Abstract
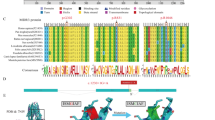
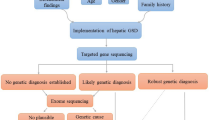
PFIC4 is a chronic liver disease which cannot be diagnosed based on clinical and biochemical findings with an unpredictable evolution. Here, we reported three consanguineous families with 9 children suffering from intrahepatic cholestasis with low GGT-activity. Three probands were chosen to undergo genetic testing. In silico analyses were conducted to assess the functional impact of the identified variant, along with variants occurring at highly conserved positions within the protein. Additionally, close clinical monitoring was carried. Targeted-NGS sequencing ruled out the diagnosis of PFIC1 and PFIC2. Subsequently, WES allowed the establishment of PFIC4 diagnosis for the three families through the identification of a homozygous TJP2 variant p. Gly532Arg classified as likely pathogenic with a structural damage predicted based on biomolecular modeling and simulation analysis. In-depth in silico analysis of 90 nsSNPs occurring in highly conserved residues in PDZ domains showed 14 ones seems to be relevant in the clinical practice. Clinically, a pronounced phenotypic variability is noted. In conclusion, our study described a homozygous missense PFIC4-related variant with a highlight on the pathogenic power of such types of variants. The clinical evaluation provided information about the importance of close monitoring to prevent liver failure and clarified the unexpected course of PFIC4.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Amer S, Hajira A. A comprehensive review of progressive familial intrahepatic cholestasis (PFIC): genetic disorders of hepatocanalicular transporters. Gastroenterol Res. 2014;7:39.
Vitale G, Mattiaccio A, Conti A, Turco L, Seri M, Piscaglia F, et al. Genetics in familial intrahepatic cholestasis: clinical patterns and development of liver and biliary cancers: a review of the literature. Cancers. 2022;14:3421.
Sambrotta M, Strautnieks S, Papouli E, Rushton P, Clark BE, Parry DA, et al. Mutations in TJP2 cause progressive cholestatic liver disease. Nat Genet. 2014;46:326–8.
Zhang J, Liu LL, Gong JY, Hao CZ, Qiu YL, Lu Y, et al. TJP2 hepatobiliary disorders: novel variants and clinical diversity. Hum Mutat. 2020;41:502–11.
Wei CS, Becher N, Friis JB, Ott P, Vogel I, Grønbæk H. New tight junction protein 2 variant causing progressive familial intrahepatic cholestasis type 4 in adults: A case report. World J Gastroenterol. 2020;26:550.
Carlton VE, Harris BZ, Puffenberger EG, Batta AK, Knisely AS, Robinson DL, et al. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet. 2003;34:91–96.
Dixon PH, Sambrotta M, Chambers J, Taylor-Harris P, Syngelaki A, Nicolaides K, et al. An expanded role for heterozygous mutations of ABCB4, ABCB11, ATP8B1, ABCC2 and TJP2 in intrahepatic cholestasis of pregnancy. Sci Rep. 2017;7. 11823.
Kornitzer GA, Alvarez F. Case report: a novel single variant TJP2 mutation in a case of benign recurrent intrahepatic cholestasis. JPGN Rep. 2021;2:e087.
Beatch M, Jesaitis LA, Gallin WJ, Goodenough DA, Stevenson BR. The tight junction protein ZO-2 contains three PDZ (PSD-95Discs-LargeZO-1) domains and an alternatively spliced region. J Biol Chem. 1996;271:25723–6.
Gonzalez-Mariscal L, Bautista P, Lechuga S, Quiros M. ZO-2, a tight junction scaffold protein involved in the regulation of cell proliferation and apoptosis. Ann N Y Acad Sci. 2012;1257:133–41.
Itoh M, Terada M, Sugimoto H. The zonula occludens protein family regulates the hepatic barrier system in the murine liver. Biochimica et Biophysica Acta (BBA)-Mol Basis Dis. 2021;1867:165994.
Shinwari K, Guojun L, Deryabina SS, Bolkov MA, Tuzankina IA, Chereshnev VA. Predicting the most deleterious missense nonsynonymous single-nucleotide polymorphisms of hennekam syndrome-causing CCBE1 gene, in silico analysis. Sci World J. 2021;2021:1–19.
Behairy MY, Abdelrahman AA, Abdallah HY, Ibrahim EEDA, Sayed AA, Azab MM. In silico analysis of missense variants of the C1qA gene related to infection and autoimmune diseases. J Taibah Univ Med Sci. 2022;17:1074–82.
Poon KS. In silico analysis of BRCA1 and BRCA2 missense variants and the relevance in molecular genetic testing. Sci Rep. 2021;11. 11114.
Mansouri M, El Haddoumi G, Bendani H, Boumajdi N, Hakmi M, Abbou H, et al. In silico analyses of all STAT3 missense variants leading to explore divergent AD-HIES clinical phenotypes. Evolut Bioinforma. 2023;19. 11769343231169374.
Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein–binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–76.
Rosina E, Pezzani L, Pezzoli L, Marchetti D, Bellini M, Pilotta A, et al. Atypical, composite, or blended phenotypes: how different molecular mechanisms could associate in double-diagnosed patients. Genes. 2022;13:1275.
Walters-Sen LC, Hashimoto S, Thrush DL, Reshmi S, Gastier-Foster JM, Astbury C, et al. Variability in pathogenicity prediction programs: impact on clinical diagnostics. Mol Genet Genom Med. 2015;3:99–110.
Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–68.
Wang D, Li J, Wang Y, Wang E. A comparison on predicting functional impact of genomic variants. NAR genomics Bioinforma. 2022;4. lqab122.
Dou J, Vorobieva AA, Sheffler W, Doyle LA, Park H, Bick MJ, et al. De novo design of a fluorescence-activating β-barrel. Nature. 2018;561:485–91.
Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19:596–604.
Idicula-Thomas S, Balaji PV. Correlation between the structural stability and aggregation propensity of proteins. silico Biol. 2007;7:225–37.
Dudola D, Hinsenkamp A, Gáspári Z. Ensemble-Based analysis of the dynamic allostery in the PSD-95 PDZ3 domain in relation to the general variability of PDZ structures. Int J Mol Sci. 2020;21:8348.
Heinemann U, Schuetz A. Structural features of tight-junction proteins. Int J Mol Sci. 2019;20:6020.
MacGowan SA, Madeira F, Britto-Borges T, Schmittner MS, Cole C, Barton GJ. Human missense variation is constrained by domain structure and highlights functional and pathogenic residues. BioRxiv. 2017;127050.
Iqbal S, Pérez-Palma E, Jespersen JB, May P, Hoksza D, Heyne HO, et al. Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants. Proc Natl Acad Sci. 2020;117:28201–11.
Lipiński P, Ciara E, Jurkiewicz D, Pollak A, Wypchło M, Płoski R, et al. Targeted next-generation sequencing in diagnostic approach to monogenic cholestatic liver disorders—single-center experience. Front Pediatr. 2020;8:414.
Tang J, Tan M, Deng Y, Tang H, Shi H, Li M, et al. Two novel pathogenic variants of TJP2 gene and the underlying molecular mechanisms in progressive familial intrahepatic cholestasis type 4 patients. Front Cell Dev Biol. 2021;9. 661599.
Zöllner J, Finer S, Linton KJ, van Heel DA, Williamson C, Dixon PH. Rare variant contribution to cholestatic liver disease in a South Asian population in the United Kingdom. Sci Rep. 2023;13. 8120.
Zhou S, Hertel PM, Finegold MJ, Wang L, Kerkar N, Wang J, et al. Hepatocellular carcinoma associated with tight-junction protein 2 deficiency. Hepatology. 2015;62:1914–6.
Rajabi S, Dastmalchi R, Dehghan MH, Eftekharian A, Aghazadeh E, Ghaderian SMH. TJP2 Gene Mutation c. G1012A May Responsible for Congenital Hearing Loss with Incomplete Penetrance in An Iranian Pedigree. J Genet Resour. 2019;5:143–8.
Zhang J, Guo S, Mei TL, Zhou J, Guan DX, Wang GL. Novel mutation of the TJP2 gene in a Chinese child with progressive cholestatic liver disease coexistent with hearing impairment. Hepatobiliary Pancreat Dis Int. 2021;20:198–200.
Gong R, Li S. Extraction of human genomic DNA from whole blood using a magnetic microsphere method. Int J Nanomed. 2014;9:3781–9.
Khabou B, Mahjoub B, Barbu V, Balhoudi N, Wardani A, Sfar MT, et al. Phenotypic variability in Tunisian PFIC3 patients harboring a complex genotype with a differential clinical outcome of UDCA treatment. Clin Chim Acta. 2018;486:122–8.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic acids Res. 2019;47:D886–D894.
Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM Jr, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–88.
Roe DR, Cheatham TE III. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem theory Comput. 2013;9:3084–95.
McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernández CX, et al. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys J. 2015;109:1528–32.
Mirza N, Bharadwaj R, Malhotra S, Sibal A. Progressive familial intrahepatic cholestasis type 4 in an Indian child: presentation, initial course and novel compound heterozygous mutation. BMJ Case Rep CP. 2020;13:e234193.
Acknowledgements
We acknowledge the Center for High Performance Computing (CHPC), South Africa, for providing computational resources to conduct the molecular simulation analysis. And Ribosite Biotechnology company for advanced molecular biology technical support and customized services. This project is carried out under the MOBIDOC scheme, funded by the EU through the SWAFY proJECT and managed by the ANPR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khabou, B., Othman, H., Guirat, M. et al. Report of a missense TJP2 variant associated to PFIC4 with a pronounced phenotypic variability: Focus on the structural effects on the protein level. J Hum Genet 70, 331–339 (2025). https://doi.org/10.1038/s10038-025-01338-w
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01338-w