Abstract
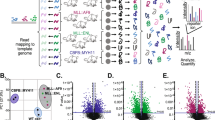
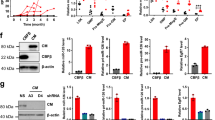
We investigated the clinical and functional role of the miR-106a-363 cluster in adult acute myeloid leukemia (AML). LAML miRNA-Seq TCGA analyses revealed that high expression of miR-106a-363 cluster members was associated with inferior survival, and miR-106a-5p and miR-20b-5p levels were significantly elevated in patients with adverse risk AML. Overexpression of the miR-106a-363 cluster and its individual members in a murine AML model significantly accelerated leukemogenesis. Proteomics analysis of leukemic bone marrow cells from these models emphasized the deregulation of proteins involved in intracellular transport, protein complex organization and mitochondrial function, driven predominantly by miR-106a-5p. These molecular alterations suggested mitochondrial activation as a potential mechanism for the observed increase in leukemogenicity. High-resolution respirometry and STED microscopy confirmed that miR-106a-5p enhances mitochondrial respiratory activity and increases mitochondrial volume. These findings demonstrate that the miR-106a-363 cluster, and particularly miR-106a-5p, contribute to AML progression through modulation of mitochondrial function and deregulation of mitochondria-coordinated pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood. 2019;133:7–17.
Thol F, Dohner H, Ganser A. How I treat refractory and relapsed acute myeloid leukemia. Blood. 2024;143:11–20.
Garciaz S, Hospital MA, Alary AS, Saillard C, Hicheri Y, Mohty B, et al. Azacitidine Plus Venetoclax for the Treatment of Relapsed and Newly Diagnosed Acute Myeloid Leukemia Patients. Cancers. 2022;14:2025.
Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med. 2018;24:1859–66.
Shivarov V, Dolnik A, Lang KM, Kronke J, Kuchenbauer F, Paschka P, et al. MicroRNA expression-based outcome prediction in acute myeloid leukemia: novel insights through cross-platform integrative analyses. Haematologica. 2016;101:e454–e6.
Abdellateif MS, Hassan NM, Kamel MM, El-Meligui YM. Bone marrow microRNA-34a is a good indicator for response to treatment in acute myeloid leukemia. Oncol Res. 2024;32:577–84.
Bhayadia R, Krowiorz K, Haetscher N, Jammal R, Emmrich S, Obulkasim A, et al. Endogenous Tumor Suppressor microRNA-193b: Therapeutic and Prognostic Value in Acute Myeloid Leukemia. J Clin Oncol. 2018;36:1007–16.
Krowiorz K, Ruschmann J, Lai C, Ngom M, Maetzig T, Martins V, et al. MiR-139-5p is a potent tumor suppressor in adult acute myeloid leukemia. Blood Cancer J. 2016;6:e508.
Petriv OI, Kuchenbauer F, Delaney AD, Lecault V, White A, Kent D, et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci USA. 2010;107:15443–8.
Itkin T, Kumari A, Schneider E, Gur-Cohen S, Ludwig C, Brooks R, et al. MicroRNA-155 promotes G-CSF-induced mobilization of murine hematopoietic stem and progenitor cells via propagation of CXCL12 signaling. Leukemia. 2017;31:1247–50.
Gentner B, Pochert N, Rouhi A, Boccalatte F, Plati T, Berg T, et al. MicroRNA-223 dose levels fine tune proliferation and differentiation in human cord blood progenitors and acute myeloid leukemia. Exp Hematol. 2015;43:858–68.e7.
Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58.
Lechman ER, Gentner B, Ng SW, Schoof EM, van Galen P, Kennedy JA, et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell. 2016;29:214–28.
Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–706.
Khuu C, Utheim TP, Sehic A. The Three Paralogous MicroRNA Clusters in Development and Disease, miR-17-92, miR-106a-363, and miR-106b-25. Science. 2016;2016:1379643.
Li M, Zhou Y, Xia T, Zhou X, Huang Z, Zhang H, et al. Circulating microRNAs from the miR-106a–363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res Treat. 2018;170:257–70.
You F, Luan H, Sun D, Cui T, Ding P, Tang H, et al. miRNA-106a Promotes Breast Cancer Cell Proliferation, Clonogenicity, Migration, and Invasion Through Inhibiting Apoptosis and Chemosensitivity. DNA Cell Biol. 2019;38:198–207.
Zhou Y, Zhang Y, Li Y, Liu L, Li Z, Liu Y, et al. MicroRNA-106a-5p promotes the proliferation, autophagy and migration of lung adenocarcinoma cells by targeting LKB1/AMPK. Exp Ther Med. 2021;22:1422.
Drobna M, Szarzyńska B, Jaksik R, Sędek Ł, Kuchmiy A, Taghon T, et al. hsa-miR-20b-5p and hsa-miR-363-3p Affect Expression of PTEN and BIM Tumor Suppressor Genes and Modulate Survival of T-ALL Cells In Vitro. Cells. 2020;9:1137.
Lim EL, Trinh DL, Ries RE, Wang J, Gerbing RB, Ma Y, et al. MicroRNA Expression-Based Model Indicates Event-Free Survival in Pediatric Acute Myeloid Leukemia. J Clin Oncol. 2017;35:3964–77.
Schneider E, Staffas A, Rohner L, Malmberg ED, Ashouri A, Krowiorz K, et al. MicroRNA-155 is a direct target of Meis1, but not a driver in acute myeloid leukemia. Haematologica. 2017;103:246–55.
Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–25.
Maetzig T, Brugman MH, Bartels S, Heinz N, Kustikova OS, Modlich U, et al. Polyclonal fluctuation of lentiviral vector-transduced and expanded murine hematopoietic stem cells. Blood. 2011;117:3053–64.
Kuchenbauer F, Mah SM, Heuser M, McPherson A, Ruschmann J, Rouhi A, et al. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood. 2011;118:3350–8.
Schaefer PM, von Einem B, Walther P, Calzia E, von Arnim CA. Metabolic Characterization of Intact Cells Reveals Intracellular Amyloid Beta but Not Its Precursor Protein to Reduce Mitochondrial Respiration. PLoS One. 2016;11:e0168157.
Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74.
Bolouri H, Farrar JE, Triche T, Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24:103–12.
Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Hothorn T, Lausen B. On the exact distribution of maximally selected rank statistics. Computational Stat Data Anal. 2001;43:121–37.
Schneider E, Pochert N, Ruess C, MacPhee L, Escano L, Miller C, et al. MicroRNA-708 is a novel regulator of the Hoxa9 program in myeloid cells. Leukemia. 2020;34:1253–65.
Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72.
Stubbins RJ, Francis A, Kuchenbauer F, Sanford D. Management of Acute Myeloid Leukemia: A Review for General Practitioners in Oncology. Curr Oncol. 2022;29:6245–59.
Stubbins RJ, Maksakova IA, Sanford DS, Rouhi A, Kuchenbauer F. Mitochondrial metabolism: powering new directions in acute myeloid leukemia. Leuk Lymphoma. 2021;62:2331–41.
Langer C, Rucker FG, Buske C, Dohner H, Kuchenbauer F. Targeted therapies through microRNAs: pulp or fiction? Ther Adv Hematol. 2012;3:97–104.
Argiropoulos B, Palmqvist L, Yung E, Kuchenbauer F, Heuser M, Sly LM, et al. Linkage of Meis1 leukemogenic activity to multiple downstream effectors including Trib2 and Ccl3. Exp Hematol. 2008;36:845–59.
Argiropoulos B, Yung E, Xiang P, Lo CY, Kuchenbauer F, Palmqvist L, et al. Linkage of the potent leukemogenic activity of Meis1 to cell-cycle entry and transcriptional regulation of cyclin D3. Blood. 2010;115:4071–82.
Wang X, Zhang X, Cao K, Zeng M, Fu X, Zheng A, et al. Cardiac disruption of SDHAF4-mediated mitochondrial complex II assembly promotes dilated cardiomyopathy. Nat Commun. 2022;13:3947.
Li K, Song K, Hou Y, Tian Y, Wang H, Sun L, et al. AHNAK Contributes to Hepatocellular Carcinoma Growth by Interacting with IGF-1R. Molecules. 2022;27:8680.
Jahic A, Iljazovic E, Hasic S, Arnautovic AC, Sabitovic D, Mesanovic S, et al. Prognostic Parameters of Acute Myeloid Leukaemia at Presentation. Med Arch. 2017;71:20–4.
Sasca D, Szybinski J, Schüler A, Shah V, Heidelberger J, Haehnel PS, et al. NCAM1 (CD56) promotes leukemogenesis and confers drug resistance in AML. Blood. 2019;133:2305–19.
Convertini P, Todisco S, De Santis F, Pappalardo I, Iacobazzi D, Castiglione Morelli MA, et al. Transcriptional Regulation Factors of the Human Mitochondrial Aspartate/Glutamate Carrier Gene, Isoform 2 (SLC25A13): USF1 as Basal Factor and FOXA2 as Activator in Liver Cells. Int J Mol Sci. 2019;20:1888.
Owusu-Ansah M, Guptan N, Alindogan D, Morizono M, Caldovic L. NAGS, CPS1, and SLC25A13 (Citrin) at the Crossroads of Arginine and Pyrimidines Metabolism in Tumor Cells. Int J Mol Sci. 2023;24:6754.
Nappi L, Thi M, Lum A, Huntsman D, Eigl BJ, Martin C, et al. Developing a Highly Specific Biomarker for Germ Cell Malignancies: Plasma miR371 Expression Across the Germ Cell Malignancy Spectrum. J Clin Oncol. 2019;37:3090–8.
Acknowledgements
We thank Edith Schneider for providing the Hoxa9/Meis1 cell line; the staff at Sahlgrenska University Hospital, especially Giti Shah Barkhordar, for assistance with biobanked samples; the Flow Core at BCCRC for cell sorting; Leonard Foster and Jason Rogalski at UBC’s Proteomics Core Facility for help with OCIAML3 proteomics; and Keng C. Chou, Department of Chemistry, UBC, for access to the STED microscope.
Funding
FK was supported by grants from Deutsche Krebshilfe grant 109420 (Max-Eder program); fellowship 2010/04 by the European Hematology Association; and by the Deutsche Forschungsgemeinschaft (DFG) (SFB 1074, project A5) and the Wilhelm Sander Stiftung (2015.153.1), Leukemia Lymphoma Society of Canada and Michael Smith Health Research BC. AR was supported by the DFG (SFB 1074, project A5) as well as the gender equality program by the DFG (SFB 1074, project Z2), a fellowship from the Canadian Institutes of Health Research, the Baustein Startförderung Program of the Medical Faculty, Ulm University and Faculty of Medicine New Faculty Research Award. LP was supported by grants from the Swedish Cancer Society (CAN2014/525), the Swedish Childhood Cancer Foundation (PR2014-0125), and Västra Götalandsregionen (ALFGBG-431881). NS, KK and SG were supported by the International Graduate School of Molecular Medicine, Ulm University, Germany. MRG and LA were supported by grants from the Canadian Institutes of Health Research (PJT-166196) and the Natural Sciences and Engineering Research Council of Canada (AWD-010517).
Author information
Authors and Affiliations
Contributions
NS and IAM performed experimental work, analyses, and prepared the manuscript. LE provided bioinformatic analysis. LA and MRG performed imaging. JB, LM, AC, DK, MY, KK, SG, NP, CR, RR, TM and SF provided experimental support. EC performed respirometry support. LP and SW performed proteomics. LF performed FACS and human assays. AR and FK conceived and designed the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. Animal experiments with H9M and H9M cells were approved by the state government of Tübingen, Germany. Animal experiments with OCIAML3 cells were performed according to animal research protocol # A19-0320 approved by the University of British Columbia Animal Care and Use Committee. All patients provided informed consent under protocols approved by the Regional Ethical Review Board in Gothenburg.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sperb, N., Maksakova, I.A., Escano, L. et al. The proto-oncogenic miR-106a-363 cluster enhances adverse risk acute myeloid leukemia through mitochondrial activation. Leukemia 39, 1090–1101 (2025). https://doi.org/10.1038/s41375-025-02558-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02558-x