Abstract
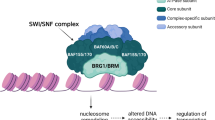
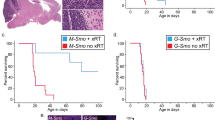
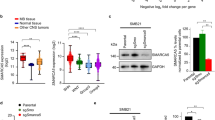
Recent large-scale genomic studies have classified medulloblastoma into four subtypes: Wnt, Shh, Group 3 and Group 4. Each is characterized by specific mutations and distinct epigenetic states. Previously, we showed that a chromatin regulator SMARCA4/Brg1 is required for Gli-mediated transcription activation in Sonic hedgehog (Shh) signaling. We report here that Brg1 controls a transcriptional program that specifically regulates Shh-type medulloblastoma growth. Using a mouse model of Shh-type medulloblastoma, we deleted Brg1 in precancerous progenitors and primary or transplanted tumors. Brg1 deletion significantly inhibited tumor formation and progression. Genome-wide expression analyses and binding experiments indicate that Brg1 specifically coordinates with key transcription factors including Gli1, Atoh1 and REST to regulate the expression of both oncogenes and tumor suppressors that are required for medulloblastoma identity and proliferation. Shh-type medulloblastoma displays distinct H3K27me3 properties. We demonstrate that Brg1 modulates activities of H3K27me3 modifiers to regulate the expression of medulloblastoma genes. Brg1-regulated pathways are conserved in human Shh-type medulloblastoma, and Brg1 is important for the growth of a human medulloblastoma cell line. Thus, Brg1 coordinates a genetic and epigenetic network that regulates the transcriptional program underlying the Shh-type medulloblastoma development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol 2006; 7: 813–820.
Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer 2012; 12: 818–834.
Rutkowski S, von Hoff K, Emser A, Zwiener I, Pietsch T, Figarella-Branger D et al. Survival and prognostic factors of early childhood medulloblastoma: an international meta-analysis. J Clin Oncol 2010; 28: 4961–4968.
Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 2012; 488: 100–105.
Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012; 488: 49–56.
Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC et al. The genetic landscape of the childhood cancer medulloblastoma. Science 2011; 331: 435–439.
Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 2012; 488: 106–110.
Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012; 488: 43–48.
Barakat MT, Humke EW, Scott MP . Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol Med 2010; 16: 337–348.
Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 2014; 25: 393–405.
Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL . Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development 2004; 131: 5581–5590.
Dahmane N, Ruiz i Altaba A . Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 1999; 126: 3089–3100.
Wallace VA . Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol 1999; 9: 445–448.
Wang VY, Zoghbi HY . Genetic regulation of cerebellar development. Nat Rev Neurosci 2001; 2: 484–491.
Briscoe J, Therond PP . The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 2013; 14: 416–429.
Fuccillo M, Joyner AL, Fishell G . Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci 2006; 7: 772–783.
Ingham PW, McMahon AP . Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001; 15: 3059–3087.
Jiang J, Hui CC . Hedgehog signaling in development and cancer. Dev Cell 2008; 15: 801–812.
Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 2008; 14: 123–134.
Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q et al. Math1 is essential for genesis of cerebellar granule neurons. Nature 1997; 390: 169–172.
Flora A, Klisch TJ, Schuster G, Zoghbi HY . Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science 2009; 326: 1424–1427.
Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 1995; 80: 949–957.
Negrini S, Prada I, D'Alessandro R, Meldolesi J . REST: an oncogene or a tumor suppressor? Trends Cell Biol 2013; 23: 289–295.
Schoenherr CJ, Anderson DJ . The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 1995; 267: 1360–1363.
Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol 2010; 12: 132–142.
Malatesta M, Steinhauer C, Mohammad F, Pandey DP, Squartrito M, Helin K . Histone acetyltransferase PCAF is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res 2013; 73: 6323–6333.
Zhan X, Shi X, Zhang Z, Chen Y, Wu JI . Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc Natl Acad Sci USA 2011; 108: 12758–12763.
Hargreaves DC, Crabtree GR . ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 2011; 21: 396–420.
Wu JI . Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim Biophys Sin (Shanghai) 2012; 44: 54–69.
Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013; 45: 592–601.
Wilson BG, Roberts CW . SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011; 11: 481–492.
Hasselblatt M, Nagel I, Oyen F, Bartelheim K, Russell RB, Schuller U et al. SMARCA4-mutated atypical teratoid/rhabdoid tumors are associated with inherited germline alterations and poor prognosis. Acta Neuropathol 2014; 128: 453–456.
Witkowski L, Carrot-Zhang J, Albrecht S, Fahiminiya S, Hamel N, Tomiak E et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet 2014; 46: 438–443.
Romero OA, Torres-Diz M, Pros E, Savola S, Gomez A, Moran S et al. MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov 2014; 4: 292–303.
Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev 2013; 27: 2648–2662.
Shi X, Zhang Z, Zhan X, Cao M, Satoh T, Akira S et al. An epigenetic switch induced by Shh signalling regulates gene activation during development and medulloblastoma growth. Nat Commun 2014; 5: 5425.
Li Q, Wang HY, Chepelev I, Zhu Q, Wei G, Zhao K et al. Stage-dependent and locus-specific role of histone demethylase Jumonji D3 (JMJD3) in the embryonic stages of lung development. PLoS Genet 2014; 10: e1004524.
Miller SA, Mohn SE, Weinmann AS . Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 2010; 40: 594–605.
Popov N, Gil J . Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics 2010; 5: 685–690.
Yang H, Xie X, Deng M, Chen X, Gan L . Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis 2010; 48: 407–413.
Moreno N, Schmidt C, Ahlfeld J, Poschl J, Dittmar S, Pfister SM et al. Loss of Smarc proteins impairs cerebellar development. J Neurosci 2014; 34: 13486–13491.
Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res 2006; 66: 10171–10178.
Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W . A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 2009; 25: 1952–1958.
Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR . An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA 2009; 106: 5187–5191.
Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 2013; 152: 248–261.
Lee EY, Ji H, Ouyang Z, Zhou B, Ma W, Vokes SA et al. Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc Natl Acad Sci USA 2010; 107: 9736–9741.
Dakubo GD, Mazerolle CJ, Wallace VA . Expression of Notch and Wnt pathway components and activation of Notch signaling in medulloblastomas from heterozygous patched mice. J Neurooncol 2006; 79: 221–227.
Fernandez LA, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev 2009; 23: 2729–2741.
Natarajan S, Li Y, Miller EE, Shih DJ, Taylor MD, Stearns TM et al. Notch1-induced brain tumor models the sonic hedgehog subgroup of human medulloblastoma. Cancer Res 2013; 73: 5381–5390.
Anne SL, Govek EE, Ayrault O, Kim JH, Zhu X, Murphy DA et al. WNT3 inhibits cerebellar granule neuron progenitor proliferation and medulloblastoma formation via MAPK activation. PLoS One 2013; 8: e81769.
Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY . In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci USA 2011; 108: 3288–3293.
Markant SL, Esparza LA, Sun J, Barton KL, McCoig LM, Grant GA et al. Targeting sonic hedgehog-associated medulloblastoma through inhibition of Aurora and Polo-like kinases. Cancer Res 2013; 73: 6310–6322.
Sengupta R, Dubuc A, Ward S, Yang L, Northcott P, Woerner BM et al. CXCR4 activation defines a new subgroup of Sonic hedgehog-driven medulloblastoma. Cancer Res 2012; 72: 122–132.
Smits M, van Rijn S, Hulleman E, Biesmans D, van Vuurden DG, Kool M et al. EZH2-regulated DAB2IP is a medulloblastoma tumor suppressor and a positive marker for survival. Clin Cancer Res 2012; 18: 4048–4058.
Uziel T, Zindy F, Sherr CJ, Roussel MF . The CDK inhibitor p18Ink4c is a tumor suppressor in medulloblastoma. Cell Cycle 2006; 5: 363–365.
Wetmore C, Eberhart DE, Curran T . Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res 2001; 61: 513–51669.
Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol 2008; 6: e256.
Ooi L, Belyaev ND, Miyake K, Wood IC, Buckley NJ . BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J Biol Chem 2006; 281: 38974–38980.
Triscott J, Lee C, Foster C, Manoranjan B, Pambid MR, Berns R et al. Personalizing the treatment of pediatric medulloblastoma: polo-like kinase 1 as a molecular target in high-risk children. Cancer Res 2013; 73: 6734–6744.
Sengupta S, Weeraratne SD, Sun H, Phallen J, Rallapalli SK, Teider N et al. alpha5-GABAA receptors negatively regulate MYC-amplified medulloblastoma growth. Acta Neuropathol 2014; 127: 593–603.
Wu JI, Lessard J, Crabtree GR . Understanding the words of chromatin regulation. Cell 2009; 136: 200–206.
Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998; 394: 203–206.
Jagani Z, Mora-Blanco EL, Sansam CG, McKenna ES, Wilson B, Chen D et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat Med 2010; 16: 1429–1433.
Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH, Mendez-Lago M et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol 2013; 125: 373–384.
Wang X, Sansam CG, Thom CS, Metzger D, Evans JA, Nguyen PT et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res 2009; 69: 8094–8101.
Dietrich N, Lerdrup M, Landt E, Agrawal-Singh S, Bak M, Tommerup N et al. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet 2012; 8: e1002494.
Hayashi S, McMahon AP . Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol 2002; 244: 305–318.
Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P . SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol 1997; 17: 5976–5986.
Chen JK, Taipale J, Young KE, Maiti T, Beachy PA . Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA 2002; 99: 14071–14076.
Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell 2008; 14: 298–311.
Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR . BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 1993; 366: 170–174.
Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 2007; 55: 201–215.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515.
Langmead B, Trapnell C, Pop M, Salzberg SL . Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10: R25.
Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 2011; 21: 1650–1658.
Acknowledgements
We thank Drs Brent Orr (St. Jude Hospital), Wenzhe Niu and Zilai Zhang, and Mou Cao for technical support and Dr Chao Xing and the University of Texas Southwestern Sequencing Facility for performing the next-generation sequencing and RNA-seq analyses. We thank Drs Lin Gan and Jane Johnson for providing the Atoh1-Cre mice and Dr Ching-Ping Chang for providing human Brg1 shRNA constructs. This work was supported by grants from March of Dimes Foundation (JW), American Cancer Society (JW), NIMH (JW) and Department of Defense Visionary postdoc fellowship (XS).
Author contributions
JW and XS designed the experiments. XS, QW and JW performed the experiments and collected the data. XS and JW analyzed the results. JG and ZX performed the main bioinformatics analyses. JW wrote the manuscript with help from all the authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Shi, X., Wang, Q., Gu, J. et al. SMARCA4/Brg1 coordinates genetic and epigenetic networks underlying Shh-type medulloblastoma development. Oncogene 35, 5746–5758 (2016). https://doi.org/10.1038/onc.2016.108
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/onc.2016.108