Abstract
Allergies are adverse immune responses to typically harmless substances, known as allergens. While allergies can involve diverse immune responses, type 2 immune responses that induce acute hypersensitivity mediated by mast cells, eosinophils and basophils are the major mechanisms underlying allergic disorders. Allergic diseases include atopic dermatitis, allergic rhinitis, food allergies and asthma. The onset and persistence of allergic disorders are influenced by genetic factors, pre-existing illnesses, age, environmental conditions and other lifestyle factors. In particular, diet and microbiomes considerably affect the incidence of various allergic diseases in the skin, lung and intestine. Individuals prone to develop allergic diseases often have impaired and skewed microbial diversification over the first year of life, and this can lead to altered levels of microbial metabolites in the intestine and inflamed tissues. Microbial metabolites, such as short-chain fatty acids, indole metabolites and bile acids, can exert specific regulatory effects on the various components of the immune system, such as barrier epithelial cells and immune cells, including dendritic cells, macrophages, T cells, B cells, innate lymphoid cells and mast cells. Microbial metabolites can also promote immune tolerance to allergenic substances by strengthening regulatory T cells. Understanding the role of these metabolites can lead to better prevention and control of allergic diseases. Here, in this review, we examine current research progress on the interactive relationship between microbial metabolites and allergic diseases and identify functionally important metabolites that affect allergic immune responses.
Similar content being viewed by others
Introduction
Type 1 hypersensitivity-associated allergies occur in response to various environmental and food-derived substances. While these responses are important for host defense by promoting elimination of parasites and immunogenic substances from the body, they have harmful effects on the body, causing rapid life-threatening anaphylaxis, breathing problems, bowel hypersensitivity or chronic inflammatory responses in tissues. In terms of common allergies, allergic rhinitis (AR) is caused by airborne environmental allergens such as pollens, house dust mites (HDM), animal dander, mold spores, cockroach debris and occupational materials1. Atopic dermatitis (AD) or eczema is commonly caused by skin exposure to HDM, animal dander from cats and dogs, and pollen2. Allergic asthma is caused by inhaled antigens such as pollen, dust mites, pet dander and cockroach proteins3. Food allergies (FAs) are frequently caused by exposure to dietary antigens in foods such as peanut proteins (Ara h 1 and Ara h 2), milk proteins (caseins and whey), egg proteins (ovalbumin and ovomucoid) and proteins from shellfish (tropomyosin)4.
Excessive exposure to allergens on the epithelial surfaces of the skin, respiratory tract and alimentary tract due to weakened barrier function can promote the development of allergic diseases. Another important factor is weakened or defective immune tolerance to allergens in foods or the environment5. One in five children in developed countries experience allergies ranging from mild hay fever and respiratory allergies to more severe anaphylactic allergies6,7,8,9. Allergic conditions such as rhinitis, asthma, dermatitis and FAs are primarily driven by type 2 cytokines (IL-4, IL-5, IL-9 and IL-13) and allergen-specific IgE10. These immune mediators increase the activity of T cells, group 2 innate lymphoid cells (ILC2), mast cells, eosinophils and basophils in affected tissues, leading to the characteristic inflammation and symptoms of these allergies The rise in allergies in developed nations over the past few decades is not entirely clear. However, a mounting body of evidence suggests a role for Westernized lifestyle factors in regulating allergic diseases.
It has been established that diet and gut microbiomes considerably affect the incidence of diverse allergies in animal models and human populations11,12,13,14,15,16,17. Microbial changes, including reduced diversity and dysbiosis, occur in the gut and affected tissues such as lungs and skin of allergic patients18,19,20. Gut microorganisms process and metabolize dietary materials and host secretions to produce a myriad of metabolites21. Major microbial metabolites are made from carbohydrates, proteins and cholesterol metabolites. More specifically, short-chain fatty acids (SCFAs) such as acetate, propionate and butyrate are produced from dietary fibers and resistant starches22. Major amino acid metabolites are produced from tryptophan (Trp), arginine (Arg) and tyrosine (Tyr)23,24,25. Primary bile acids (BAs) are made from cholesterol in the liver and are modified (that is, dehydroxylated) by gut microorganisms to become secondary BAs26. In addition, there are many other metabolites with positive or negative functions in regulating the host immune system27,28.
Microbial metabolites affect barrier tissue cells and immune cells via specific receptors21. Microbial metabolites also serve as nutrients for tissue cells and immune cells, increasing their cellular energy levels, affecting cellular state, differentiation and functions29. These metabolites can also regulate important enzymes such as histone deacetylases (HDACs)30. These effects can profoundly affect cellular activities and gene expressions to control barrier cells and immune cells involved in allergic immune responses (Fig. 1). The purpose of this review is to assess and summarize the current research development on the roles of gut microbial metabolites in regulating allergies in experimental and clinical settings.
Microbial dysbiosis is a risk factor, occurring even before the onset of allergy pathogenesis. Microbial dysbiosis accompanies altered production of microbial metabolites, reducing the production of beneficial metabolites such as SCFAs, indole derivatives and BA metabolites, among others. At the same time, the production of harmful microbial metabolites or products from pathogenic microorganisms is increased. These changes weaken the barrier function in the skin, respiratory tract and intestine, increasing environmental or food antigen exposures to the immune system in the tissues. This process is called antigenic sensitization. Due to heightened expression of allergic alarmin cytokines such as TSLP, IL-25 and IL-33, the recruitment and tissue population of immune cells such as ILC2, eosinophils, basophils and mast cells are increased over time. DCs differentiated under these conditions would migrate to secondary lymphoid tissues, where they promote the differentiation of Th2, Tfh and Tc2 cells, which collectively induce allergen-specific IgE production by B cells and drive allergic tissue inflammation. Allergen-specific IgE molecules bind to the high-affinity receptor FcεRI on mast cells for their sensitization and ultimate activation by allergens and other inflammatory signals in tissues. Following repeated exposure to allergens, these processes are eventually amplified to pathogenic levels and cause chronic allergic diseases.
Microbial dysbiosis in allergies
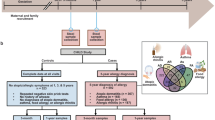
The full colonization of the human intestine in early life is a gradual process that continues over the first few years after birth31. Colonization of the lungs starts to occur at the same time within the second week of life and is mostly completed by the first few years of life32. The microbiome is required to support barrier tissue immunity and influences the fitness of the immune system33. The gut microbiome is diverse with 500–1,000 species, the size and composition of which can change depending on host conditions and diets34. Healthy individuals without allergic diseases or symptoms tend to have a more diverse microbiome in the gut, and this is associated with better working immune systems with high levels of immune tolerance35,36. Meanwhile, allergic individuals often exhibit reduced microbial diversity, especially in early life. In microbial dysbiosis, decreased levels of beneficial bacteria and/or increased levels of harmful bacteria occur (Fig. 2). Allergy-associated dysbiosis is thought to be caused, in part, by genetic variations, inflammatory responses in affected tissues, unhealthy lifestyles including poor diet choice, or changes in diet due to allergy symptoms. More specifically, aversion behaviors, occurring after allergic episodes, can contribute to changes in the gut microbiome by altering diet choice and lifestyle.
In major allergic conditions, the microbiomes in the intestine and affected tissues such as the skin and respiratory system are altered, increasing certain pathogenic strains while decreasing beneficial metabolite-producing microorganisms, leading to microbial metabolite dysbiosis. This can amplify pathological immune responses, leading to persistent allergies. Listed above are the metabolites produced by microbes frequently altered in allergies182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207. While simplified here, the microbes and their metabolites may have complex roles, both exacerbating and ameliorating allergic pathogenesis depending on tissues and diseases.
Microbial colonization of the skin occurs immediately at birth, but the cutaneous microbial community increases its diversity with age over the first year of life37,38,39. The early diversification or maturation of the microbial community is important for lowering the AD incidence in children in later years40,41. Thus, delayed maturation or dysbiosis of microbiome can occur in apparently normal babies without any diseases. However, these babies are more likely to develop AD later in life. Microorganisms on the skin and in the gut affect AD pathogenesis. Staphylococcus aureus is often the dominant microbial species associated with AD. S. aureus overgrowth can disrupt lipid metabolism and compromise the skin barrier. By contrast, colonization of Staphylococcus epidermidis and Roseomonas mucosa is beneficial in that they inhibit S. aureus colonization and affect immune responses to prevent AD development. Also increased in AD are Escherichia coli, Clostridium difficile and Sutterella species. Overgrowth of C. difficile and E. coli in infants with AD has been linked to decreased beneficial bacteria, abnormal gut barrier function and a loss of immune tolerance41. Bacteroidetes, Akkermansia, Bifidobacterium and Faecalibacterium are often found in lower proportions in patients with AD. In general, AD is associated with reduced levels of butyrate-producing bacteria42.
FAs manifest in various symptoms in respiratory, digestive and skin tissue systems. Microbial changes also occur in FAs43,44. For example, increased Streptococcaceae, Lachnospiraceae and Leuconostocaceae have been observed in children with egg allergies. By contrast, increased levels of Clostridia and Firmicutes are linked to the resolution of milk allergy45. However, a caveat is that some Clostridia species, such as Ruminococcus gnavus, are positively associated with FA46,47,48,49. Increased levels of Dorea, Dialister, Haemophilus and Clostridium species were found in individuals with FA who were sensitive to milk, egg, peanut, soy and wheat, while Leuconostoc, Veillonella and Weissella were underrepresented in infants who developed FAs later50,51. Decreased numbers of Bacteroidetes species were observed in food-sensitized children52. Young infants with lower levels of Lactobacillus and Bifidobacterium species were more likely to develop FAs to egg white, cow’s milk and airborne allergens53,54. Decreased abundance of certain bacteria, such as Bifidobacterium longum, Bacteroides dorei and Bacteroides vulgatus, has been observed in allergic patients. By contrast, an increase in S. aureus has been linked to FAs to eggs and milk55. Increased levels of Clostridium and Anaerobacter species have been observed in infants with IgE-mediated FA56. FA has also been associated with increased abundance of Enterobacterales species57. Faecalibacterium prausnitzii and R. gnavus were increased in patients with FAs as well as respiratory allergies46. Despite the association of lower fecal diversity in infancy and allergy incidence, higher microbial diversity is paradoxically associated with increased risk of developing allergic diseases in adults58. Therefore, diversity by itself is not a strong indicator of allergic diseases later in life. A fecal microbiome analysis in healthy and food-allergic twins revealed increased abundance of Phascolarctobacterium faecium and Ruminococcus bromii in healthy versus FA twins59. Importantly, Phascolarctobacterium faecium produces SCFAs, and Ruminococcus bromii can process resistant starch60,61.
In the lungs, it is estimated that healthy individuals have 103–105 bacteria per gram of the tissue, largely composed of Proteobacteria, Firmicutes, and Bacteroidetes species. Early disruptions in the intestinal microbiome can potentially increase the risk of developing AR later in life62. Microbial dysbiosis in the gut may exacerbate nasal inflammation and rhinitis symptoms. It has been reported that Ruminococcus species were enriched in a mouse model of AR compared with control animals. It appears that the microbial changes associated with asthma are highly diverse depending on cohort63. At this point, the impact of many of these microbial groups or species on respiratory allergies remains largely speculative.
Asthma subtypes include neutrophilic, eosinophilic, mixed granulocytic and paucigranulocytic asthma, depending on histological features. It is also classified as allergic, non-allergic, occupational, seasonal and exercise-induced depending on triggers and disease modes. Eosinophilic and neutrophilic asthma are relatively strongly linked to changes in the airway microbiome64,65. In patients with asthma, there is a shift in the lung microbiota toward increased diversity, while the gut microbiota shows decreased diversity66,67. For example, the Proteobacteria group was increased in abundance in the respiratory system of asthma patients. More specifically, Haemophilus, Moraxella and Neisseria species have been associated with more frequent viral infections and severe asthma symptoms68. Prevotella buccalis, Alkanindiges hongkongensis and Gardnerella vaginalis were overrepresented in the nasal microbiota of individuals with asthma compared to controls. Colonization with Bacteroides fragilis strains has been proposed as a potential predictor of possible asthma development11,69. The bacterial genera Veillonella, Faecalibacterium, Lachnospira and Rothia were underrepresented in fecal samples of 3-month-old children with asthma70. Early colonization with these microorganisms was associated with increased blood eosinophil counts and total IgE levels71. Reduced intestinal abundance of Bacteroidetes and Bifidobacterium was also observed in those with allergic airway diseases72. A decreased abundance of Lachnospira species and Clostridium neonatale in infants’ stool was correlated with asthma development at 4 years of age73, whereas C. difficile colonization was associated with allergy development later in life74.
Major microbial metabolites and their impact on host cells
The normal gut microbiome, composed of approximately a thousand species, can produce a myriad of metabolites when there are enough nutrient materials in the colon. Because each microbial species has a finite number of genes, it can produce only a limited range of metabolites. In the case of dysbiosis, where a small number of microbial species become dominant, this can substantially limit the production of certain metabolites. Moreover, diet and health status are key regulators of microbiomes and help determine the mass and diversity of microbial metabolites. In most allergic conditions, reduced or altered production of SCFAs has been observed75,76. Moreover, alterations in the metabolism of Trp and BAs are linked to dysregulated immune responses in allergic individuals77,78,79.
Carbohydrates that reach the colon are degraded and fermented by the microbiome to SCFAs80,81. Because starches are mostly digested and absorbed in the upper alimentary tract, it is mainly the hard-to-digest complex carbohydrate polymers, such as dietary fibers and resistant starches, that are available for microorganisms to make SCFAs in the colon. Butyrate, propionate and acetate are the major SCFAs in the mammalian colon. SCFAs regulate cells by several different pathways. The first way to affect host cells is by activating G-protein-coupled receptors (that is, GPR41, GPR109A, GPR43 and Olfr78) on host cells80,81. This activation will trigger Gαi/o proteins (GPR41 and GPR109A) and Gαq/11 and Gαi/o proteins (GPR43)82,83,84. Second, SCFAs are used as an energy source after transportation into cells and conversion to acetyl-CoA29. SCFAs can also affect metabolic pathways such as glycolysis and oxidative phosphorylation. The third way to regulate host cells is to inhibit type 1 and 2 HDACs for gene expression regulation85,86. SCFAs regulate both innate and adaptive immune cells, including macrophages, dendritic cells (DCs), ILCs, B cells and T cells29,86,87,88,89. More specifically, SCFAs increase IL-10 production to promote immune tolerance and enhance immunity by boosting the numbers of effector T cells, including Th1, Th17 and effector CD8+ T cells, in the intestine80,86. SCFAs increase antibody production by inhibiting HDACs and metabolically support B cell differentiation to IgA- or IgG-producing cells29. SCFAs increase the activity of both ILC2 and ILC3 during active immune responses88. It is also reported that SCFAs suppress ILC2 in the lungs during experimental asthma development90.
Primary BAs are made in the liver from cholesterol and play important roles in digestion and nutrient absorption, particularly of fats and fat-soluble vitamins91. Primary BAs are typically conjugated with amino acids such as glycine or taurine in the liver. They are stored in the gallbladder before secretion into the duodenum. In the colon, they are modified by gut microorganisms to become secondary BAs. The modifications include deconjugation (removing glycine or taurine), dihydroxylation, oxidation and epimerization92. Major primary BAs are cholic acid and chenodeoxycholic acid. The most abundant secondary BAs in the intestine are deoxycholic acid (DCA) and lithocholic acid (LCA)93. Both primary and secondary BAs are reabsorbed into the ileum and returned to the liver. Farnesoid X receptor (FXR) and the Takeda G-protein-coupled receptor 5 (TGR5) are the major receptors for BAs91. FXR is highly expressed by hepatocytes, enterocytes, pancreatic β-cells, kidney cells, adrenal gland cells and immune cells such as macrophages, DCs and natural killer T cells94. TGR5 is highly expressed by the gallbladder epithelium, bile duct cholangiocytes, intestinal stem and epithelial cells, Kupffer cells, liver sinusoidal endothelial cells and neuron cells95,96,97,98,99. Among immune cells, TGR5 is expressed by monocytes and macrophages. Other major receptors for BAs include pregnane X receptor (PXR) and S1PR2100,101. Within the immune system, BAs control gut microorganisms with their antimicrobial activity, support the intestinal barrier function by regulating epithelial cells, and regulate macrophages and DCs for their functions in cytokine production and T cell activation102,103. TGR5 activation by DCA and LCA decreases the production of IL-12 and TNFα, and FXR activation by isoallo-LCA promotes Treg cells at the expense of Th1 and Th17 cells104.
Amino acids are also degraded and modified by microorganisms. Among them, microbial metabolites from Trp (indole derivatives), Arg (nitric oxide and ornithine), branched-chain amino acids, Phe (for example, phenylacetylglutamine and phenylacetic acid) and Tyr (tyramine, 4-hydroxyphenylpyruvate and p-cresol) have been known to have important immune regulatory functions21. Indole metabolites regulate cells via the aryl hydrocarbon receptor (AhR)105. AhR activation by indole derivatives can affect Th1 versus Th2 immune responses, potentially suppressing Th2 polarization in allergic immune responses106. Ornithine is a precursor of polyamines, which have tissue-repair and anti-inflammatory properties107. Phenylacetylglutamine has been shown to activate α2A, α2B and β2-adrenergic receptors108. Phenylacetic acid is implicated in promoting AhR signaling109. Phenylpropanoic acid activates GPR109B, and cadaverine triggers HRH4110.
Key immunological features that affect allergic immune responses in the skin, respiratory and intestinal tissues
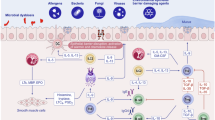
The skin, lungs and intestines possess unique immune systems tailored to their specific needs and microbial exposures. These organs have unique combinations of tissue and immune cells and develop general as well as tissue-specific immune responses, which may lead to pathological allergic conditions (Fig. 3).
The three barrier tissues or organs have shared yet distinct structures and cell compositions. The skin immune system consists of multiple layers of keratinocytes containing LCs, with underlying layers housing other immune cells such as dermal DCs, ILC2s, T cells, mast cells and others. The intestinal immune system is composed of a single layer of epithelial cells in villi in the small intestine and crypts in the colon at various differentiation stages or fates for distinct functions. Tuft cells that sense succinate from microorganisms and goblet cells that produce mucin and transport protein antigens from the lumen are good examples. The intestine has T cells in the epithelial layer (i.e., intestinal epithelial lymphocytes) and lamina propria. ILC3 plays a critical role in supporting epithelial barrier function and immunity, which are also supported by Th17 cells. Specialized DCs such as CX3CR1+ DCs can sense the presence of luminal antigens, and RORγt+ DCs (also called Thesis cells) are important for the generation of RORγt+ FoxP3+ T cells that protect the intestine from developing inflammatory diseases. B cells actively produce IgA, which is secreted to the intestinal lumen to bind commensal bacteria and potential pathogens. Lungs are composed of alveoli that are formed by a single layer of type I AECs (also called pneumocytes) and type 2 AECs that function as stem cells and secrete surfactants. AMs patrol the alveolar space to clean up any particles in the breathed air. ILC2 are the dominant ILCs in the lungs. Several DC types and macrophages monitor antigens in the alveolar and interstitium. T cells, DCs and ILCs are in the interstitium and mucosal-associated lymphoid tissues in the lungs. The presence of Treg cells is important to keep these tissues from developing excessive inflammatory responses. Upon barrier breach due to genetic, physical and chemical reasons, inflammatory cytokines such as TSLP, IL-25 and IL-33 are produced to initiate pro-allergic Th2 type inflammatory responses characterized by IL-4, IL-5, IL-9 and IL-13. Due to compromised barrier functions, allergens are introduced and captured by antigen-presenting cells such as DCs, which migrate to secondary lymphoid tissues to activate and differentiate T cells. Because of the cytokine milieu rich in type 2 cytokines, allergen-specific Th2 cells are generated in increased numbers. Th2 cells further increase the production of IL-4, IL-5, IL-9 and IL-13, which skew B cell differentiation into IgE producers. The cytokines increase the numbers of mast cells, basophils and eosinophils, and allergen-specific IgE sensitizes them by binding to the FcεRI complex on mast cells. Whenever there are allergen challenges through the weakened barriers, these allergic granulocytes undergo activation to degranulate or secrete a battery of allergy mediators such as histamine, neutral proteases (tryptases and chymases), proteoglycans (heparin), cytokines (TNFα, IL-4, IL-6, IL-13 and TSLP) and lipid mediators (prostaglandins and leukotrienes). Moreover, mast cells are activated by various inflammatory stimuli, such as changes in pressure, temperature, complements, IgG, pathogen-associated molecule patterns, neuropeptides, cytokines (SCF, IL-33, IL-25 and TSLP) and extracellular ATP in tissues. Microbial metabolites have several different regulatory effects on allergic immune responses. First, the barrier cells are strengthened for their functions by microbial metabolites such as SCFAs. Second, T and B cells are regulated by microbial metabolites such as SCFAs and indole metabolites to produce protective Treg cells and IgA, but not Th2 and IgE. Moreover, the allergy-associated granulocytes, including mast cells, are regulated by the microbial metabolites especially by SCFAs and TUDCA/UDCA. Other BAs have generally stimulating effects on immune cells.
The skin is a bona fide barrier tissue with keratinocytes in the epidermis, sweat glands and hair follicles. The epidermis acts as the first line of defense in the skin, producing antimicrobial peptides and communicating with other immune cells through cytokines and chemokines111. It is also activated by pathogenic microorganisms to initiate allergic immune responses. For example, S. aureus induces IL-33 production in keratinocytes112. Langerhans cells (LCs), a specialized type of dendritic cell (DC), are present in the epidermis. LCs are regulated by host and microbial metabolites. The indole derivative, indole-3-aldehyde (IAId), activates AhR and suppresses the function of LCs, leading to IL-10 production and decreased CD4+ T cell proliferation113. In addition, there are conventional DCs in the dermis layer (dDCs). These dDCs capture and present antigens to T lymphocytes after migration to lymphoid tissues. The skin is a nonlymphoid tissue and therefore harbors only memory, effector or tissue-resident, but not naive, types of T cells. In particular, a significant number of memory T cells, including CD8+ tissue resident memory T cells (called TRM cells) reside in the epidermis, whereas many CD4+ T cells are found in the dermis114. These cells provide rapid and localized immunity upon reinfection. dDCs also play a role in antigen presentation and support T cell responses115. The majority of dDCs are IRF4+ DC2, which can readily induce Th2 cells, a property shared with LCs. In addition, basophils, ILC2, mast cells and eosinophils, which are found in the dermis of the skin, can also promote Th2 cells in the skin116.
The lungs also have unique cell types such as alveolar macrophages (AMs) and airway epithelial cells (AECs), which form a complex tissue network embedded with innate immune cells117. The mucociliary system traps and clears pathogens in the upper respiratory tract. AECs and AMs secrete antimicrobial peptides, cytokines and chemokines, initiating a rapid, nonspecific response to pathogens. These cells express Toll-like receptors, which recognize microbial components and trigger immune cell activation and cytokine production. The lungs have many ILCs in the adventitial cuffs around airways and vessels in the alveolar capillary beds, among which ILC2 is the dominant cell type118,119,120,121. The lungs also have T lymphocytes, IgA-producing B lymphocytes and specialized DCs122. TRM cells in the lungs can mount rapid responses to previously encountered pathogens123. The role of IgA in neutralizing pathogens in the airways is important in the respiratory tract124. The propensity to develop Th2-type allergic immune responses in the lungs is probably largely due to the nature of the antigens (for example, pollen, dust mites, animal dander and fungal spores), Th2-biased epithelial alarmin responses (TSLP, IL-25 and IL-33) and the activity of ILC2s. It has been shown that SCFAs have positive effects on fighting respiratory infections125.
Unlike the skin, the intestine has a single layer of epithelial cells that serves as a barrier and is responsible for processing food into absorbable nutrients. Therefore, there is a need to develop stronger immune tolerance to food antigens. Another factor is the high load of gut microorganisms in the colon as well as the ileum. The microorganisms are part of the digestion system to process hard-to-digest materials to extract additional nutrients and synthesize certain metabolites important for the host. The intestinal microbiome is significantly greater than those on the lungs and skin in metabolite production, leading to higher and more diverse production of metabolites. These metabolites are transported via blood vessels to most parts of the body, including the skin and lungs. An important task of intestines is to maintain a balance between defending against pathogens and tolerating food antigens and commensal microbiota. Intestinal epithelial cells produce enzymes, defensins and mucins126. In particular, antimicrobial peptides are produced by Paneth cells, which are specialized epithelial cells in the crypts. The intestine has gut-associated secondary lymphoid tissue such as Peyer’s patches and colonic patches for activation of T and B cells. As in the lungs, IgA is an important immunological factor for the intestine, binding and keeping pathogens and the gut microbiome from invading127. ILCs play important roles in directing intestinal epithelial cells to form a strong physical and chemical barrier by inducing the expression of epithelial tight junction proteins and antimicrobial peptides128. The intestine is rich in production of IL-33, IL-25, TSLP, IL-4, IL-9, IL-5 and IL-13 by epithelial cells, T cells, basophils, ILC2 and/or Tuft cells129. Activation of this pathway is key in allergic immune responses in the intestine. The intestinal epithelial cells require microbial metabolites for the maintenance of intestinal barrier and immunological functions29,86,87,130. Intestinal Tuft cells are activated by succinate, produced by parasites, such as Polymorphus minutus and Trypanosoma brucei. This is mediated by the succinate receptor 1 (SUCNR1) on Tuft cells to produce IL-25, which promotes ILC2 expansion131. Butyrate inhibits the activity of HDAC3, a HDAC enzyme that is crucial for stem cell differentiation into Tuft cells132. Thus, succinate and butyrate can differentially regulate type 2 immunity to combat parasites, which could be dysregulated to promote allergic immune responses.
Regulation of AD by microbial metabolites
Microbial metabolites, produced by bacteria in the skin and gut, play a significant role in regulating AD pathogenesis. In particular, SCFAs and indole derivatives can modulate immune responses, lipid metabolism and skin barrier function, impacting AD development and severity133.
SCFAs are believed to reach the skin through the bloodstream or to be produced locally within the skin. Propionate is a sebum-derived metabolite produced in the skin134. Butyrate can be metabolized by keratinocytes to produce acetyl-CoA. SCFAs promote the production of ceramides, which are utilized to strengthen the skin barrier135. Moreover, SCFAs promote keratinocyte metabolism and differentiation and help form the stratum corneum (that is, the outside layer of the skin)135. SCFAs can support the immune cells in the skin, such as macrophages, ILCs, T cells and B cells29,86,88,136. SCFAs can inhibit the inflammatory response triggered by inflammatory skin bacteria such as Propionibacterium acnes137. HDAC inhibition by butyrate increases the expression of transglutaminase-1 (TGM1) and filaggrin (FLG)138. SCFAs can increase cellular metabolism including mitochondrial tricarboxylic acid cycle for energy production in skin cells29,135. Thus, SCFAs, whether produced in the skin or the intestine, can strengthen the skin barrier and help maintain skin immune homeostasis. This can decrease the sensitization of the skin immune system by various environmental antigens. SCFAs have anti-inflammatory effects, and SCFA production is associated with a lower risk of AD139. In contrast to the largely protective roles of SCFAs, the level of six-carbon SCFA caproic acid was increased in patients with AD139. Thus, caproic acid may have a proinflammatory effect on the skin.
The level of IAId, a tryptophan metabolite that triggers AhR, was found to be significantly decreased on both lesional and nonlesional skin of patients with AD140. IAId is produced by skin commensal bacteria, and it has been shown to negatively regulate skin inflammation in patients with AD. A related AhR ligand, indole-3-carbinol (I3C), has been shown to reduce skin inflammation and the levels of allergen-specific IgE and TSLP141.
Dermatitis development is associated with elevated levels of BAs in the skin, which can further contribute to inflammation and pruritus (that is, itch)142. BA secretion is increased in people consuming a Western style diet with high levels of fat and meat and low levels of dietary fibers, and this is associated with an increased incidence of dermatitis with features of Th2 and Th17 inflammation, both of which are involved in AD pathogenesis142. BA sequestrants, such as cholestyramine, when used topically, have been shown to reduce epidermal thickening and levels of cutaneous inflammatory cytokines in a mouse model of dermatitis142. While BAs are associated with increased AD symptoms, the secondary BA ursodeoxycholic acid (UDCA) appears to alleviate AD-associated inflammation143. More specifically, UDCA decreases the activities of MAPK and NF-kB and the production of MCP-1, MDC, TARC and IL-6 by keratinocytes and mast cells. In addition, a microbial metabolite associated with atopy is 12,13-diHOME (12,13-dihydroxy-9Z-octadecenoic acid), a fatty acid derivative produced by microorganisms that can promote macrophage expression of inflammatory cytokines (IL-1β, TNFα and IL-6), B cell proliferation and IgE production144.
Regulation of FA by microbial metabolites
FA incidence has risen to high levels in the past several decades in developed countries, with 5–8% of the population affected depending on the country and age group145. Children have a higher incidence of FA than adults. The high prevalence of this disorder in developed countries and in younger populations implies the potential importance of diet, the microbiome and immunological maturation. Patients with FA react to certain foods (including the nine most common allergens: peanuts, nuts, milk, eggs, wheat, soy, sesame, crab and shrimp) that are introduced to multiple organs such as the gastrointestinal tract, respiratory system and skin146. The etiology of FA remains unknown, but it is caused by overzealous or excessive immune responses to food-derived protein antigens, which are primarily mediated by type 2 immune responses involving Th2 cells, IgE production and mast cell activation. Defective barrier functions in the skin and the intestinal tract are also suspected to increase lymphocyte activation in response to food antigens147,148.
Studies have linked low SCFA levels in early life to an increased risk of FA. In particular, butyrate has shown promise in protecting against FA. Butyrate strengthens the intestinal barrier in part by suppressing stress-mediated Notch signaling149. The levels of major SCFAs in feces were significantly reduced in allergic individuals43. P. copri, a producer of acetate and propionate, was highly increased in the healthy group compared with the FA group. P. copri was generally increased in the gut when a diet rich in fibers was consumed150 or in Th1-associated diseases such as colitis and rheumatoid arthritis151,152. Another group reported decreased butyrate in allergic patients, and oral administration of butyrate ameliorated some of the FA-associated immune responses149. Butyrate decreased ovalbumin-induced experimental anaphylaxis in mice. Another important function of butyrate is to suppress IgE-mediated mast cell activation153. HDAC inhibition is a mechanism behind the suppression of mast cells by butyrate. The signaling molecules upon triggering of the IgE receptor FcεR1, such as Bruton tyrosine kinase (BTK), spleen tyrosine kinase (SYK) and linker for activation of T cells (LAT), were found to be downregulated by butyrate in their expression in mast cells. HDAC inhibition can increase histone acetylation and boost the expression of many genes. Another study found that SCFAs activated GPR109A (a receptor for butyrate and niacin) on mast cells and increased the expression of prostaglandin E2 (PGE2), leading to inhibition of mast cells154. Moreover, SCFAs can contribute to the development and function of regulatory T cells to support oral tolerance to food antigens155.
Indole metabolites are also linked to suppressed FAs. Bifidobacterium breve M-16V promotes the production of indole derivatives such as indole-3-propionic acid (IPA) by other microorganisms, and its supplementation ameliorated cow’s milk allergy156. In this regard, IPA supplementation decreased allergy symptoms, which was mediated by AhR activation by IPA. Another ligand of AhR, I3C, which is abundant in cruciferous plants, decreased ovalbumin-specific IgG1 and peanut allergy symptoms157.
Primary BAs, including chenodeoxycholic acid, can increase immunological sensitization to food antigens. This appears to be mediated by increasing the expression of retinoic acid-regulated genes and the production of food allergen-specific antibodies (IgE and IgG1)79. More studies are required to better understand the interaction and functions of BA and retinoic acid pathways. Peanut oral immunotherapies have been developed to treat patients with peanut allergy. Mixtures of primary and secondary BAs (that is, cholic acid, chenodeoxycholic acid and UDCAs) support colonic FOXP3+ Treg cells expressing the transcription factor RORγ158. These Treg cells have the potential to suppress inflammatory immune responses159. A specific profile of fecal BAs before tolerance-inducing treatment predicted the efficacy of the oral peanut immunotherapy160. Thus, BAs have complex functions in regulating intestinal immune responses including those for FAs. An interesting feature of the microbiome or microbial metabolites associated with oral immunotherapy failure is the increased amino acid utilization, which could degrade and therefore decrease amino acid-conjugated secondary BAs. Certain BA-producing microbial species such as R. gnavus were elevated in food-allergic patients who were responding to oral immunotherapy46. Despite some exceptions, many BAs appear to exert pro-allergic effects on FA pathogenesis.
Impact of microbial metabolites on respiratory allergies
It has been reported that SCFAs, particularly butyrate and propionate, can protect people from developing asthma. Butyrate downregulated GATA3 expression and suppressed ILC2-driven airway hyperresponsiveness161. Studies have linked decreased levels of fecal SCFAs in infants to increased risks of developing asthma161,162,163,164. Increased levels of butyrate and propionate in baby stools have been associated with decreased asthma development76. SCFAs regulate immune responses by activating GPCRs and inhibiting HDACs86,87, which impact inflammation, remodeling and hyperresponsiveness in airways. SCFAs regulate the proliferation and function of ILC2, reducing the production of asthma-related cytokines such as IL-5 and IL-13161. SCFAs induce the differentiation of Treg cells and suppressive myeloid cells for suppression of allergic airway diseases86,165. In addition, SCFAs suppress Th2 cell activity by altering DC function and migration, making them less inflammatory and more tolerogenic166. It has been reported that SCFAs can reduce the survival, adhesion and migration of eosinophils, further contributing to the attenuation of airway inflammation167. Intestinal helminth infection unexpectedly reduced the severity of allergic airway inflammation in part by increasing SCFA production168. Helminth infection altered the intestinal microbiome to increase SCFA production, and SCFAs increased Treg activity in GPR41-dependent manner. Another function of SCFAs is to suppress asthma, in part, by decreasing IL-13-producing Tfh cells that skew B cell production of IgE169. Yet, another function of butyrate is to suppress mast cell activation153. Moreover, F. prausnitzii administration increased Lachnoclostridium abundance and SCFA production, alleviating HDM-induced allergic asthma170.
Reduced levels of SCFAs were found in patients with AR75. Antibiotics, such as vancomycin, can decrease SCFA-producing bacteria and fecal SCFA levels in mice, exacerbating AR severity171. Administration of butyrate in a conjugated form to starch increased colonic butyrate concentration, decreased serum allergen-specific-IgE, IL-4 and IL-5, and improved clinical activity in a mouse model of AR172. These results indicate that microbial SCFAs are effective suppressors of AR.
Reduced levels of Trp have been associated with increased expression of TSLP and subsequent asthma inflammation173. In a mouse model of ovalbumin-induced asthma, the disease severity was alleviated by administration of Trp metabolites, such as kynurenine (KYN), indole-3-lactic acid (ILA), indole acetic acid (IAA) and I3C174. Another indole derivative, IPA, reduces mitochondrial respiration and superoxide production in lung tissues, which decreases allergic airway inflammation175. Trp metabolites, such as I3C, KYN and ILA, also reduced allergic asthma in a mouse model174. These metabolites activate AhR, which can influence the Th1/Th2 balance, favoring Th1-dominant responses and potentially suppressing Th2 responses106. The aminos acid Tyr has also been linked to the regulation of lung inflammation in allergic asthma. Increased p-cresol sulfate (a Tyr metabolite), generally considered a toxic metabolite, is unexpectedly associated with low asthma risks in children176.
BAs are also implicated in asthma development. Specific BAs, such as glycocholic acid (a glycine conjugate of cholic acid) and glycoursodeoxycholic acid (a glycine conjugate of UDCA), were increased in individuals with asthma177. Tauroursodeoxycholic acid (TUDCA) can act on airway cells and attenuate allergen-induced airway inflammation (that is, IgE production, mucus secretion, IL-4, IL-5 and IL-13) and lung hyperresponsiveness178. This was studied using an HDM-induced airway inflammation model. The mechanism appears to be mediated by inhibiting the unfolded protein response, a cellular process activated under stress conditions such as inflammation. Thus, BAs are ambivalent in regulating asthma responses, either promoting or suppressing asthmatic activity depending on the type of BAs.
Concluding remarks
The information discussed in this review highlights the strong association between allergic diseases and microbial dysbiosis. Microorganisms, such as S. aureus in the skin, Proteobacteria in the lungs and Bacteroidales in the intestine, which are overrepresented in allergic diseases are generally pathogenic. Typically, Clostridiales, Bifidobacterium and Lactobacillum are decreased in most major allergy diseases, and these microorganisms are thought to produce beneficial SCFAs, BAs and/or indole metabolites. The first few years of life after birth seem to be critical in the maturation of the immune system and prevention of microbial dysbiosis and allergic diseases later in life. The information discussed in this review raises the question of whether various allergic disorders are treatable with intervention strategies using dietary modifications, prebiotics, probiotics and microbial metabolites.
Although many microorganisms have been identified as altered in adults or infants at increased risk of developing allergies, it remains necessary to pinpoint the functionally important species that, if administered, could reduce allergy pathogenesis. While certain probiotic strains have been shown to ameliorate experimental allergies, it is important to identify natural microorganisms that can suppress allergy symptoms in a sustained manner. Identifying specific microbial species and their metabolites that most effectively promote immune tolerance is also crucial for developing targeted therapeutic strategies.
It is exciting to witness the development of new allergy therapies, including antigen-based desensitization immunotherapies that induce antigen-specific Treg cells, monoclonal antibodies targeting IL-4, IL-5 or IL-13, and BTK inhibitors that act on B cells and mast cells179. However, these approaches are limited in efficacy or require ongoing use to be effective. More definitive or permanent approaches to stop allergic diseases will require the immune system to gain stable tolerance against allergens. Microbial therapies, including microbial metabolite approaches, hold promise in making these approaches effective. Perhaps certain microorganisms and metabolites may be used as an adjunct strategy to increase the efficacy and duration of therapeutically induced immune tolerance. Microbial metabolites are potentially good approaches to decrease allergic immune responses or train immune cells not to become allergic. In this regard, concentrations and combinations of metabolites that are effective in treating or preventing allergic patients need to be experimentally determined in human participants. There are expected side effects of utilizing microorganism- or metabolite-based therapies. Major examples are potential infections and inflammatory responses, particularly in the urogenital system180,181. These issues should be carefully considered when devising therapeutic strategies with microbial or microbial metabolite-based therapies.
References
Wu, A. C., Dahlin, A. & Wang, A. L. The role of environmental risk factors on the development of childhood allergic rhinitis. Children 8, 708 (2021).
Tamagawa-Mineoka, R. & Katoh, N. Atopic Dermatitis: identification and management of complicating factors. Int. J. Mol. Sci. 21, 2671 (2020).
Nolte, H., Backer, V. & Porsbjerg, C. Environmental factors as a cause for the increase in allergic disease. Ann. Allergy Asthma Immunol. 87, 7–11 (2001).
Francis, O. L., Wang, K. Y., Kim, E. H. & Moran, T. P. Common food allergens and cross-reactivity. J. Food Allergy 2, 17–21 (2020).
Sogkas, G. et al. Cellular and molecular mechanisms breaking immune tolerance in inborn errors of immunity. Cell Mol. Immunol. 18, 1122–1140 (2021).
Bloom, B., Cohen, R. A. & Freeman, G. Summary health statistics for U.S. children: National Health Interview Survey, 2010. Vital Health Stat. 10, 1–80 (2011).
van de Veen, W., Wirz, O. F., Globinska, A. & Akdis, M. Novel mechanisms in immune tolerance to allergens during natural allergen exposure and allergen-specific immunotherapy. Curr. Opin. Immunol. 48, 74–81 (2017).
Ptaschinski, C. & Gibbs, B. F. Early-life risk factors which govern pro-allergic immunity. Semin. Immunopathol 46, 9 (2024).
Lukacs, N. W. & Hogan, S. P. Food allergy: begin at the skin, end at the mast cell?. Nat. Rev. Immunol. 25, 783–797 (2025).
Molofsky, A. B. & Locksley, R. M. The ins and outs of innate and adaptive type 2 immunity. Immunity 56, 704–722 (2023).
Cobos-Uribe, C. & Rebuli, M. E. Understanding the functional role of the microbiome and metabolome in asthma. Curr. Allergy Asthma Rep 23, 67–76 (2023).
McGowan, E. C. & Keet, C. A. Prevalence of self-reported food allergy in the National Health and Nutrition Examination Survey (NHANES) 2007–2010. J. Allergy Clin. Immunol. 132, 1216–1219 e1215 (2013).
Trompette, A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166 (2014).
Yu, R., Yang, B., Cai, L., Lu, X. & Wang, X. Excess free fructose beverages and allergy in children and adolescents: results from NHANES 2005-2006. Ann. Fam. Med. 16, 408–418 (2018).
Han, Y. Y. et al. The dietary inflammatory index and current wheeze among children and adults in the United States. J. Allergy Clin. Immunol. Pract. 6, 834–841 e832 (2018).
Melo, B., Rezende, L., Machado, P., Gouveia, N. & Levy, R. Associations of ultra-processed food and drink products with asthma and wheezing among Brazilian adolescents. Pediatr. Allergy Immunol. 29, 504–511 (2018).
Augustine, T., Kumar, M., Al Khodor, S. & van Panhuys, N. Microbial dysbiosis tunes the immune response towards allergic disease outcomes. Clin. Rev. Allergy Immunol. 65, 43–71 (2023).
Penders, J., Stobberingh, E. E., van den Brandt, P. A. & Thijs, C. The role of the intestinal microbiota in the development of atopic disorders. Allergy 62, 1223–1236 (2007).
Melli, L. C., do Carmo-Rodrigues, M. S., Araujo-Filho, H. B., Sole, D. & de Morais, M. B. Intestinal microbiota and allergic diseases: a systematic review. Allergol. Immunopathol. 44, 177–188 (2016).
Azad, M. B. et al. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin. Immunol. 9, 15 (2013).
Kim, C. H. Immune regulation by microbiome metabolites. Immunology 154, 220–229 (2018).
Wong, J. M., De Souza, R., Kendall, C. W., Emam, A. & Jenkins, D. J. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 40, 235–243 (2006).
Xue, C. et al. Tryptophan metabolism in health and disease. Cell Metab. 35, 1304–1326 (2023).
Nuse, B., Holland, T., Rauh, M., Gerlach, R. G. & Mattner, J. L-Arginine metabolism as pivotal interface of mutual host-microbe interactions in the gut. Gut Microbes 15, 2222961 (2023).
Dodd, D. et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 (2017).
Sayin, S. amaI. et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013).
Krautkramer, K. A., Fan, J. & Backhed, F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 19, 77–94 (2021).
Takeuchi, T., Nakanishi, Y. & Ohno, H. Microbial metabolites and gut immunology. Annu. Rev. Immunol. 42, 153–178 (2024).
Kim, M., Qie, Y., Park, J. & Kim, C. H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20, 202–214 (2016).
Rooks, M. G. & Garrett, W. S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352 (2016).
Fahur Bottino, G. et al. Early life microbial succession in the gut follows common patterns in humans across the globe. Nat. Commun. 16, 660 (2025).
Nathan, A. M. et al. Colonization of the newborn respiratory tract and its association with respiratory morbidity in the first 6 months of life: a prospective cohort study. Int. J. Infect. Dis. 122, 712–720 (2022).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Dapa, T. & Xavier, K. B. Effect of diet on the evolution of gut commensal bacteria. Gut Microbes 16, 2369337 (2024).
Burger, E. & Gallo, R. L. Host-microbiome interactions in the holobiome of atopic dermatitis. J. Allergy Clin. Immunol. 151, 1236–1238 (2023).
Sbihi, H. et al. Thinking bigger: How early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy 74, 2103–2115 (2019).
Capone, K. A., Dowd, S. E., Stamatas, G. N. & Nikolovski, J. Diversity of the human skin microbiome early in life. J. Invest. Dermatol. 131, 2026–2032 (2011).
Hoskinson, C. et al. Delayed gut microbiota maturation in the first year of life is a hallmark of pediatric allergic disease. Nat. Commun. 14, 4785 (2023).
Bunyavanich, S. et al. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 138, 1122–1130 (2016).
Durack, J. et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun. 9, 707 (2018).
Kim, J. E. & Kim, H. S. Microbiome of the skin and gut in atopic dermatitis (AD): understanding the pathophysiology and finding novel management strategies. J. Clin. Med. 8, 444 (2019).
Nylund, L. et al. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy 70, 241–244 (2015).
Goldberg, M. R. et al. Microbial signature in IgE-mediated food allergies. Genome Med. 12, 92 (2020).
Bunyavanich, S. & Berin, M. C. Food allergy and the microbiome: current understandings and future directions. J. Allergy Clin. Immunol. 144, 1468–1477 (2019).
Davis, E. C., Jackson, C. M., Ting, T., Harizaj, A. & Jarvinen, K. M. Predictors and biomarkers of food allergy and sensitization in early childhood. Ann. Allergy Asthma Immunol. 129, 292–300 (2022).
De Filippis, F. et al. Specific gut microbiome signatures and the associated pro-inflamatory functions are linked to pediatric allergy and acquisition of immune tolerance. Nat. Commun. 12, 5958 (2021).
Berni Canani, R. et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 10, 742–750 (2016).
Fieten, K. B. et al. Fecal microbiome and food allergy in pediatric atopic dermatitis: a cross-sectional pilot study. Int. Arch. Allergy Immunol. 175, 77–84 (2018).
Stefka, A. T. et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl Acad. Sci. USA 111, 13145–13150 (2014).
Savage, J. H. et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 73, 145–152 (2018).
Tanaka, M. et al. Signatures in the gut microbiota of Japanese infants who developed food allergies in early childhood. FEMS Microbiol. Ecol. https://doi.org/10.1093/femsec/fix099 (2017).
Chen, C. C., Chen, K. J., Kong, M. S., Chang, H. J. & Huang, J. L. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr. Allergy Immunol. 27, 254–262 (2016).
Bjorksten, B., Naaber, P., Sepp, E. & Mikelsaar, M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin. Exp. Allergy 29, 342–346 (1999).
Bisgaard, H. et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 128, 646–652 (2011).
Tsilochristou, O. et al. Association of Staphylococcus aureus colonization with food allergy occurs independently of eczema severity. J. Allergy Clin. Immunol. 144, 494–503 (2019).
Ling, Z. et al. Altered fecal microbiota composition associated with food allergy in infants. Appl. Environ. Microbiol. 80, 2546–2554 (2014).
Azad, M. B. et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin. Exp. Allergy 45, 632–643 (2015).
Panzer, A. R. et al. The impact of prenatal dog keeping on infant gut microbiota development. Clin. Exp. Allergy 53, 833–845 (2023).
Bao, R. et al. Fecal microbiome and metabolome differ in healthy and food-allergic twins. J. Clin. Invest. 131, e141935 (2021).
Ogata, Y. et al. Complete genome sequence of Phascolarctobacterium faecium JCM 30894, a succinate-utilizing bacterium isolated from human feces. Microbiol. Resour. Announc. 8, e01487–18 (2019).
Ze, X., Duncan, S. H., Louis, P. & Flint, H. J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543 (2012).
Wan, J. et al. Alterations in the gut microbiome of young children with airway allergic disease revealed by next-generation sequencing. J. Asthma Allergy 16, 961–972 (2023).
Kim, Y. J. & Bunyavanich, S. Microbial influencers: the airway microbiome’s role in asthma. J. Clin. Invest. 135, e184316 (2025).
Earl, C. S., An, S. Q. & Ryan, R. P. The changing face of asthma and its relation with microbes. Trends Microbiol. 23, 408–418 (2015).
Barcik, W., Boutin, R. C. T., Sokolowska, M. & Finlay, B. B. The role of lung and gut microbiota in the pathology of asthma. Immunity 52, 241–255 (2020).
Dickson, R. P. & Huffnagle, G. B. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog. 11, e1004923 (2015).
Segal, L. N. et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 1, 16031 (2016).
Durack, J. et al. Distinct associations of sputum and oral microbiota with atopic, immunologic, and clinical features in mild asthma. J. Allergy Clin. Immunol. 146, 1016–1026 (2020).
Vael, C., Nelen, V., Verhulst, S. L., Goossens, H. & Desager, K. N. Early intestinal Bacteroides fragilis colonisation and development of asthma. BMC Pulm. Med. 8, 19 (2008).
Arrieta, M. C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7, 307ra152 (2015).
Bisgaard, H. et al. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 357, 1487–1495 (2007).
Liwen, Z., Yu, W., Liang, M., Kaihong, X. & Baojin, C. A low abundance of Bifidobacterium but not Lactobacillius in the feces of Chinese children with wheezing diseases. Medicine 97, e12745 (2018).
Stiemsma, L. T. et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin. Sci. 130, 2199–2207 (2016).
Lee, S. H., Gong, Y. N. & Ryoo, E. Clostridium difficile colonization and/or infection during infancy and the risk of childhood allergic diseases. Korean J. Pediatr. 60, 145–150 (2017).
Zhou, M. S. et al. Altered diversity and composition of gut microbiota in patients with allergic rhinitis. Microb. Pathog. 161, 105272 (2021).
Roduit, C. et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 74, 799–809 (2019).
Hu, Q. et al. Tryptophan metabolite-regulated Treg responses contribute to attenuation of airway inflammation during specific immunotherapy in a mouse asthma model. Hum. Vaccin. Immunother. 16, 1891–1899 (2020).
van der Sluijs, K. F. et al. Systemic tryptophan and kynurenine catabolite levels relate to severity of rhinovirus-induced asthma exacerbation: a prospective study with a parallel-group design. Thorax 68, 1122–1130 (2013).
Wu, R. et al. The bile acid-activated retinoic acid response in dendritic cells is involved in food allergen sensitization. Allergy 77, 483–498 (2022).
Kim, C. H., Park, J. & Kim, M. Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Netw. 14, 277–288 (2014).
Kim, C. H. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell Mol. Immunol. 18, 1161–1171 (2021).
Brown, A. J. et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 278, 11312–11319 (2003).
Nilsson, N. E., Kotarsky, K., Owman, C. & Olde, B. Identification of a free fatty acid receptor, FFA2R, expressed on leukocytes and activated by short-chain fatty acids. Biochem. Biophys. Res. Commun. 303, 1047–1052 (2003).
Le Poul, E. et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 278, 25481–25489 (2003).
Cuisset, L., Tichonicky, L. & Delpech, M. A protein phosphatase is involved in the inhibition of histone deacetylation by sodium butyrate. Biochem. Biophys. Res. Commun. 246, 760–764 (1998).
Park, J. et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR–S6K pathway. Mucosal Immunol. 8, 80–93 (2015).
Kim, M. H., Kang, S. G., Park, J. H., Yanagisawa, M. & Kim, C. H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145, 396–406 (2013).
Sepahi, A., Liu, Q., Friesen, L. & Kim, C. H. Dietary fiber metabolites regulate innate lymphoid cell responses. Mucosal Immunol. 14, 317–330 (2021).
Chun, E. et al. Metabolite-sensing receptor Ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity 51, 871–884 (2019).
Thio, C. L., Chi, P. Y., Lai, A. C. & Chang, Y. J. Regulation of type 2 innate lymphoid cell-dependent airway hyperreactivity by butyrate. J. Allergy Clin. Immunol. 142, 1867–1883 e1812 (2018).
Ticho, A. L., Malhotra, P., Dudeja, P. K., Gill, R. K. & Alrefai, W. A. Bile acid receptors and gastrointestinal functions. Liver Res. 3, 31–39 (2019).
Fleishman, J. S. & Kumar, S. Bile acid metabolism and signaling in health and disease: molecular mechanisms and therapeutic targets. Signal Transduct. Target. Ther. 9, 97 (2024).
Taylor, S. A. & Green, R. M. Bile acids, microbiota, and metabolism. Hepatology 68, 1229–1231 (2018).
Chiang, J. Y. L. & Ferrell, J. M. Discovery of farnesoid X receptor and its role in bile acid metabolism. Mol. Cell Endocrinol. 548, 111618 (2022).
Sorrentino, G. et al. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology 159, 956–968 (2020).
Thomas, C. et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 10, 167–177 (2009).
Perino, A. et al. TGR5 reduces macrophage migration through mTOR-induced C/EBPbeta differential translation. J. Clin. Invest. 124, 5424–5436 (2014).
Qi, Y., Duan, G., Wei, D., Zhao, C. & Ma, Y. The bile acid membrane receptor tgr5 in cancer: friend or foe?. Molecules 27, 5292 (2022).
Duboc, H., Tache, Y. & Hofmann, A. F. The bile acid TGR5 membrane receptor: from basic research to clinical application. Dig. Liver Dis. 46, 302–312 (2014).
Staudinger, J. L. et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl Acad. Sci. USA 98, 3369–3374 (2001).
Liu, R. et al. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology 60, 908–918 (2014).
Wang, X. J., Chen, B. Y., Yang, B. W., Yue, T. L. & Guo, C. F. Short communication: chemical structure, concentration, and pH are key factors influencing antimicrobial activity of conjugated bile acids against lactobacilli. J. Dairy Sci. 104, 1524–1530 (2021).
Jia, H., He, X., Jiang, T. & Kong, F. Roles of bile acid-activated receptors in monocytes-macrophages and dendritic cells. Cells 14, 920 (2025).
Paik, D. et al. Human gut bacteria produce Tau(Eta)17-modulating bile acid metabolites. Nature 603, 907–912 (2022).
Heath-Pagliuso, S. et al. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry 37, 11508–11515 (1998).
Negishi, T. et al. Effects of aryl hydrocarbon receptor signaling on the modulation of TH1/TH2 balance. J. Immunol. 175, 7348–7356 (2005).
Xuan, M. et al. Polyamines: their significance for maintaining health and contributing to diseases. Cell Commun. Signal 21, 348 (2023).
Saha, P. P. et al. Gut microbe-generated phenylacetylglutamine is an endogenous allosteric modulator of beta2-adrenergic receptors. Nat. Commun. 15, 6696 (2024).
Hu, J. et al. Gut microbiota-derived 3-phenylpropionic acid promotes intestinal epithelial barrier function via AhR signaling. Microbiome 11, 102 (2023).
Colosimo, D. A. et al. Mapping interactions of microbial metabolites with human G-protein-coupled receptors. Cell Host Microbe 26, 273–282 (2019).
Nguyen, A. V. & Soulika, A. M. The dynamics of the skin’s immune system. Int. J. Mol. Sci. 20, 1811 (2019).
Luo, C. H. et al. Staphylococcus aureus exacerbates dermal IL-33/ILC2 axis activation through evoking RIPK3/MLKL-mediated necroptosis of dry skin. JCI Insight 9, e166821 (2024).
Liu, X. et al. Activation of aryl hydrocarbon receptor in Langerhans cells by a microbial metabolite of tryptophan negatively regulates skin inflammation. J. Dermatol. Sci. 100, 192–200 (2020).
Lefevre, M. A., Vocanson, M. & Nosbaum, A. Role of tissue-resident memory T cells in the pathophysiology of allergic contact dermatitis. Curr. Opin. Allergy Clin. Immunol. 21, 355–360 (2021).
Nestle, F. O., Di Meglio, P., Qin, J. Z. & Nickoloff, B. J. Skin immune sentinels in health and disease. Nat. Rev. Immunol. 9, 679–691 (2009).
Kumar, S., Jeong, Y., Ashraf, M. U. & Bae, Y. S. Dendritic cell-mediated Th2 immunity and immune disorders. Int. J. Mol. Sci. 20, 2159 (2019).
Kopf, M., Schneider, C. & Nobs, S. P. The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 16, 36–44 (2015).
Verma, M., Can, U. I. & Reinhardt, R. L. ILC2 diversity, location, and function in pulmonary disease. Immunol. Rev. 332, e70036 (2025).
Kim, C. H., Hashimoto-Hill, S. & Kim, M. Migration and tissue tropism of innate lymphoid cells. Trends Immunol. 37, 68–79 (2016).
Drake, L. Y. & Kita, H. Group 2 innate lymphoid cells in the lung. Adv. Immunol. 124, 1–16 (2014).
Shirley, S. et al. A vasculature-resident innate lymphoid cell population in mouse lungs. Nat. Commun. 16, 3718 (2025).
Naderi, W., Schreiner, D. & King, C. G. T-cell–B-cell collaboration in the lung. Curr. Opin. Immunol. 81, 102284 (2023).
Macedo, B. G., Masuda, M. Y. & Borges da Silva, H. Location versus ID: what matters to lung-resident memory T cells?. Front. Immunol. 15, 1355910 (2024).
Bertrand, Y., Sanchez-Montalvo, A., Hox, V., Froidure, A. & Pilette, C. IgA-producing B cells in lung homeostasis and disease. Front. Immunol. 14, 1117749 (2023).
Antunes, K. H. et al. Airway-delivered short-chain fatty acid acetate boosts antiviral immunity during rhinovirus infection. J. Allergy Clin. Immunol. 151, 447–457 (2023).
Grondin, J. A., Kwon, Y. H., Far, P. M., Haq, S. & Khan, W. I. Mucins in intestinal mucosal defense and inflammation: learning from clinical and experimental studies. Front. Immunol. 11, 2054 (2020).
Bemark, M., Pitcher, M. J., Dionisi, C. & Spencer, J. Gut-associated lymphoid tissue: a microbiota-driven hub of B cell immunity. Trends Immunol. 45, 211–223 (2024).
Diefenbach, A., Gnafakis, S. & Shomrat, O. Innate lymphoid cell–epithelial cell modules sustain intestinal homeostasis. Immunity 52, 452–463 (2020).
Stanbery, A. G., Shuchi, S., Jakob von, M., Tait Wojno, E. D. & Ziegler, S. F. TSLP, IL-33, and IL-25: not just for allergy and helminth infection. J. Allergy Clin. Immunol. 150, 1302–1313 (2022).
Ornelas, A., Dowdell, A. S., Lee, J. S. & Colgan, S. P. Microbial metabolite regulation of epithelial cell-cell interactions and barrier function. Cells 11, 944 (2022).
Fung, C. et al. Tuft cells mediate commensal remodeling of the small intestinal antimicrobial landscape. Proc. Natl Acad. Sci. USA 120, e2216908120 (2023).
Eshleman, E. M. et al. Microbiota-derived butyrate restricts tuft cell differentiation via histone deacetylase 3 to modulate intestinal type 2 immunity. Immunity 57, 319–332 (2024).
Kim, H. B. et al. Skin microbiome dynamics in atopic dermatitis: understanding host–microbiome interactions. Allergy Asthma Immunol. Res. 17, 165–180 (2025).
Qiu, Z. et al. A dysregulated sebum-microbial metabolite-IL-33 axis initiates skin inflammation in atopic dermatitis. J. Exp. Med. 219, e20212397 (2022).
Trompette, A. et al. Gut-derived short-chain fatty acids modulate skin barrier integrity by promoting keratinocyte metabolism and differentiation. Mucosal Immunol. 15, 908–926 (2022).
Trompette, A. et al. Dietary fiber confers protection against flu by shaping Ly6c− patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity 48, 992–1005 (2018).
Wang, Y. et al. A precision microbiome approach using sucrose for selective augmentation of Staphylococcus epidermidis fermentation against Propionibacterium acnes. Int. J. Mol. Sci. 17, 1870 (2016).
Leon Carrion, S., Sutter, C. H. & Sutter, T. R. Combined treatment with sodium butyrate and PD153035 enhances keratinocyte differentiation. Exp. Dermatol. 23, 211–214 (2014).
Blicharz, L. et al. Severity of atopic dermatitis is associated with gut-derived metabolites and leaky gut-related biomarkers. Sci. Rep. 15, 26146 (2025).
Yu, J. et al. A tryptophan metabolite of the skin microbiota attenuates inflammation in patients with atopic dermatitis through the aryl hydrocarbon receptor. J. Allergy Clin. Immunol. 143, 2108–2119(2019).
Kang, Y. M. et al. Indole-3-carbinol alleviates allergic skin inflammation via periostin/thymic stromal lymphopoietin suppression in atopic dermatitis. Chin. Med. 19, 177 (2024).
Jena, P. K. et al. Long-term Western diet intake leads to dysregulated bile acid signaling and dermatitis with Th2 and Th17 pathway features in mice. J. Dermatol. Sci. 95, 13–20 (2019).
Kim, E. J. et al. Ursodeoxycholic acid alleviates atopic dermatitis-associated inflammatory responses in HaCaT and RBL-2H3 cells and DNCB/DFE-treated mice. Life Sci. 344, 122560 (2024).
Lin, D. L. et al. 12,13-diHOME promotes inflammatory macrophages and epigenetically modifies their capacity to respond to microbes and allergens. J. Immunol. Res. 2024, 2506586 (2024).
Prescott, S. L. et al. A global survey of changing patterns of food allergy burden in children. World Allergy Organ J. 6, 21 (2013).
Valenta, R., Hochwallner, H., Linhart, B. & Pahr, S. Food allergies: the basics. Gastroenterology 148, 1120–1131 (2015).
Izadi, N., Luu, M., Ong, P. Y. & Tam, J. S. The role of skin barrier in the pathogenesis of food allergy. Children 2, 382–402 (2015).
Ferraro, V. A., Zanconato, S. & Carraro, S. The epithelial barrier hypothesis in food allergies: the state of the art. Nutrients 17, 1014 (2025).
Shi, J. et al. Butyrate alleviates food allergy by improving intestinal barrier integrity through suppressing oxidative stress-mediated Notch signaling. iMeta 4, e70024 (2025).
Tett, A. et al. The Prevotella copri complex comprises four distinct clades underrepresented in Westernized populations. Cell Host Microbe 26, 666–679 (2019).
Larsen, J. M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 151, 363–374 (2017).
Scher, J. U. et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013).
Folkerts, J. et al. Butyrate inhibits human mast cell activation via epigenetic regulation of FcepsilonRI-mediated signaling. Allergy 75, 1966–1978 (2020).
Nagata, K. et al. Butyrate, valerate, and niacin ameliorate anaphylaxis by suppressing ige-dependent mast cell activation: roles of GPR109A, PGE2, and epigenetic regulation. J. Immunol. 212, 771–784 (2024).
Tan, J. et al. Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep. 15, 2809–2824 (2016).
Shao, H. et al. Bifidobacterium breve M-16V alleviates cow’s milk allergy in a mouse model via gut microbiota-derived indole-3-propionic acid-aryl hydrocarbon receptor signaling axis. Allergy https://doi.org/10.1111/all.16684 (2025).
Hammerschmidt-Kamper, C. et al. Indole-3-carbinol, a plant nutrient and AhR-ligand precursor, supports oral tolerance against OVA and improves peanut allergy symptoms in mice. PLoS ONE 12, e0180321 (2017).
Song, X. et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020).
Bhaumik, S., Mickael, M. E., Moran, M., Spell, M. & Basu, R. RORgammat promotes Foxp3 expression by antagonizing the effector program in colonic regulatory T cells. J. Immunol. 207, 2027–2038 (2021).
Ozcam, M. et al. Gut microbial bile and amino acid metabolism associate with peanut oral immunotherapy failure. Nat. Commun. 16, 6330 (2025).
Lewis, G. et al. Dietary fiber-induced microbial short chain fatty acids suppress ILC2-dependent airway inflammation. Front. Immunol. 10, 2051 (2019).
Cheng, H. Y. et al. Evaluation of stool short chain fatty acids profiles in the first year of life with childhood atopy-related outcomes. Front. Allergy 3, 873168 (2022).
Lee-Sarwar, K. A., Lasky-Su, J., Kelly, R. S., Litonjua, A. A. & Weiss, S. T. Gut microbial-derived metabolomics of asthma. Metabolites 10, 97 (2020).
Chiu, C. Y. et al. Gut microbial-derived butyrate is inversely associated with IgE responses to allergens in childhood asthma. Pediatr. Allergy Immunol. 30, 689–697 (2019).
Huang, M. T. et al. Short-chain fatty acids ameliorate allergic airway inflammation via sequential induction of PMN-MDSCs and Treg cells. J. Allergy Clin. Immunol. Glob. 2, 100163 (2023).
Cait, A. et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 11, 785–795 (2018).
Theiler, A. et al. Butyrate ameliorates allergic airway inflammation by limiting eosinophil trafficking and survival. J Allergy Clin. Immunol. 144, 764–776 (2019).
Zaiss, M. M. et al. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 43, 998–1010 (2015).
Yu, B. et al. Microbiota-derived butyrate alleviates asthma via inhibiting Tfh13-mediated IgE production. Signal Transduct. Target Ther. 10, 181 (2025).
Hu, W. et al. Both living and dead Faecalibacterium prausnitzii alleviate house dust mite-induced allergic asthma through the modulation of gut microbiota and short-chain fatty acid production. J. Sci. Food Agric. 101, 5563–5573 (2021).
Chen, Z. et al. Vancomycin-induced gut microbiota dysbiosis aggravates allergic rhinitis in mice by altered short-chain fatty acids. Front. Microbiol. 13, 1002084 (2022).
Wang, J., Wen, L., Wang, Y. & Chen, F. Therapeutic effect of histone deacetylase inhibitor, sodium butyrate, on allergic rhinitis in vivo. DNA Cell Biol. 35, 203–208 (2016).
Miao, Y. et al. Impaired tryptophan metabolism by type 2 inflammation in epithelium worsening asthma. iScience 27, 109923 (2024).
Wang, H. et al. Gut microbiota-derived tryptophan metabolites alleviate allergic asthma inflammation in ovalbumin-induced mice. Foods 13, 1336 (2024).
Perdijk, O. et al. Antibiotic-driven dysbiosis in early life disrupts indole-3-propionic acid production and exacerbates allergic airway inflammation in adulthood. Immunity 57, 1939–1954 (2024).
Kelly, R. S. et al. Plasma metabolite profiles in children with current asthma. Clin. Exp. Allergy 48, 1297–1304 (2018).
Manni, M. L. et al. Nitroalkene fatty acids modulate bile acid metabolism and lung function in obese asthma. Sci Rep. 11, 17788 (2021).
Nakada, E. M. et al. Conjugated bile acids attenuate allergen-induced airway inflammation and hyperresponsiveness by inhibiting UPR transducers. JCI Insight 4, e98101 (2019).
Hamideh, N. & Wong, L. S. Y. Emerging therapeutics in the management of food allergy. Curr. Probl. Pediatr. Adolesc. Health Care 55, 101732 (2025).
Park, J., Goergen, C. J., HogenEsch, H. & Kim, C. H. Chronically elevated levels of short-chain fatty acids induce t cell-mediated ureteritis and hydronephrosis. J. Immunol. 196, 2388–2400 (2016).
Doron, S. & Snydman, D. R. Risk and safety of probiotics. Clin. Infect. Dis. 60, S129–S134 (2015).
Wicaksono, D. P., Washio, J., Abiko, Y., Domon, H. & Takahashi, N. Nitrite production from nitrate and its link with lactate metabolism in oral Veillonella spp. Appl. Environ. Microbiol. 86, e01255–20 (2020).
Lopez-Siles, M., Duncan, S. H., Garcia-Gil, L. J. & Martinez-Medina, M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 11, 841–852 (2017).
Ottman, N. et al. Genome-scale model and omics analysis of metabolic capacities of akkermansia muciniphila reveal a preferential mucin-degrading lifestyle. Appl. Environ. Microbiol. 83, e01014–e01017 (2017).
Oliveira, A. S., Saraiva, L. M. & Carvalho, S. M. Staphylococcus epidermidis biofilms undergo metabolic and matrix remodeling under nitrosative stress. Front. Cell Infect. Microbiol. 13, 1200923 (2023).
Liu, X. et al. Transcriptomics and metabolomics reveal the adaption of Akkermansia muciniphila to high mucin by regulating energy homeostasis. Sci. Rep. 11, 9073 (2021).
Zhang, S. M. & Huang, S. L. The commensal anaerobe Veillonella dispar reprograms its lactate metabolism and short-chain fatty acid production during the stationary phase. Microbiol. Spectr. 11, e0355822 (2023).
Rios-Covian, D., Salazar, N., Gueimonde, M. & de Los Reyes-Gavilan, C. G. Shaping the metabolism of intestinal bacteroides population through diet to improve human health. Front. Microbiol. 8, 376 (2017).
Liu, M. J. et al. Recent findings in Akkermansia muciniphila-regulated metabolism and its role in intestinal diseases. Clin. Nutr. 41, 2333–2344 (2022).
Distler, W. & Kroncke, A. The lactate metabolism of the oral bacterium Veillonella from human saliva. Arch. Oral Biol. 26, 657–661 (1981).
Myles, I. A. et al. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight 3, e120608 (2018).
Hosseinkhani, F. et al. The contribution of gut bacterial metabolites in the human immune signaling pathway of non-communicable diseases. Gut Microbes 13, 1–22 (2021).
Sen, A. et al. Comprehensive analysis of metabolites produced by co-cultivation of Bifidobacterium breve MCC1274 with human iPS-derived intestinal epithelial cells. Front. Microbiol. 14, 1155438 (2023).
Bocchio, F. et al. Compendium of Bifidobacterium-based probiotics: characteristics and therapeutic impact on human diseases. Microbiome Res. Rep. 4, 2 (2025).
Tang, H., Huang, W. & Yao, Y. F. The metabolites of lactic acid bacteria: classification, biosynthesis and modulation of gut microbiota. Microb. Cell 10, 49–62 (2023).
Devika, N. T. & Raman, K. Deciphering the metabolic capabilities of Bifidobacteria using genome-scale metabolic models. Sci. Rep. 9, 18222 (2019).
Candeliere, F. et al. The metabolism of Leuconostoc genus decoded by comparative genomics. Microorganisms 12, 1487 (2024).
Fuochi, V. et al. Metabolic characterization of supernatants produced by Lactobacillus spp. with in vitro anti-Legionella activity. Front Microbiol 10, 1403 (2019).
Ponsetto, P., Sasal, E. M., Mazzoli, R., Valetti, F. & Gilardi, G. The potential of native and engineered Clostridia for biomass biorefining. Front. Bioeng. Biotechnol. 12, 1423935 (2024).
Zhang, S. M., Hung, J. H., Yen, T. N. & Huang, S. L. Mutualistic interactions of lactate-producing lactobacilli and lactate-utilizing Veillonella dispar: lactate and glutamate cross-feeding for the enhanced growth and short-chain fatty acid production. Microb. Biotechnol. 17, e14484 (2024).
Zuniga, M., Monedero, V. & Yebra, M. J. Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium species. Front. Microbiol. 9, 1917 (2018).
Zaplana, T., Miele, S. & Tolonen, A. C. Lachnospiraceae are emerging industrial biocatalysts and biotherapeutics. Front. Bioeng. Biotechnol. 11, 1324396 (2023).
Fernandez-Cantos, M. V., Babu, A. F., Hanhineva, K. & Kuipers, O. P. Identification of metabolites produced by six gut commensal Bacteroidales strains using non-targeted LC–MS/MS metabolite profiling. Microbiol. Res. 283, 127700 (2024).
Russell, D. A., Ross, R. P., Fitzgerald, G. F. & Stanton, C. Metabolic activities and probiotic potential of bifidobacteria. Int. J. Food Microbiol. 149, 88–105 (2011).
Catlett, J. L. et al. Metabolic feedback inhibition influences metabolite secretion by the human gut symbiont Bacteroides thetaiotaomicron. mSystems 5, e00252-20 (2020).
Oliphant, K. & Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 7, 91 (2019).
Abdugheni, R. et al. Metabolite profiling of human-originated Lachnospiraceae at the strain level. eMeta 1, e58 (2022).
Acknowledgements
This study was supported, in part, by the National Institute of Health (R01AI173179 to CHK) and the Mary H Weiser Food Allergy Center. C.H.K. is Kenneth and Judy Betz Professor of Food Allergy Research. J.R.B. is supported by the Ruth Dow Doan Professorship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, C.H., Baker, J.R. Regulation of allergies across the body by microbial metabolites. Exp Mol Med (2026). https://doi.org/10.1038/s12276-026-01642-1
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s12276-026-01642-1