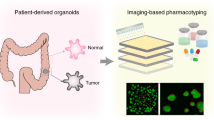
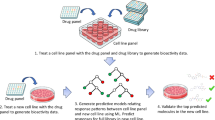
Abstract
Primary central nervous system lymphoma (PCNSL) exhibits substantial intratumoural and intertumoural heterogeneity, complicating the development of effective treatment methods. Existing in vitro models fail to simulate the cellular and mutational diversity of native tumours and require prolonged generation times. Therefore, we developed a culture method for patient-derived PCNSL organoids (CLOs) and evaluated the organoids through extensive molecular characterisation, histopathological analysis, single-nucleus RNA sequencing, bulk RNA sequencing and whole-exome sequencing. These CLOs accurately mimicked the histological attributes, gene expression landscapes and mutational profiles of their original tumours. Single-nucleus RNA sequencing also revealed that CLOs maintained cell-type heterogeneity and the molecular signatures of their original tumours. CLOs were generated within 2 weeks, demonstrating rapid development and reliability. Therapeutic profiling was performed on three selected CLOs treated with four standard drugs. The CLOs exhibited specific sensitivity to methotrexate, and resistance to dexamethasone, ibrutinib and rituximab, suggesting that CLOs may be valuable tools for reflecting drug sensitivities. Taken together, these results emphasise that CLOs effectively emulate the key characteristics of PCNSL, increasing the understanding of the genetic landscape of this complex disease. CLOs provide a rapid and reliable platform for exploring individualised treatment strategies, potentially accelerating the transition of research findings to clinical practice.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The WES, RNA-seq, and snRNA-seq raw sequence data have been deposited in the Genome Sequence Archive [62] of the National Genomics Data Centre [63], China National Centre for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA005755; HRA009621; https://ngdc.cncb.ac.cn/gsa-human). Any additional information required to reanalyse the data is available from the lead contact upon request.
References
Ferreri AJM, Calimeri T, Cwynarski K, Dietrich J, Grommes C, Hoang-Xuan K, et al. Primary central nervous system lymphoma. Nat Rev Dis Prim. 2023;9:29.
Chen T, Liu Y, Wang Y, Chang Q, Wu J, Wang Z, et al. Evidence-based expert consensus on the management of primary central nervous system lymphoma in China. J Hematol Oncol. 2022;15:136.
Schaff LR, Grommes C. Primary central nervous system lymphoma. Blood. 2022;140:971–9.
DeAngelis LM, Yahalom J, Thaler HT, Kher U. Combined modality therapy for primary CNS lymphoma. J Clin Oncol. 1992;10:635–43.
Rubenstein JL, Hsi ED, Johnson JL, Jung S-H, Nakashima MO, Grant B, et al. Intensive chemotherapy and immunotherapy in patients with newly diagnosed primary CNS lymphoma: CALGB 50202 (Alliance 50202). J Clin Oncol. 2013;31:3061–8.
Glass J, Gruber ML, Cher L, Hochberg FH. Preirradiation methotrexate chemotherapy of primary central nervous system lymphoma: long-term outcome. J Neurosurg. 1994;81:188–95.
Mulazzani M, Fräßle SP, von Mücke-Heim I, Langer S, Zhou X, Ishikawa-Ankerhold H, et al. Long-term in vivo microscopy of CAR T cell dynamics during eradication of CNS lymphoma in mice. Proc Natl Acad Sci USA. 2019;116:24275–84.
Qiu Y, Li Z, Pouzoulet F, Vishnu P, Copland JA, Knutson KL, et al. Immune checkpoint inhibition by anti-PDCD1 (anti-PD1) monoclonal antibody has significant therapeutic activity against central nervous system lymphoma in an immunocompetent preclinical model. Br J Haematol. 2018;183:674–8.
Takashima Y, Hayano A, Yamanaka R. Metabolome analysis reveals excessive glycolysis via PI3K/AKT/mTOR and RAS/MAPK signaling in methotrexate-resistant primary CNS lymphoma-derived cells. Clin Cancer Res. 2020;26:2754–66.
Tateishi K, Miyake Y, Kawazu M, Sasaki N, Nakamura T, Sasame J, et al. A hyperactive RelA/p65-hexokinase 2 signaling axis drives primary central nervous system lymphoma. Cancer Res. 2020;80:5330–43.
Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403.
Isbell LK, Tschuch C, Doostkam S, Waldeck S, Andrieux G, Shoumariyeh K, et al. Patient-derived xenograft mouse models to investigate tropism to the central nervous system and retina of primary and secondary central nervous system lymphoma. Neuropathol Appl Neurobiol. 2023;49:e12899.
Liu N, Jiang C, Yao X, Fang M, Qiao X, Zhu L, et al. Single-cell landscape of primary central nervous system diffuse large B-cell lymphoma. Cell Discov. 2023;9:55.
Xia Y, Sun T, Li G, Li M, Wang D, Su X, et al. Spatial single cell analysis of tumor microenvironment remodeling pattern in primary central nervous system lymphoma. Leukemia. 2023;37:1499–510.
Hernández-Verdin I, Kirasic E, Wienand K, Mokhtari K, Eimer S, Loiseau H, et al. Molecular and clinical diversity in primary central nervous system lymphoma. Ann Oncol. 2023;34:186–99.
Fukumura K, Kawazu M, Kojima S, Ueno T, Sai E, Soda M, et al. Genomic characterization of primary central nervous system lymphoma. Acta Neuropathol. 2016;131:865–75.
Braggio E, Van Wier S, Ojha J, McPhail E, Asmann YW, Egan J, et al. Genome-wide analysis uncovers novel recurrent alterations in primary central nervous system lymphomas. Clin Cancer Res. 2015;21:3986–94.
Mulazzani M, Huber M, Borchard S, Langer S, Angele B, Schuh E, et al. APRIL and BAFF: novel biomarkers for central nervous system lymphoma. J Hematol Oncol. 2019;12:102.
Corsini NS, Knoblich JA. Human organoids: New strategies and methods for analyzing human development and disease. Cell. 2022;185:2756–69.
Xu H, Jiao D, Liu A, Wu K. Tumor organoids: applications in cancer modeling and potentials in precision medicine. J Hematol Oncol. 2022;15:58.
Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204.e22.
Kawasaki K, Toshimitsu K, Matano M, Fujita M, Fujii M, Togasaki K, et al. An organoid biobank of neuroendocrine neoplasms enables genotype-phenotype mapping. Cell. 2020;183:1420–1435.e21.
van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–45.
Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90.
Li S, Xia Z, Cao J, Zhang J, Chen B, Chen T, et al. Proposed new prognostic model using the systemic immune-inflammation index for primary central nervous system lymphoma: A prospective-retrospective multicohort analysis. Front Immunol. 2022;13:1039862.
Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82.
Li Q, Ma J, Ma Y, Lin Z, Kang H, Chen B. Improvement of outcomes of an escalated high-dose methotrexate-based regimen for patients with newly diagnosed primary central nervous system lymphoma: a real-world cohort study. Cancer Manag Res. 2021;13:6115–22.
Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125.
Zylber-Katz E, Gomori JM, Schwartz A, Lossos A, Bokstein F, Siegal T. Pharmacokinetics of methotrexate in cerebrospinal fluid and serum after osmotic blood-brain barrier disruption in patients with brain lymphoma. Clin Pharm Ther. 2000;67:631–41.
Lionakis MS, Dunleavy K, Roschewski M, Widemann BC, Butman JA, Schmitz R, et al. Inhibition of B cell receptor signaling by ibrutinib in primary CNS lymphoma. Cancer Cell. 2017;31:833–843.e5.
Jackson RK, Liebich M, Berry P, Errington J, Liu J, Parker C, et al. Impact of dose and duration of therapy on dexamethasone pharmacokinetics in childhood acute lymphoblastic leukaemia-a report from the UKALL 2011 trial. Eur J Cancer. 2019;120:75–85.
Yonezawa A, Otani Y, Kitano T, Mori M, Masui S, Isomoto Y, et al. Concentration and glycoform of rituximab in plasma of patients with B cell Non-Hodgkin’s lymphoma. Pharm Res. 2019;36:82.
Xia W, Hu B, Li H, Shi W, Tang Y, Yu Y, et al. Deep learning for automatic differential diagnosis of primary central nervous system lymphoma and glioblastoma: multi-parametric magnetic resonance imaging based convolutional neural network model. J Magn Reson Imaging. 2021;54:880–7.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
li shengjie, Li D, Xia Z, Wu J, Ren J, Li Y et al. Genomic landscape and molecular subtypes of primary central nervous system lymphoma. medRxiv 2024; 2024.10.22.24315961.
Wang Y, Bae T, Thorpe J, Sherman MA, Jones AG, Cho S, et al. Comprehensive identification of somatic nucleotide variants in human brain tissue. Genome Biol. 2021;22:92.
Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–59.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8:469–77.
Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Kanehisa M. Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–14.
Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36:421–7.
McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–97.
Radke J, Ishaque N, Koll R, Gu Z, Schumann E, Sieverling L, et al. The genomic and transcriptional landscape of primary central nervous system lymphoma. Nat Commun. 2022;13:2558.
Ji AL, Rubin AJ, Thrane K, Jiang S, Reynolds DL, Meyers RM, et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. 2020;182:497–514.e22.
Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–4.
Abbasi J. Patient-derived organoids predict cancer treatment response. JAMA. 2018;319:1427.
Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–35.
Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–6.
Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020;26:17–26.e6.
Pina-Oviedo S, Bellamy WT, Gokden M. Analysis of primary central nervous system large B-cell lymphoma in the era of high-grade B-cell lymphoma: detection of two cases with MYC and BCL6 rearrangements in a cohort of 12 cases. Ann Diagn Pathol. 2020;48:151610.
Ting C-Y, Chang K-M, Kuan J-W, Sathar J, Chew L-P, Wong O-LJ, et al. Clinical significance of BCL2, C-MYC, and BCL6 genetic abnormalities, Epstein-Barr virus infection, CD5 protein expression, germinal center B cell/non-germinal center B-cell subtypes, co-expression of MYC/BCL2 proteins and co-expression of MYC/BCL2/BCL6 proteins in diffuse large B-cell lymphoma: a clinical and pathological correlation study of 120 patients. Int J Med Sci. 2019;16:556–66.
Chapuy B, Roemer MGM, Stewart C, Tan Y, Abo RP, Zhang L, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood. 2016;127:869–81.
Los-de Vries GT, Stathi P, Rutkens R, Hijmering NJ, Luijks JACW, Groenen PJTA, et al. Large B-cell Lymphomas of Immune-Privileged Sites Relapse via Parallel Clonal Evolution from a Common Progenitor B Cell. Cancer Res. 2023;83:1917–27.
Brunn A, Nagel I, Montesinos-Rongen M, Klapper W, Vater I, Paulus W, et al. Frequent triple-hit expression of MYC, BCL2, and BCL6 in primary lymphoma of the central nervous system and absence of a favorable MYC(low)BCL2 (low) subgroup may underlie the inferior prognosis as compared to systemic diffuse large B cell lymphomas. Acta Neuropathol. 2013;126:603–5.
Montesinos-Rongen M, Akasaka T, Zühlke-Jenisch R, Schaller C, Van Roost D, Wiestler OD, et al. Molecular characterization of BCL6 breakpoints in primary diffuse large B-cell lymphomas of the central nervous system identifies GAPD as novel translocation partner. Brain Pathol. 2003;13:534–8.
Chen T, Chen X, Zhang S, Zhu J, Tang B, Wang A, et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteom Bioinforma. 2021;19:578–83.
CNCB-NGDC Members and Partners. Database resources of the national genomics data center, China national center for bioinformation in 2022. Nucleic Acids Res. 2022;50:D27–D38.
Acknowledgements
We extend their gratitude to Yiyin Zhang from KingMed Diagnostics in Shanghai for his invaluable assistance with data processing. They also wish to express their sincere thanks to the colleagues at Huashan Hospital of Fudan University and Fudan University Shanghai Cancer Centre for their crucial support during the specimen collection process. Appreciation is also extended to ShanghaiTech University for providing the research platform.
Funding
The study was funded by the National Natural Science Foundation of China (82302582), Shanghai Municipal Health Commission Project (20224Y0317), Youth Medical Talents—Clinical Laboratory Practitioner Programme (2022-65), and Industry-University-Research Innovation Fund for Chinese Universities (2023JQ006).
Author information
Authors and Affiliations
Contributions
WJC, and SJL conceptualized and designed this study. SJL, YZL, JNW, and JR performed most experiments. ZGX, CXL, and SJL performed partial experiments. SJL, CXL, and JR finished the acquisition and analysis of data. SJL, YZL, and JNW prepared figures, performed the statistical analysis, and wrote original draft. WJC, and SJL reviewed and supervised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, S., Ren, J., Wu, J. et al. Establishment and molecular characterisation of patient-derived organoids for primary central nervous system lymphoma. Leukemia 39, 1169–1183 (2025). https://doi.org/10.1038/s41375-025-02562-1
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02562-1