 (2010).
Henault, J. et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 37, 986–997 (2012).
Lee, I. H. et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 336, 225–228 (2012).
Monastyrska, I. et al. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy 9, 164–174 (2012).
Reggiori, F. et al. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7, 500–508 (2010).
Al-Younes, H. M. et al. Autophagy-independent function of MAP-LC3 during intracellular propagation of Chlamydia trachomatis. Autophagy 7, 814–828 (2011).
Ma, J., Becker, C., Lowell, C. A. & Underhill, D. M. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J. Biol. Chem. 287, 34149–34156 (2012).
Py, B. F., Lipinski, M. M. & Yuan, J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy 3, 117–125 (2007).
Acknowledgements
John H. Brumell holds the Pitblado Chair in Cell Biology, a joint University-Hospital Chair between the University of Toronto, the Hospital for Sick Children and the Sickkids Foundation. Research into autophagy by the Brumell lab has been supported by an Investigators in Pathogenesis Award from the Burroughs Wellcome Fund and an operating grant from The Arthritis Society of Canada. Infrastructure for the Brumell laboratory was provided by the Canadian Foundation for Innovation and the Ontario Innovation Trust. Ju Huang holds a postdoctoral fellowship from the Canadian Institutes of Health Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Cytoplasm-to-vacuole targeting pathway
-
(Cvt pathway). A constitutive biosynthetic pathway that occurs under nutrient-rich conditions and that delivers precursor aminopeptidase I into the vacuole for maturation.
- Trans-Golgi network
-
(TGN). A network of tubular and vesicular structures at the trans face (that is, the side responsible for export) of the Golgi apparatus.
- E1-like enzyme
-
A protein enzyme that, like a ubiquitin-activating enzyme (E1) in the ubiquitylation reaction, catalyses the first step in the covalent conjugation of a ubiquitin-like molecule to the target protein.
- E2-like enzyme
-
A protein enzyme that, like a ubiquitin-conjugating enzyme (E2) in the ubiquitylation reaction, catalyses the second step in covalent conjugation of a ubiquitin-like molecule to the target protein.
- SNARE proteins
-
(Soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins). A family of membrane proteins that mediate membrane fusion during vesicle fusion and exocytosis. Vesicle-membrane SNAREs (v-SNAREs) localize on the membranes of transporting vesicles, whereas target-membrane SNAREs (t-SNAREs) localize on the target membrane.
- E3-like enzyme
-
An enzyme that functions similarly to the ubiquitin ligase (E3), which transfers the ubiquitin from E2 to the substrate by catalysing the covalent attachment of ubiquitin to a lysine residue in the substrate.
- Guanine nucleotide exchange factor
-
(GEF). Proteins that bind to small GTPases and catalyse the release of a GDP molecule from, and then the binding of a GTP molecule to, the GTPase substrate to activate the GTPase.
- Toll-like receptor
-
(TLR). A protein that belongs to a family of transmembrane protein receptors, usually found in immune cells. It recognizes specific microorganisms and induces immune responses.
- Dendritic cells
-
Immune cells that process and present antigens on their surfaces.
- Fc receptor
-
An immune cell surface protein receptor that recognizes the Fc region of antibodies and activates phagocytosis of antibody-tagged microorganisms.
- Reactive oxygen species
-
(ROS). Oxygen free radicals that harbour unpaired electrons and are highly unstable and reactive.
- Type I interferons
-
(Type I IFNs). A group of cytokines that have antiviral functions. In humans, this group consists of IFNα, IFNβ and IFNω. Type I IFNs all bind to the IFNα receptor.
- STING
-
(Stimulator of interferon genes). An endoplasmic reticulum-resident transmembrane protein that is involved in the initiation of type I interferon production by cytosolic double-stranded DNA in cells.
- Coronin
-
A eukaryotic actin-binding protein that is involved in many actin-mediated cellular processes, such as phagocytosis.
Rights and permissions
About this article
Cite this article
Huang, J., Brumell, J. Bacteria–autophagy interplay: a battle for survival. Nat Rev Microbiol 12, 101–114 (2014). https://doi.org/10.1038/nrmicro3160
Published:
Issue date:
DOI: https://doi.org/10.1038/nrmicro3160
This article is cited by
-
Selenomethionine Inhibits NF-κB-mediated Inflammatory Responses of Bovine Mammary Epithelial Cells Caused by Klebsiella pneumoniae by Increasing Autophagic Flux
Biological Trace Element Research (2024)
-
Evaluation of the mTORC activity in the presence of Toxoplasma gondii and azathioprine in human monocyte cell line
BMC Microbiology (2023)
-
Autophagy promotes jasmonate-mediated defense against nematodes
Nature Communications (2023)
-
Porphyromonas gingivalis, a periodontal pathogen, impairs post-infarcted myocardium by inhibiting autophagosome–lysosome fusion
International Journal of Oral Science (2023)
-
NRF2 and STAT3: friends or foes in carcinogenesis?
Discover Oncology (2023)