Abstract
Triple negative breast cancer (TNBC) is responsible for significant number of breast cancer-associated deaths because of lacking of successful molecular-targeted therapy. To explore a therapeutic target for TNBC, we performed a siRNA-mediated knockdown screening and identified Polo-like kinase 1 (PLK1) as a potential therapeutic target for TNBC. Knockdown of PLK1 as well as a small compound inhibitor for PLK1, BI-2536, induced G2/M arrest and created polyploid cell population, shown by increased DNA content and nuclear size. Inhibition of PLK1 eventually triggered apoptosis in multiple TNBC cell lines. In addition, we confirmed that PLK1 was significantly overexpressed in the tissues from TNBC patients compared with the tissues of normal mammary glands and benign breast tumors. Our data indicated that PLK1 plays a pivotal role in the regulation of mitosis of TNBC cells. Although future in vivo studies are warranted, targeting PLK1 by a selective inhibitor such as BI-2536 can be an attractive molecular-targeted therapy for TNBC.
Similar content being viewed by others
Introduction
Breast cancer is the leading cause of cancer-associated deaths in women worldwide, affecting more than 1 million new cases and half a million deaths every year. Clinicopathological characterization categorizes the disease into five classical subtypes, luminal A, luminal B (HER2 negative), luminal B (HER2 positive), HER2 positive, and triple negative breast cancer (TNBC). TNBC, defined by a lack of immunohistochemical expression of estrogen receptor (ER), progesterone receptor (PgR) and HER2 receptor, accounts for approximately 15% of all breast cancers [1, 2], and is commonly observed in premenopausal African-American women [3]. In Asian population, TNBC accounts for approximately 10% of all breast cancer [4]. TNBC is a highly aggressive type of breast cancer and often causes metastasis in early stage. However, because TNBC does not express hormone receptors or HER2, the patients with TNBC do not benefit from hormonal therapy or anti-HER2 therapy. It is also well-known that TNBC is sensitive to the conventional chemotherapy at first, but it has higher recurrence rate and thus results in poor prognosis [5, 6]. Although the recent report which re-analyzed the dataset of gene expression profiles of TNBC enabled subcategorization of the TNBC into 6 subtypes [7], the molecular-targeted therapy for TNBC, highly effective in other types of breast cancer, still remains to be investigated.
Genome-wide association studies have shown that defects in DNA repairing machineries contribute to the pathogenesis of breast cancer [8]. When severe genomic DNA damages including double-strand breaks (DSBs) and single-strand breaks (SSBs) occur, intrinsic DNA damage repairing pathways cooperatively fix the damage to suppress undesirable mutations. In higher eukaryotes, DSBs are fixed by two major DNA repairing mechanisms, the homologous recombination (HR) and the non-homologous end joining (NHEJ) pathways. BRCA1 is a key modulator protein required for efficient repair of DSB through HR. Following DSB, serine/threonine kinases Ataxia Telangiectasia Mutated (ATM) and Ataxia Telangiectasia and Rad3-Related (ATR) are activated and they phosphorylate BRCA1 [9]. Polo-like knase-1 (PLK1) also phosphorylates BRCA1 and mediates recruitment of BRCA1 to DSBs, resulting in promotion of HR [10]. BRCA2 protein is also involved in the HR. The implications of these DNA repairing machineries in breast cancer has been shown by robust association of mutations in BRCA1 and BRCA2 genes with hereditary breast cancer patients. In the patients with mutations of BRCA1/2 genes, the non-functional DNA repairing system due to lack of BRCA1/2 fails to fix cancerous mutations and thus they cause breast cancer.
On the other hand, the major chemotherapy used for treatment of TNBC, including anthracyclines and platinum compounds, also utilizes DNA damages to inhibit tumor growth. These chemotherapeutic compounds induce severe damage in genomic DNA. If the degree of DNA damage is overwhelming to the repairing machinery, the accumulation of DNA damage is monitored by cell cycle checkpoints, such as G1/S checkpoint and the G2/M checkpoint, resulting in inhibition of DNA replication and impairment of segregation of chromosome at mitosis, respectively. If further irreversible DNA damages accumulate, they trigger apoptosis. Therefore, the molecule indispensable in the DNA repairing pathways may be a strong candidate of a new drug target for TNBC.
In this study, we screened 24 genes involved in the DNA repair using the siRNA-mediated cell viability assay and found that knockdown of PLK1 triggered apoptosis in multiple TNBC cell lines. We also confirmed that both knockdown of PLK1 as well as a small compound inhibitor for PLK1, BI-2536, induced G2/M arrest and created polyploid cell population, and eventually induced apoptosis. In addition, overexpression of PLK1 in TNBC tissues was confirmed by immunohistochemical analysis. Our data shown here indicate that PLK1 regulates mitosis of TNBC cells the function of PLK1 is indispensable for survival of TNBC in vitro. Although further in vivo studies and sophisticated clinical trials are necessary, inhibition of PLK1 should be considered as a probable option of molecular-targeted therapy for TNBC.
materials and methods
Cell lines and reagents
TNBC cell lines, MDA-MB-231, Hs578T, MDA-MB-436, MDA-MB-468 and HCC1937 cell lines were obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA) in December 2015. MDA-MB-453 cell line was obtained from RIKEN BRC Cell Bank (Tsukuba, Ibaraki, Japan) in July 2015. HCC38 and HCC1806 cell lines were generous gifts from Department of Gastroenterological surgery at Yokohama City University Hospital in July 2015. HCC38, HCC1806 and HCC1937 cells were cultured in RPMI-1640 medium (Gibco, Waltham, MA) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin at 37 °C in 5% CO2. Hs578T was cultured in DMEM supplemented with 0.01 mg/ml bovine insulin (Sigma, St. Louis, MO), 10% FBS and antibiotics at 37 °C in 5% CO2. MDA-MB-231, MDA-MB-436, MDA-MB-453 and MDA-MB-468 were cultured in Leibovitz’s L-15 medium (Gibco) supplemented with 10% FBS, 1% GlutaMAX (Gibco) and antibiotics at 37 °C in 100% room air. BI-2536 was purchased from Selleck chemicals (Houston, TX).
siRNA library
The RNAi library was purchased from Dharmacon (Lafayette, CO, USA). The targeted genes were shown in Table 1. The siRNA for each gene contains four different siRNA to obtain better knockdown efficiency.
siRNA screening
MDA-MB-436, MDA-MB-468 and HCC1806 were seeded (8000 cells/well) in a 96-well plate. The cells were transfected with 10 nM of each siRNA using Lipofectamine RNAi MAX (Invitrogen, Carlsbad, CA). Forty-eight hours after transfection, the cell viability was assessed by Cell-titer glo 2.0 (Promega, Fitchburg, WI) following the manufacturer’s instruction.
Immunohistochemistry (IHC)
All samples were obtained from patients treated at Tokyo Medical University Hospital (Tokyo, Japan) under an approved Institutional Review Board protocol (TS2018-0071). Samples were fixed in 10% neutral buffered formalin followed by embedding in paraffin. Sections were deparaffinized and rehydrated to distilled water before staining. Endogenous peroxidase activity was quenched by incubation with 0.5% H2O2/methanol for 10 min, and heat-induced epitope retrieval was performed. Immunohistochemistry was done with anti-PLK1 antibody (Abcam, Cambridge, MA) and visualized using the EnVision DAB kit (DAKO, Glostrup, Denmark) according to the manufacturer’s instructions.
Scoring of IHC staining
All samples (TNBCs, normal mammary glands, benign tumors, luminal A breast cancer, luminal B breast cancer and HER-2 positive breast cancer, N = 5 for each group) were observed with the BZ-9000 Fluorescence Microscope (KEYENCE, Osaka, Japan). To evaluate the intensity and proportion of cells positive for PLK1, we applied a semi-quantitative scale of the Quickscore method [11]. First, the staining intensity (0 = negative, 1 = weak, 2 = intermediate, 3 = strong) and the proportion of the cells immune-positive (1 = 0–4%, 2 = 5–19%, 3 = 20–39%, 4 = 40–59%, 5 = 60–79%, 6 = 80–100%) were scored, then each score was multiplied to obtain the Quickscore. Either nucleus or cytoplasmic signal above background was counted as positive.
Western blot analysis
MDA-MB-231, MDA-MB-436, MDA-MB-468, HCC38 and HCC1937 were grown in 100 mm cell culture dishes and lysed in M-PER mammalian protein extraction reagent (Thermo Fisher Scientific) containing phosphatase inhibitors and protease inhibitors (Roche Diagnostics, Basel, Switzerland). Each sample containing 30 μg of proteins was applied to SDS-PAGE and transferred to PVDF membranes. After blocking in Blocking One solution (Nacalai Tesque, Kyoto, Japan), the membrane was incubated overnight with anti-GAPDH antibody (Cell Signaling Technology, Danvers, MA) or anti-PLK1 mouse monoclonal antibody (Abcam) at 4 °C. After incubation with horseradish peroxidase-labeled secondary antibodies, protein expressions were detected with ChemiDoc imaging system (Biorad, Hercules, CA) using Amersham ECL Select Western Blotting Detection Reagents (GE Healthcare, Chicago, IL). The images were acquired with Image Lab Software (Biorad).
Immunocytochemistry
For staining of endogenous PLK1, MDA-MB-436 cells were fixed with 4% paraformaldehyde-PBS for 15 min and permeabilized with 0.5% Triton X-100 for 5 min at room temperature. After blocking with Blocking One solution (Nacalai Tesque) for 1 h, the cells were incubated for overnight with anti-PLK1 mouse monoclonal antibody (Abcam) at 4 °C. The cells were washed in PBS twice and incubated with Alexa-Fluor 488-conjugated goat anti-mouse IgG (Abcam) for 1 h. Nuclei were counterstained with 4′,6′-diamidino-2-phenylinodole (DAPI). For staining of phosphorylated histone H2AX (γ-H2AX), MDA-MB-231 cells, treated with 10 μM of bleomycin or 10 nM of BI-2536 for 6 h, were immunostained with anti-γ-H2AX antibody (Cell signaling). For imaging of spindle assembly, MDA-MB-231 cells and MDA-MB-436 cells expressing GFP-tubulin were treated with DMSO or 10 nM of BI-2536 for 6 h or 24 h.
Cell viability assay
To examine the effect of siRNA-mediated knockdown on the cell viability, TNBC cell lines including HCC1806, HCC1937, Hs578T, MDA-MB-231, MDA-MB-436, MDA-MB-453, and MDA-MB-468, seeded (5000 or 8,000 cells/well) in 96-well plates, were transfected with 10 nM of negative control siRNA or siRNA against PLK1 using Lipofectamine RNAiMAX (Invitrogen). To examine the effect of BI-2536, a selective inhibitor for PLK1, HCC1806, HCC1937, Hs578T, MDA-MB-231, MDA-MB-436, MDA-MB-453, and MDA-MB-468 were incubated with DMSO or BI-2536 (from 0.25 to 100 nM) for 48 h. Cell viability was assessed by Calcein-AM solution (Dojindo, Tokyo, Japan) or Cell-titer glo (Promega). The viability was measured with the microplate reader infinite F200 PRO (TECAN, Männedorf, Switzerland).
Cell cycle analysis
MDA-MB-436 cells and MDA-MB-231 cells were seeded (1 × 105 cells/well) in 6-well plates and transfected with 10 nM of negative control siRNA or siRNA against PLK1. Forty-eight h after transfection, the cells were collected, washed with PBS containing 2% FBS and then re-suspended in 70% ice-cold ethanol to fix and permeabilize the cells. The fixed cells were washed in PBS and re-suspended in PBS containing 100 μg/ml RNase A and 20 μg/ml propidium iodide. After incubation for 30 min at room temperature, samples were analyzed by fluorescence-activated cell sorter (FACS). All experiments were repeated three times independently. FlowJo software (TreeStar Inc., Ashland, OR) was used for data analysis.
Apoptosis analysis using Annexin V-7AAD staining
MDA-MB-436 cells and MDA-MB-231 cells were seeded (1 × 105 cells/well) in 6-well plates and treated with 10 nM BI-2536 for 24 h. The cells were collected, washed twice with 2% FBS-PBS and re-suspended in a binding buffer. Then cells were incubated with PE-Annexin V (BD Pharmingen, Franklin Lakes, NJ) and AAD (BD Pharmingen) for 15 min at room temperature in the dark. Samples were analyzed by FACS. All experiments were repeated five times independently.
Transmission electron microscopy
MDA-MB-231 cells treated with or without 10 nM of BI-2536 were fixed in 4% paraformaldehyde/2.5% glutaraldehyde-PBS for 1 h. After wash with PBS, the samples were treated with 1.5% osmium tetroxide-PBS and embedded in the epoxy resin. The embedded samples were then ultrathin sectioned and imaged by LEM-1200EX transmission electron microscopy (JEOL, Tokyo, Japan).
Results
siRNA library screening identified PLK1 as a therapeutic target of TNBC cell lines
To identify the gene involved in the DNA damage repair and indispensable for survival of TNBC cell lines, we searched literatures and chose 24 genes related to the DNA damage repair and create a library of siRNAs (Fig. 1a and Table 1). The siRNA screening was performed in 3 different TNBC cell lines, MDA-MB-436 (a mutation in BRCA1), HCC1806 (mutations in both BRCA1/2) and MDA-MB-468 (wild type BRCA1). The cell viability assay showed that knockdown of PLK1 decreased number of viable cells in all three TNBC lines examined (Fig. 1b–d).
Inhibition of PLK1 decreases the viability of various TNBC cell lines
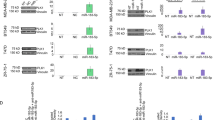
It has been shown that TNBC can be subclassified into 6 subtypes by gene expression profiles (BL1; basal-like 1, BL2; basal-like 2, IM; immunomodulatory, M; mesenchymal-like, MSL; mesenchymal stem-like, LAR; luminal androgen receptor) [7]. Therefore, we next examined if the cytotoxic phenotype by inhibition of PLK1 was specific to the cell lines tested in the screening above, or it is commonly shared by various type of TNBC lines. To confirm this, we first performed the immunoblot analysis of PLK1 in various TNBC cells lines including MDA-MB-231, MDA-MB-436, MDA-MB-468, HCC38 and HCC1937 and confirmed that all TNBC line endogenously expressed high level of PLK1 (Fig. 2a). We also confirmed that transfection of siRNA targeting PLK1 effectively decreased the expression of endogenous PLK1 in TNBC cell lines (Fig. 2b). The immunocytochemical analysis revealed that the expression of PLK1 was highly enhanced in the mitotic cells and it localized to kinetochores and spindles as reported previously (Fig. 2c) [12]. Next, we examined whether inhibition of PLK1 induces cell death in different types of TNBC. We tried seven TNBC lines with the different gene expression profiles and confirmed that all TNBC lines were vulnerable to depletion of PLK1 and the cell viability was decreased by transfection with siRNA targeting PLK1 (Fig. 2d), suggesting that PLK1 is essential for survival of TNBC cells.
a Immunoblot analysis of PLK1 and GAPDH in TNBC cell lines. b Immunoblot analysis of PLK1 and GAPDH in TNBC lines transfected with control siRNA (si-control) or siRNA targeting PLK1 (siPLK1). c Immunocytochemical analysis of PLK1 in MDA-MB-436 cells. The most right panel shows a pseudocolor image of PLK1. A white arrow indicates accumulation of PLK1 in the spindle. d The cell viability of TNBC lines transfected with si-control or siRNA targeting PLK1. Data were mean ± S.D. from 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
BI-2536, a selective inhibitor for PLK1, triggered apoptosis in TNBC lines
PLK1 is reported to be involved in mitosis and also highly expressed in cancer tissues [13,14,15,16,17,18,19,20], therefore its selective inhibitor could be a potential anti-cancer drug. The high throughput screen identified BI-2536 as a small molecule inhibitor for PLK1, and it has been shown that BI-2536 prevents tumor growth in vitro and in vivo [21]. To test the effect of pharmacological inhibition of PLK1 on TNBC cells, we treated MDA-MB-231 cells (wild type BRCA1) and MDA-MB-436 cells (a mutation in BRCA1) with different dose of BI-2536 and performed the cell viability assays (Fig. 3a–d). The cell viability was decreased in a dose-dependent fashion in both cell lines regardless of BRCA genotype. Flowcytometric analyses with Annexin-V and 7-aminoactinomycin D (7-AAD) staining indicated that treatment with BI-2536 triggered apoptotic cell death in MDA-MB-231 cells and MDA-MB-436 cells shown by increase of number of cells with both early apoptotic phase (Annexin-V positive, 7-AAD negative) and late apoptotic phase (Annexin-V positive, 7-AAD positive) (Fig. 3e–h). Electron microscopic images showed that treatment with BI-2536 caused fragmentation of the nucleus in MDA-MB-231 cells, also indicating that inhibition of PLK1 triggers apoptosis in TNBC (Fig. 3i). We further examined the effect of BI-2536 on a variety of TNBC lines of different subtypes and confirmed that BI-2536 induced cell death in all TNBC lines examined (Fig. 3j).
a Dose-response curve of BI-2536 on MDA-MB-231 cells. Y-axis indicates relative cell viability and X-axis shows concentration of BI-2536 (nM). Statistical significance was calculated by one-way ANOVA followed by Dunnett’s test. *p < 0.05, **p < 0.01, ***p < 0.001. b Microscopic images of MDA-MB-231 cells treated with DMSO or 5 nM of BI-2536. c Dose-response curve of BI-2536 on MDA-MB-436 cells. Y-axis indicates relative cell viability and X-axis shows concentration of BI-2536 (nM). Statistical significance was calculated by one-way ANOVA followed by Dunnett’s test. *p < 0.05, **p < 0.01, ***p < 0.001. d Microscopic images of MDA-MB-436 cells treated with DMSO or 5 nM of BI-2536. e, f Flowcytometric analysis of MDA-MB-231 cells treated with DMSO or 10 nM BI-2536, stained with PE-Annexin V and 7-AAD. g, h Flowcytometric analysis of MDA-MB-436 cells treated with DMSO or 10 nM BI-2536, stained with PE-Annexin V and 7-AAD. i Transmission electron microscopic images of MDA-MB-231 cells treated with or without 10 nM BI-2536. The arrows show fragmented nucleus. Scale bars show 2 μM. j Effects of BI-2536 on the survival of TNBC lines. Data were mean ± S.D. from 3 independent experiments. Statistical significance was calculated by Student t test. *p < 0.05, **p < 0.01, ***p < 0.001
Inhibition of PLK1 increases polyploidy and size of the nucleus in TNBC cells
Because it has been reported that PLK1 drives transition of G2/M and its role to establish the bipolar spindle is indispensable [22], inhibition of PLK1 results in a fatal error of mitosis and increases number of polyploid cells [23, 24]. To investigate whether inhibition of PLK1 increases polyploid cells in TNBCs, we demonstrated the propidium iodide (PI) staining to quantify the DNA content in TNBC cells. For this end, we tested the effect of depletion of PLK1 in both MDA-MB-231 cells (wild type BRCA1) and MDA-MB-436 cells (a mutation in BRCA1), and siRNA-mediated knockdown of PLK1 increased the rate of cells harboring 4n DNA compared with the cells transfected with si-control regardless of BRCA genotype (Fig. 4a–d). Treatment with BI-2536 also drastically increased the rate of tetraploid cells in TNBC lines (Fig. 4e–h). It has been shown that accumulation of DNA damage induces G2/M arrest and thus many anti-cancer genotoxic compounds induce G2/M arrest as a result of DNA damage [25]. To examine whether the inhibition of cell cycle by BI-2536 is dependent on DNA damage, we compared BI-2536 with a traditional genotoxic drug, bleomycin. When we treated the TNBC cells with BI-2536 or bleomycin, both BI-2536 and bleomycin increased the cell population with 4n DNA content, but the effect was much more drastic with BI-2536 (Fig. 4f, h). We also evaluated the damage of DNA by staining with anti-phosphorylated histone 2AX (γ-H2AX) antibody, a marker for DSB. After 6 h incubation, bleomycin caused severe DNA damages, shown by increased staining with γ-H2AX antibody whereas BI-2536 did not, suggesting that bleomycin increased 4n population via inducing DSB followed by arrest at G2/M checkpoint but BI-2536 increased 4n polyploid cells independent of DNA damage (Fig. 4i). Because PLK1 is known to contribute to bipolar spindle formation, we next examined whether inhibition of PLK1 disrupts spindle assembly, resulting in incomplete segregation of chromosome. To visualize bipolar spindle, we overexpressed GFP-fused tubulin in MDA-MB-231 cells and MDA-MB-436 cells. When treated with BI-2536, abnormal monopolar spindle was formed and separation of chromosome was inhibited (Fig. 4J). The increased DNA content might result in the larger nuclear size and thus we next measured the size of nucleus in BI-2536 treated MDA-MB231 cells. As Fig. 4k, l show, the treated cells have larger nuclear size compared with control cells, also suggesting the increment of DNA content.
a, b Flowcytometric analysis to quantify the DNA content in MDA-MB-231 cells transfected with control siRNA (si-control) or siRNA targeting PLK1 (siPLK1). Statistical significance was calculated by Student t test. c, d Flowcytometric analysis to quantify the DNA content in MDA-MB-436 cells transfected with control siRNA (si-control) or siRNA targeting PLK1 (siPLK1). Statistical significance was calculated by Student t test. e, f Flowcytometric analysis to quantify the DNA content in MDA-MB-231 cells treated with DMSO, 10 μM of Bleomycin or 10 nM of BI-2536. Statistical significance was calculated by one-way ANOVA followed by Dunnett’s test. g, h Flowcytometric analysis to quantify the DNA content in MDA-MB-436 cells treated with DMSO, 10 μM of Bleomycin or 10 nM of BI-2536. Statistical significance was calculated by one-way ANOVA followed by Dunnett’s test. i Immunocytochemistry of MDA-MB-231 cells treated with 10 μM of bleomycin or 10 nM of BI-2536 for 6 h, stained with anti- γ-H2AX antibody. Arrows show γ-H2AX positive cells. j Confocal imaging of bipolar spindle formation of MDA-MB-231 cells and MDA-MB-436 cells expressing GFP-tubulin, treated with DMSO or 10 nM of BI-2536 for 6 h (MDA-MB-436) or for 24 h (MDA-MB-231). The nucleus was visualized with DAPI. k Microscopic images of MDA-MB-231 cells treated with DMSO or 10 nM BI-2536. The nuclei were visualized with DAPI. Scale bars show 20 μm. l A box plot showing the size of nuclei of MDA-MB-231 cells treated with DMSO or 10 nM BI-2536. The size of nuclei was determined by measuring major axis of each nucleus. N = 23 for each group. Statistical significance was calculated by Student t test. All data were mean ± S.D. from at least 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
PLK1 is overexpressed in TNBC
To investigate the clinical relevance of the PLK1 to TNBC, we assessed the expression of PLK1 in samples of normal mammary glands (n = 5), benign mammary tumors (n = 5), luminal A breast cancer (n = 5), luminal B breast cancer (n = 5), HER-2 positive breast cancer (n = 5) and TNBC patients (n = 5). The immunohistochemical analyses showed that PLK1 was hardly observed in the benign tumors or in normal mammary glands, on the other hand, PLK1 was highly expressed in cancer tissues, especially in TNBC (Fig. 5a, d). The high magnification image of TNBC tissue showed that especially cancer cells at mitotic stages with condensed chromosomes had intense signals of PLK1 (Fig. 5d). We also immunohistochemically analyzed the breast cancer tissues with anti-Ki67 antibody because PLK1 was highly expressed in cells at mitotic phases thus we hypothesized that the expression of PLK1 might correlate with the Ki67, a cell proliferation marker. The immunohistochemical analysis with anti-Ki67 antibody showed that the breast cancer tissues, especially TNBC was immunopositive with anti-Ki67 antibody (Fig. 5e).
a Immunohistochemistry of PLK1 in normal mammary gland. b Immunohistochemistry of PLK1 in benign tumors. c Immunohistochemistry of PLK1 in luminal A, luminal B, HER-2 positive or TNBC tissues (# shows each patient). d The high magnification image of TNBC tissue immunostained with anti-PLK1 antibody. Arrows show condensed chromosomes. e Immunohistochemistry of Ki67 in luminal A, luminal B, HER-2 positive or TNBC tissues. f A box plot of semi-quantified Quickscore of PLK1 in normal mammary glands, benign tumor (three fibroadenoma cases, one benign plyllodes case and one intraductal papilloma case), luminal A, luminal B, HER-2 positive and TNBC. g A box plot of quantified Ki67-positive cells in the luminal A, luminal B, HER-2 positive and TNBC tissues. h The correlation of the Quickscore of PLK1 and rate of Ki67-positive cells. Statistical significance was calculated by one-way ANOVA followed by Tukey’s test. *p < 0.05, **p < 0.01. N = 5
To evaluate PLK1 expression level in detail, we semi-quantified PLK1 staining by the Quickscore [11], obtained by multiplying the rate of positive cells and intensity of signals (Please see the material and method section.). The mean of the Quickscore of PLK1 expression was 5.4 for TNBC, 4.8 for Luminal B, 3.2 for HER-2, 2.4 for Luminal A, 0.6 for benign breast tumors and 0 for normal mammary glands (Fig. 5f) and the Quickscore of TNBC was significantly higher than those of normal mammary glands and benign tumors. There was no significant difference in the Quickscore between the normal mammary glands and benign tumors. We also quantified the Ki67-psotive cells in the breast cancer tissues, and TNBC tended to have higher positivity of Ki67 (Fig. 5g). We next investigated whether the Quickscore of PLK1 and the Ki67-positivity correlates. As expected, there was a good correlation between them (r = 0.633), and it might be a part of reasons why PLK1 was highly expressed in TNBC tissues (Fig. 5c, f).
Discussion
Breast cancer is one of the biggest threats for women health worldwide. Recent advances in development of molecular targeting therapies including anti-HER2 therapy and hormonal therapy have drastically improved the outcome and quality-of-life of patients with breast cancer. However, TNBC, diagnosed by a lack of hormone receptors and HER2 receptor, is still enigmatic breast cancer with worst prognosis due to its high recurrence rate after conventional chemotherapy and deficiency of molecular-targeting therapy [5]. To identify a therapeutic target for TNBC, we performed siRNA-mediated screening and identified PLK1 as a possible therapeutic target for TNBC. Depletion of PLK1 by siRNA-mediated knockdown or BI-2536, a selective inhibitor for PLK1, decreased viability of various TNBC lines through inhibition of mitosis. Furthermore, immunohistochemical analyses showed that PLK1 was highly expressed in the tissues from TNBC patients compared with benign breast tumors or normal mammary gland, proposing PLK1 as a possible therapeutic target for TNBC.
TNBC is a heterogeneous cancer and has a wide variety of genetic mutations. A recent study classified the TNBC into 6 subtypes by the gene expression profile [7]. To find a new therapeutic approach to improve prognosis of TNBC, a series of signal transduction pathways such as the Notch signaling pathway, Wnt/β-catenin pathway and Hedgehog pathway, have been pre-clinically and clinically studied [26, 27]. However, still there is no validated molecular-targeted therapy for TNBC yet and thus TNBC has the poorest outcome among breast cancer. The RNAi library screening is a powerful tool to identify potential therapeutic targets for diseases with unmet medical needs [28,29,30]. In this study, we performed siRNA-mediated screening of 24 genes, involving in the DNA repair and BRCA1 signaling, to identify the gene whose expression is indispensable for survival of TNBC and identified PLK1 as such a gene because knockdown of PLK1 triggered cell death in the screened TNBC lines, MDA-MB-436 (a mutation in BRCA1), HCC1806 (mutations in both BRCA1/2) and MDA-MB-468 (wild type BRCA1). Although we examined three lines with different BRCA features for the initial screen, no gene other than PLK1 was essential for survival of TNBC when depleted by siRNA. We also examined the effect of siRNA against PLK1 and BI-2536 on the viability of TNBC lines with various genetic signatures because inhibition of PLK1 might be effective in the specific type of TNBC cells [31, 29, 32], but BI-2536 was effective for all the TNBC lines examined. The absence of BRCA function results in failure of HR and thus the repair of undesirable mutations in genomic DNA is dependent on the NHEJ pathway or poly (ADP ribose) polymerase (PARP)-dependent SSB-repairing pathway. Actually, PARP inhibitors such as olaparib and talazoparib are in clinical trials for BRCA-negative TNBC [33, 34]. However, these compounds are supposed to be ineffective for BRCA-positive TNBCs which accounts for more than 50% of TNBC. As we demonstrated here, inhibition of PLK1 efficiently inhibited growth of all TNBC lines examined regardless of BRCA1-genotype, suggesting that inhibition of PLK1 may be a broader therapeutic for TNBC than inhibition of PARP1. In addition, the expression of PLK1 was correlated with cellular proliferation, implying that inhibition of PLK1 may be especially effective for the aggressive type of TNBC. Further in vivo experiments using TNBC animal models will help to show the superiority of activity and broader applicability of PLK1 inhibition.
PLK family members are known to be involved in mitosis and cytoskeletal reorganization in normal and malignant cells [35]. PLKs family consists of 5 members, PLK1, −2, −3, −4 and −5, and PLK1 is the most investigated member of the PLK family which plays an important role in mitosis, especially in G2/M phase [36]. Most importantly, PLK1 drives mitotic events including centrosome disjunction, activation of cyclin and cyclin-dependent kinases, spindle assembly and chromosome separation [37, 38]. PLK1 is also known to phosphorylate BRCA1 to facilitate the recruitment to the DSB site [10]. Clinically, PLK1 is highly overexpressed in many types of cancer including glioma, head and neck cancer, ovarian cancer, prostate cancer, non-small cell lung cancer and breast cancer [13,14,15,16,17,18,19,20]. Consistent with these previous studies, the immunohistochemical analyses we performed here also indicated that PLK1 is highly overexpressed in breast cancer, especially in TNBC, but not in benign tumors or normal mammary gland. In breast cancer, overexpression of PLK1 is reported to correlate with poor prognosis [39]. It is also shown that expression level of PLK1 is particularly higher in TNBC than other subtypes (Fig. 5c, f) [40, 41], and higher tumor grade of TNBC (poorer differentiation and higher genome instability, resulting in aggressive tumor growth) correlates with PLK1 expression [42]. Therefore, PLK1-targeting therapy in TNBC is rational and worth being considered.
BI-2536 is a potent and selective PLK1 inhibitor (IC50 = 0.83 nM), and its robust anti-tumor activity has been confirmed in vitro and in vivo [21]. When given intravenously once or twice per week, BI-2536 is well-tolerated and inhibits growth of diverse xenograft solid tumor models such as colon cancer, pancreas cancer, non-small cell lung cancer, prostate cancer and osteosarcoma [21, 43, 44]. These favorable preclinical data prompted clinical trials of BI-2536 for several types of cancer including breast cancer [45]. Although phase I trials showed that BI-2536 has acceptable safety profiles, its effectiveness was not validated so far in phase II trials yet, thus further sophistication of the design or modification of protocol may be required [45,46,47,48]. The immunohistochemical analysis showed that PLK1 was highly expressed in the TNBC cells at mitotic phase and the expression level was correlated with the rate of Ki67-positivity (Fig. 5d, h), suggesting that the criteria based on the results of Ki67 immunostaining, which are routinely performed for diagnosis of breast cancer, may help to choose appropriate TNBC patient group for treatment with PLK1 inhibitors. For further improvement of clinical outcome by inhibition of PLK1 may be achieved by different types of PLK1 inhibitors. BI-2536 was identified as a ATP- competing compound for PLK family proteins [21], and other types of PLK1 inhibitors have been under clinically investigated [49].
In summary, we identified PLK1 as a therapeutic target for TNBC through siRNA-mediated screen and demonstrated that inhibition of PLK1 by siRNA-mediated knockdown or a chemical inhibitor promoted cell cycle arrest and apoptosis in multiple TNBC lines. We also confirmed that PLK1 was highly overexpressed in the TNBC tissue. These our in vitro data will be further strengthened by a future study using TNBC animal model. The lack of druggable target is the problem for poor prognosis of TNBC, and the effectiveness and unique action profile of PLK1 inhibition, in addition to relatively specific expression to TNBC tissue, suggest that PLK1 is a promising molecular target for TNBC.
References
Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer. 2009;9(Suppl 2):S73–81.
Chacon RD, Costanzo MV. Triple-negative breast cancer. Breast Cancer Res. 2010;12(Suppl 2):S3.
Morris GJ, Naidu S, Topham AK, Guiles F, Xu Y, McCue P, et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and End Results database. Cancer. 2007;110:876–84.
Iqbal J, Ginsburg O, Rochon PA, Sun P, Narod SA. Differences in breast cancer stage at diagnosis and cancer-specific survival by race and ethnicity in the United States. J Am Med Assoc. 2015;313:165–73.
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34.
Sharma P. Biology and management of patients with triple-negative breast cancer. Oncologist. 2016;21:1050–62.
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–6.
Chabalier-Taste C, Brichese L, Racca C, Canitrot Y, Calsou P, Larminat F. Polo-like kinase 1 mediates BRCA1 phosphorylation and recruitment at DNA double-strand breaks. Oncotarget. 2016;7:2269–83.
Detre S, Saclani Jotti G, Dowsett MA. “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–8.
Liu X, Zhou T, Kuriyama R, Erikson RL. Molecular interactions of Polo-like-kinase 1 with the mitotic kinesin-like protein CHO1/MKLP-1. J Cell Sci. 2004;117:3233–46.
Dietzmann K, Kirches E, von B, Jachau K, Mawrin C. Increased human polo-like kinase-1 expression in gliomas. J Neurooncol. 2001;53:1–11.
Donizy P, Halon A, Surowiak P, Kaczorowski M, Kozyra C, Matkowski R. Augmented expression of Polo-like kinase 1 is a strong predictor of shorter cancer-specific overall survival in early stage breast cancer at 15-year follow-up. Oncol Lett. 2016;12:1667–74.
Knecht R, Elez R, Oechler M, Solbach C, Von Ilberg C, Strebhardt K. Prognostic significance of polo-like kinase (PLK) expression in squamous cell carcinomas of the head and neck. Cancer Res. 1999;59:2794–7.
Weichert W, Denkert C, Schmidt M, Gekeler V, Wolf G, Köbel M, et al. Polo-like kinase isoform expression is a prognostic factor in ovarian carcinoma. Br J Cancer. 2004;90:815.
Weichert W, Kristiansen G, Winzer K-J, Schmidt M, Gekeler V, Noske A, et al. Polo-like kinase isoforms in breast cancer: expression patterns and prognostic implications. Virchows Archiv. 2005;446:442–50.
Wilko W, Mathias S, Volker G, Carsten D, Carsten S, Klaus J, et al. Polo‐like kinase 1 is overexpressed in prostate cancer and linked to higher tumor grades. Prostate. 2004;60:240–5.
Wolf G, Elez R, Doermer A, Holtrich U, Ackermann H, Stutte HJ, et al. Prognostic significance of polo-like kinase (PLK) expression in non-small cell lung cancer. Oncogene. 1997;14:543–9.
Zhang Y, Wu Z, Liu D, Wang M, Xiao G, Wang P, et al. Augmented expression of polo-like kinase 1 indicates poor clinical outcome for breast patients: a systematic review and meta-analysis. Oncotarget. 2017;8:57723–32.
Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–22.
Donaldson MM, Tavares AAM, Hagan IM, Nigg EA, Glover DM. The mitotic roles of Polo-like kinase. J Cell Sci. 2001;114:2357–8.
Liu X, Erikson RL. Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci USA. 2003;100:5789–94.
Li J, Wang R, Kong Y, Broman MM, Carlock C, Chen L, et al. Targeting Plk1 to enhance efficacy of olaparib in castration-resistant prostate cancer. Mol Cancer Ther. 2017;16:469–79.
Swift LH, Golsteyn RM. Genotoxic anti-cancer agents and their relationship to dna damage, mitosis, and checkpoint adaptation in proliferating cancer cells. Intl J Mol Sci. 2014;15:3403–31.
Jamdade VS, Sethi N, Mundhe NA, Kumar P, Lahkar M, Sinha N. Therapeutic targets of triple-negative breast cancer: a review. Br J Pharmacol. 2015;172:4228–37.
O’Toole SA, Beith JM, Millar EK, West R, McLean A, Cazet A, et al. Therapeutic targets in triple negative breast cancer. J Clin Pathol. 2013;66:530–42.
MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nature Cell Biol. 2005;7:591.
Hu K, Law JH, Fotovati A, Dunn SE. Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells. Breast Cancer Res. 2012;14:R22.
Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ, et al. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24:182–96.
Lerner RG, Grossauer S, Kadkhodaei B, Meyers I, Sidorov M, Koeck K, et al. Targeting a Plk1-controlled polarity checkpoint in therapy-resistant glioblastoma-propagating cells. Cancer Res. 2015;75:5355–66.
Danovi D, Folarin A, Gogolok S, Ender C, Elbatsh AM, Engstrom PG, et al. A high-content small molecule screen identifies sensitivity of glioblastoma stem cells to inhibition of polo-like kinase 1. PLoS ONE. 2013;8:e77053.
Robson M, Im S-A, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. New Eng. J Med. 2017;377:523–33.
Litton J, Rugo H, Ettl J, Hurvitz S, Gonçalves A, Lee K, et al. Abstract GS6-07: EMBRACA: A phase 3 trial comparing talazoparib, an oral PARP inhibitor, to physician’s choice of therapy in patients with advanced breast cancer and a germline BRCA mutation. Cancer Res. 2018;78:7445.
Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9:643.
Barr FA, Silljé HHW, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429.
Glover DM, Hagan IM, Tavares AA. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 1998;12:3777–87.
Nigg EA. Polo-like kinases: positive regulators of cell division from start to finish. Curr Opin Cell Biol. 1998;10:776–83.
King SI, Purdie CA, Bray SE, Quinlan PR, Jordan LB, Thompson AM, et al. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast Cancer Res. 2012;14:R40.
Maire V, Nemati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013;73:813–23.
Ha GH, Kim DY, Breuer EK, Kim CK. Combination treatment of polo-like kinase 1 and Tankyrase-1 inhibitors enhances anticancer effect in triple-negative breast cancer cells. Anticancer Res. 2018;38:1303–10.
Loddo M, Kingsbury SR, Rashid M, Proctor I, Holt C, Young J, et al. Cell-cycle-phase progression analysis identifies unique phenotypes of major prognostic and predictive significance in breast cancer. Br J Cancer. 2009;100:959–70.
Liu X, Choy E, Harmon D, Yang S, Yang C, Mankin H, et al. Inhibition of polo-like kinase 1 leads to the suppression of osteosarcoma cell growth in vitro and in vivo. Anticancer Drugs. 2011;22:444–53.
Wissing MD, Mendonca J, Kortenhorst MSQ, Kaelber NS, Gonzalez M, Kim E, et al. Targeting prostate cancer cell lines with polo-like kinase 1 inhibitors as a single agent and in combination with histone deacetylase inhibitors. FASEB J. 2013;27:4279–93.
Schöffski P, Blay J-Y, De Greve J, Brain E, Machiels J-P, Soria J-C, et al. Multicentric parallel phase II trial of the polo-like kinase 1 inhibitor BI 2536 in patients with advanced head and neck cancer, breast cancer, ovarian cancer, soft tissue sarcoma and melanoma. The first protocol of the European Organization for Research and Treatment of Cancer (EORTC) Network Of Core Institutes (NOCI). Eur J Cancer. 2010;46:2206–15.
Vose JM, Young A, Friedberg JW, Waller EK, Cheson BD, Trommeshauser D, et al. Phase I dose-escalation trial of bi 2536, a polo-like kinase 1 inhibitor, in relapsed and refractory Non-Hodgkin’s lymphoma. Blood. 2008;112:233.
Frost A, Mross K, Steinbild S, Hedbom S, Unger C, Kaiser R, et al. Phase i study of the Plk1 inhibitor BI 2536 administered intravenously on three consecutive days in advanced solid tumours. Curr Oncol. 2012;19:e28–35.
Mross K, Dittrich C, Aulitzky WE, Strumberg D, Schutte J, Schmid RM, et al. A randomised phase II trial of the Polo-like kinase inhibitor BI 2536 in chemo-naïve patients with unresectable exocrine adenocarcinoma of the pancreas—a study within the Central European Society Anticancer Drug Research (CESAR) collaborative network. Br J Cancer. 2012;107:280–6.
Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–66.
Acknowledgements
We appreciate the members of department of Molecular Pathology, Tokyo Medical University and department of Breast Oncology and Surgery, Tokyo Medical University Hospital for their supports and helpful suggestions.
Funding
This work was partly supported by the grants from the JSPS KAKENHI Grant number (17K16520 to A. U., 16H06247 to K. K., 17H04067 to M. Kuroda). This work was also supported in part by Japan Agency for Medical Research and Development (AMED), Strategic Research Foundation Grant-aided Project for Private Universities from the Ministry of Education, Culture, Sports, Science and Technology of Japan and Project for Development of Innovative Research on Cancer Therapeutics.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
We obtained the written forms of informed consent from all the patients regarding the use of their clinical data, following the ethical guideline of Tokyo Medical University. The data from patients were analyzed anonymously.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ueda, A., Oikawa, K., Fujita, K. et al. Therapeutic potential of PLK1 inhibition in triple-negative breast cancer. Lab Invest 99, 1275–1286 (2019). https://doi.org/10.1038/s41374-019-0247-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41374-019-0247-4
This article is cited by
-
Therapeutic Potential of PLK1, KIF4A, CDCA5, UBE2C, CDT1, SKA3, AURKB, and PTTG1 Genes in Triple-Negative Breast Cancer: Correlating Their Expression with Sensitivity to GSK 461364 and IKK 16 Drugs
Biochemical Genetics (2025)
-
CHFR expression and sensitivity of breast cancer cells to antimitotic agents in vitro and in the I-SPY trial
npj Breast Cancer (2025)
-
Exploring Prognostic Gene Factors in Breast Cancer via Machine Learning
Biochemical Genetics (2024)
-
Exploration of subtype-specific perturbations in breast cancer
Journal of Proteins and Proteomics (2024)
-
HN1L/AP-2γ/PLK1 signaling drives tumor progression and chemotherapy resistance in esophageal squamous cell carcinoma
Cell Death & Disease (2022)