Abstract
Unbiased kinome-wide CRISPR screening identified DYRK1A as a potential therapeutic target in KMT2A-rearranged (KMT2A-R) B-acute lymphoblastic leukemia (ALL). Mechanistically, we demonstrate that DYRK1A is regulated by the KMT2A fusion protein and affects cell proliferation by regulating MYC expression and ERK phosphorylation. We further observed that pharmacologic DYRK1A inhibition markedly reduced human KMT2A-R ALL cell proliferation in vitro and potently decreased leukemia proliferation in vivo in drug-treated patient-derived xenograft mouse models. DYRK1A inhibition induced expression of the proapoptotic factor BIM and reduced the expression of BCL-XL, consequently sensitizing KMT2A-R ALL cells to BCL2 inhibition. Dual inhibition of DYRK1A and BCL2 synergistically decreased KMT2A-R ALL cell survival in vitro and reduced leukemic burden in mice. Taken together, our data establishes DYRK1A as a novel therapeutic target in KMT2A-R ALL and credential dual inhibition of DYRK1A and BCL2 as an effective translational therapeutic strategy for this high-risk ALL subtype.

Similar content being viewed by others
Introduction
Rearrangements in KMT2A (formerly MLL) occur in ~75% of acute lymphoblastic leukemia (ALL) cases in infants less than 12 months of age, 1–2% of older children, and 10% of adults [1,2,3]. KMT2A-rearranged (KMT2A-R) ALL is a high-risk disease associated with frequent chemoresistance and poor clinical outcomes in most patients with a survival rate of <75% in children and <35% in infants and adults. We thus focused in these studies on identifying new biological drivers and therapeutic vulnerabilities in this high-risk ALL subtype.
The use of small molecule inhibitors targeting activated and disease-driving kinases has demonstrated significant clinical success in lymphoid malignancies. For instance, targeting the BCR::ABL1 oncogene in patients with Philadelphia chromosome-positive (Ph+) ALL has significantly improved their event-free and overall survival [4, 5]. Similarly, BTK inhibition with ibrutinib significantly improved the survival of patients with chronic lymphocytic leukemia [6]. Therapeutic targeting of FLT3 and other non-kinase dependencies in KMT2A-R ALL, including DOT1L and menin, have demonstrated robust activity in preclinical studies and have been translated to the clinic [7,8,9,10,11]. However, incomplete therapeutic activity and/or failure to improve clinical outcomes have been observed with several of these drugs [12,13,14]. The rapid development of resistance mutations in inhibitor-treated patients despite initial response has also been reported [15], highlighting a persistent scientific knowledge gap and unmet medical need for better therapies.
Given the success of various small molecule inhibitors in specific genetic subtypes of ALL and acute myeloid leukemia (AML) and the improved specificity of next-generation kinase inhibitors [16,17,18,19,20], we performed a kinome-wide domain-specific CRISPR screen in 3 high-risk subtypes of ALL and found that dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) is specifically required for the survival of KMT2A-R ALL. DYRK1A is a dual-specificity kinase that autophosphorylates a tyrosine residue for activation [21], but otherwise primarily targets serine/threonine residues on substrates for phosphorylation. The protein has a proposed, but still incompletely defined, role in cell cycle regulation that may vary depending on cell type. Significant research has characterized the role of DYRK1A in neuronal development, and a smaller body of work has also implicated its role in normal B cell differentiation and in Down syndrome (DS)-associated ALL and CRLF2-R Ph-like ALL [22,23,24,25].
Herein, we demonstrate an unanticipated direct regulation of DYRK1A via oncogenic KMT2A rearrangements in ALL and a specific requirement of DYRK1A for leukemia proliferation. We also report that DYRK1A is required to suppress the proapoptotic molecule BIM and that DYRK1A inhibition consequently renders KMT2A-R ALL cells sensitive to BCL2 inhibition. Overall, these data offer new insights into the mechanisms underlying KMT2A fusion-driven leukemogenesis and may have translational therapeutic implications. This is particularly relevant given that early-stage DYRK1A inhibitors are currently under clinical investigation for osteoarthritis (NCT05603754) and atopic dermatitis (NCT05382819) [26, 27].
Materials and methods
Primary human ALL specimens and cell lines
Diagnostic bone marrow specimens from children and adults with ALL were obtained via informed consent on Institutional Review Board-approved research protocols of the Children’s Hospital of Philadelphia (CHOP) and University of Pennsylvania (UPenn) in accordance with the Declaration of Helsinki (Table S1) as previously described [28, 29]. Human KMT2A-R and non-KMT2A-R ALL cell lines (Table S2) were obtained via collaborators or purchased from the DSMZ biorepository (Braunschweig, Germany), validated by short tandem repeat analysis, and confirmed as Mycoplasma-free every 6 months [30,31,32,33].
Human ALL cells harvested from murine spleens of some PDX models were cultured on OP9 stroma in Minimum Essential Medium (MEMα; Life Technologies) with GlutaMAX containing 20% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 1 mM sodium pyruvate. Cell lines were cultured in Roswell Park Memorial Institute medium ([RPMI] Life Technologies; Carlsbad, CA) with GlutaMAX containing 10 or 20% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin (hereafter referred to as ‘B cell medium’) at 37 °C in a humidified incubator with 5% CO2 and maintained in culture for fewer than 3 months to minimize infectious contamination and genetic drift.
Retroviral and lentiviral transduction
Transfections of 293FT cells with retroviral and lentiviral constructs were performed using lipofectamine 2000 with Opti-MEM media (Invitrogen; Carlsbad, CA). Viral supernatants were produced by co-transfecting 293FT cells with the viral gag-pol and packaging vectors together with the Cas9 or sgRNA constructs (Table S3). Cultivation was performed in high glucose Dulbecco’s modified Eagle’s medium ([DMEM] Invitrogen) with GlutaMAX containing 10% fetal bovine serum, 100 IU/mL penicillin, 100 μg/mL streptomycin, 25 mM HEPES, 1 mM sodium pyruvate, and 0.1 mM non-essential amino acids. Regular media were replaced after 16 h by growth media containing 3 mM caffeine. After 24 h, the viral supernatant was harvested and filtered through a 0.45 μm filter. Retroviral transductions were performed by loading the viral supernatant on 50 μg/mL retronectin-coated (Takara) non-tissue culture-treated 6-well plates prior to centrifugation (2000 × g, 90 min at 32 °C) two times. Subsequently, 1–2 × 106 cells were loaded and transduced per well by centrifugation at 600 × g for 30 min and maintained for 72 h at 37 °C with 5% CO2 prior to transfer into culture flasks. Lentiviral transductions were performed by loading viral supernatant on 50 μg/mL retronectin-coated non-tissue culture-treated 6-well plates with 1–2 × 106 cells. Cells were centrifuged ×30 min at 600 × g and maintained for 16 h at 37 °C with 5% CO2 prior to replacing media with fresh RPMI and GlutaMAX containing 20% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin. Cells were then cultured at 37 °C with 5% CO2 prior to analysis.
Results
KMT2A-R ALL is dependent on DYRK1A expression for survival
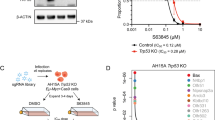
We performed a domain-specific kinome-wide CRISPR screen in genetically diverse high-risk ALL cell lines SEM (KMT2A::AFF1) and HAL-01 (TCF3::HLF) and TVA-1 (ETV6::ABL1) cells previously immortalized from a Ph-like ALL patient-derived xenograft (PDX) model [30] to identify novel druggable targets in high-risk ALL (Fig. 1A). As expected, we identified dependencies upon (1) FLT3 in the FLT3 wild-type overexpressing SEM cell line, (2) the checkpoint inhibitors ATR and CHK1 in the HAL-01 ALL cell line, and (3) ABL1 in TVA-1 cells, validating the specificity of our screens (Fig. 1B and Supplementary Fig. 1A). Unexpectedly, sgRNAs targeting DYRK1A were reduced in SEM cells compared to the other tested samples (Fig. 1C). DYRK1A is not a common essential gene validated via the Cancer Dependency MAP website, indicating that targeting DYRK1A may represent a target with a unique therapeutic window (Supplementary Fig. 1B). Five different DYRK family members are known, but only DYRK1A was required for KMT2A-AFF1 cell survival (Fig. 1D). The importance of DYRK1A was validated in a second CRISPR screen consisting of 14 kinases that we identified in our initial screen (Fig. 1E). Taken together, our screening results demonstrate a unique dependency of KMT2A::AFF1 cells upon DYRK1A.
A Overview of the performed domain-specific kinome-wide library screen. SEM (KMT2A::AFF1), HAL-01 (TCF3::HLF), and TVA-1 (ETV6::ABL1) cells were virally transduced with a lentiviral Cas9-Puro construct. After puromycin selection, the cells were transduced with a GFP-tagged domain-specific kinome-wide CRISPR library consisting of 550 kinases, 100 negative controls, and 50 positive controls (6 guide RNAs (sgRNA) per kinase). Cells were collected at 2 time points for sequencing; 3 days after the library transduction, which served as input control, and 3 weeks after the transduction which served as the experimental readout. B Shown is the fold depletion of the individual sgRNAs in the tested samples. Highlighted are known positive controls for each individual ALL subtype. C Shown is the fold depletion of DYRK1A among the different ALL subtypes. D Shown is the fold depletion of each DYRK family member in SEM cells. E Cas9-puro-selected SEM cells were lentivirally transduced with a smaller sgRNA-GFP library. The library included two sgRNAs targeting one of 14 previously identified targets including 21 negative controls. Data are represented as individual values with mean ± SEM bars. *P < 0.05; **P < 0.01; ***P < 0.001 by t-test.
KMT2A directly binds to the DYRK1A promoter and regulates its expression
Given this specific identification of DYRK1A as a potential driver in KMT2A-R ALL via our CRISPR screen, we then hypothesized that oncogenic KMT2A rearrangements may directly regulate DYRK1A expression. Concordant with our hypothesis, analysis of previously published ChIP-Seq data of KMT2A::AFF1 ALL cells demonstrated that both KMT2A and AFF1 directly bind to the promoter region of DYRK1A in ALL cell lines SEM and RS4;11 (Fig. 2A) [34, 35]. For the ChIP-Seq experiment, antibodies binding to the N-terminal region of KMT2A and the C-terminal region of AFF1 were used. Given that KMT2A-R ALL is often a monoallelic balanced translocation with wild-type KMT2A and AFF1 still present in the cells, we then analyzed a ChIP-Seq data set that used human KMT2A-R leukemia cells generated via overexpression of a KMT2A-Aff1-flag construct [36]. Using a FLAG antibody, we identified oncogene-specific binding partners and confirmed that KMT2A-Aff1 directly binds to the DYRK1A promoter region, validating that KMT2A fusions can interact directly with the DYRK1A promoter (Fig. 2B). The binding of the KMT2A fusion construct to MYC is shown as a control.
A ChIP-Seq (Chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) tracks on the DYRK1A promoter region using antibodies specific for the KMT2A N- and the AFF1 C-terminal domain(s) were used. The Y-axis represents the number of reads for peak summit normalized by the total number of reads per track. Data from two KMT2A-AFF1 cell lines SEM (GSE83671) [34] and RS4;11 (GSE38403) [35] are shown. B ChIP-Seq tracks on the DYRK1A and MYC promoter regions using a FLAG-specific antibody in KMT2A-Aff1-FLAG-transformed human ALL cells. The Y-axis represents the number of reads for peak summit normalized by the total number of reads per track (GSE84116) [36]. C ChIP-Seq tracks on the DYRK1A and MYC promoter regions using KMT2A N-terminal-, menin-, and H3K79me2-specific antibodies. Two KMT2A-R cell lines (MOLM13/AML and RS4;11/ALL) were either treated with control or the Menin inhibitor VTP50469 as previously described (GSE127508) [10]. D Western blot analysis of DYRK1A, MYC, and β-actin in three KMT2A-R ALL cell lines (SEM, KOPN8, RS4;11) and the IgG-MYC rearranged ALL cell line MN60. Each cell line was treated with either control or the menin inhibitor VTP50469 (100 nM) for 72 h.
To validate that KMT2A fusions not only bind to the promoter region but also directly affect DYRK1A transcription, we next analyzed ChIP-Seq data from the MOLM13 (KMT2A::MLLT3) AML cell line and the RS4;11 (KMT2A::AFF1) ALL cell line [10]. Cells were treated with vehicle or the menin inhibitor VTP50469 (a prolog of the clinical drug revumenib) [10] for 3 days. As expected, menin inhibition prevented the binding of menin, an important regulatory unit that is required for the transcriptional regulation of KMT2A fusion’s target genes, to the DYRK1A promoter. MYC was analyzed as a positive control demonstrating reduced binding of menin to the MYC promoter site and H3K79me2 was analyzed demonstrating the binding of DYRK1A is associated with an active promoter region (Fig. 2C). Of note, other DYRK family members are not regulated via KMT2A-R with the exception of DYRK2 where minimal binding activity was observed (Supplementary Fig. 2C). We next analyzed gene expression data from SEM cells treated with either the DOT1L inhibitor EPZ004777, the menin inhibitor MI-2-2, or a combination of both drugs. Our analysis shows that DYRK1A was specifically downregulated following menin inhibition, while DOT1L inhibition only marginally reduced DYRK1A expression levels (Supplementary Fig. 1C). We concluded that KMT2A fusions transcriptionally regulate DYRK1A via menin, but not epigenetically via DOT1L.
To validate this finding, we first performed RT-PCR analysis of control and VTP50469-treated SEM cells, which showed that in vitro menin inhibition led to a significant reduction in DYRK1A expression and other known transcriptional targets (Supplementary Fig. 1D). We then used CRISPR-mediated deletion of KMT2A in both KMT2A-R and wild-type (WT) KMT2A ALL cell lines. Our results clearly demonstrate that deletion of both KMT2A-R and KMT2A-WT decreased DYRK1A expression levels, indicating that both KMT2A-R and KMT2A-WT regulate DYRK1A (Supplementary Fig. 1E).
DYRK1A is highly expressed in KMT2A-R ALL and high expression levels are retained during relapse
We next tested the DYRK1A protein levels across different subtypes of ALL and AML and, interestingly, observed increased DYRK1A expression in KMT2A-R ALL compared to the tested KMT2A wild-type leukemia cell lines and PDX cases (Supplementary Fig. 1F, G). We then confirmed our cell line findings in cryopreserved cells from established infant and childhood KMT2A-R ALL PDX models created from diagnosis (black) and relapse (red) biospecimens and compared them to non-KMT2A-R ALL [37]. These data demonstrate that the high DYRK1A protein levels are retained at relapse and are overall elevated in KMT2A-R ALL compared to Ph-like ALL (Supplementary Fig. 1H). Finally, we performed a Western blot analysis of KMT2A-R ALL cell lines treated with control or 72 h of VTP50469 and observed a reduction in DYRK1A protein levels over time (Fig. 2D). However, menin inhibition did not fully abrogate the DYRK1A protein expression (Fig. 2D) and only affected cell proliferation of KMT2A-R ALL cells after multiple days of VTP50469 exposure (Supplementary Fig. 2A). In addition, apoptosis was induced only when high concentrations of VTP50469 were used (Supplementary Fig. 2B), consistent with previous reports [10]. Overall, these results suggest that KMT2A fusions in ALL regulate DYRK1A RNA and protein expression levels via direct transcriptional regulation that requires menin binding.
DYRK1A and MYC negatively regulate each other
Viral overexpression of DYRK1A has also been shown to mediate MYC phosphorylation (Ser62 and Thr58) and consequent MYC degradation in non-KMT2A-R AML [38], suggesting that DYRK1A may negatively regulate MYC also in ALL. To validate further the observed DYRK1A-mediated regulation of MYC, we used an orthogonal approach by inhibiting DYRK1A and conducting a phosphoproteomics study. This analysis showed reduced phosphorylation at Ser62 and Thr58 at both two hours and eight hours compared to control cells (Fig. 3A). To confirm this finding, we performed a Western blot analysis to assess MYC expression levels following treatment with the DYRK1A inhibitor EHT1610 (without modifying KMT2A fusion activity [39]). As expected, this treatment led to significant MYC upregulation/accumulation across all tested KMT2A-R ALL cell lines (Fig. 3B). Based on these findings we conclude that DYRK1A regulates MYC levels independently of the KMT2A fusion oncogene. To determine if a regulatory feedback mechanism exists between DYRK1A and MYC, we analyzed DYRK1A expression levels at different stages during B cell development including transgenic mouse model systems with overexpression of Bcl6 or Myc/Bcl6 or deletion of Lig4/tp53, three distinct models of B cell lymphoma [40]. Notably, tumor cells isolated from the Bcl6 transgenic and the Myc/Bcl6 transgenic mice displayed reduced Dyrk1a mRNA expression levels (Fig. 3C and Supplementary Fig. 3A), indicating that Dyrk1a may be negatively regulated either by Bcl6 or Myc in these models.
A Phophoproteomics analysis in HB11;19 cells treated with 5 µM GNF2133 for 2 h and 8 h. B Western blot analysis of MYC and β-actin in KMT2A-R ALL cell lines treated either with control or EHT1610 (5 µM/72 h). C Dyrk1a gene expression levels in a comprehensive panel of purified developmentally defined normal murine B cells and genetically distinct murine lymphoma models (GSE26408) [40]. D Chip-Seq experiment on the MYC-inducible B cell malignancy model cell line P493-6. Gene tracks of Myc binding at the Dyrk1a promoter region at 0 h (top), 1 h (middle), and 24 h (bottom) are shown (GSE36354) [43]. E Myc and Dyrk1a gene expression levels in P493-6 cells before and after MYC inactivation. Myc inactivation was induced via tetracycline treatment for 48 h (GSE120246). F Myc and Dyrk1a gene expression levels in Eμ-myc transgenic mice. Data are represented as individual values with mean ± SEM bars. *P < 0.05; **P < 0.01; ***P < 0.001 by t-test.
MYC, but not BCL6, negatively regulates DYRK1A
We next investigated whether BCL6 acts as a negative regulator of DYRK1A, given its role as a transcriptional repressor [41]. Using a meta-analysis of ChIP-Seq data from the ICN13 (KMT2A::AFF1) ALL PDX model, as well as the RS4;11 and SEM ALL cell lines (KMT2A::AFF1) [42], we found that BCL6 does not bind to the DYRK1A promoter (Supplementary Fig. 3B). Furthermore, genetic deletion of Bcl6 in murine Bcr-Abl1 ALL-like cells only resulted in moderate upregulation of Dyrk1a in one out of three probe sets (Supplementary Fig. 3C). Induced Bcl6 deletion in an ALL-like mouse knockout model system with overexpression of Tcf3-Pbx1 did not change RNA expression levels of DYRK1A, further validating that BCL6 does not regulate DYRK1A transcript levels (Supplementary Fig. 3D). Overall, we conclude that Bcl6 does not directly regulate DYRK1A.
To then test if MYC regulates DYRK1A transcriptionally, we analyzed ChIP-Seq data of a human MYC-driven B cell malignancy model (P493-6) [43] in which MYC expression can be induced via removal of doxycycline from the cell culture medium [43]. Activation of MYC expression resulted in increased binding of MYC to the DYRK1A promoter in a time-dependent manner (Fig. 3D). To assess further if MYC negatively regulates DYRK1A, we performed loss-of-function analyses by examining gene expression data of P493-6 before and after 48 h of doxycycline-induced MYC deletion, which resulted in significant upregulation of DYRK1A (Fig. 3E). We validated MYC-mediated negative regulation of Dyrk1a using gene expression data of Myc-expressing and Myc-deleted BCR-ABL1 transformed mouse ALL-like cells [44, 45] (Supplementary Fig. 4A). We also performed a gain-of-function meta-analysis of an Eμ-myc transgenic lymphoma/leukemia model [46, 47] (Fig. 3F) and observed that increased Myc levels correlate with decreased Dyrk1a levels during in vivo leukemia progression, further confirming a negative regulation of Dyrk1a via Myc.
Taken together, our analyses demonstrate that DYRK1A and MYC are closely connected and negatively regulate each other. Dyrk1a genetic deletion or DYRK1A pharmacologic inhibition resulted in increased Myc/MYC levels in our preclinical models, while genetic deletion or overexpression of MYC mediate increased or decreased DYRK1A expression, respectively.
KMT2A-R ALL cells are sensitive to pharmacologic inhibition of DYRK1A
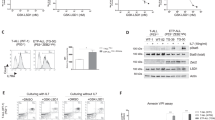
To evaluate DYRK1A as a potential target for the treatment of ALL, we first tested the extent to which in vitro pharmacologic inhibition of DYRK1A could induce apoptosis or cell cycle arrest in KMT2A-R ALL cells. Given that DYRK1A is important for Down Syndrome-associated ALL and the closely related CRLF2-R ALL [24, 25], we therefore excluded these ALL types from our analysis. We first used the DYRK1A inhibitor EHT1610 (Fig. 4A) for our translational studies. Interestingly, in vitro exposure to EHT1610 specifically reduced cell proliferation of KMT2A-R ALL compared to non-KMT2A-R ALL cell lines (Fig. 4B). It has been described in B- and T-cells that DYRK1A inhibition results in an accumulation of cells in the cell cycle S-phase [24]. To evaluate if DYRK1A inhibition affects cell proliferation or induces apoptosis specifically in KMT2A-R ALL, we performed a cell cycle analysis before and after EHT1610 treatment, which resulted in a significant reduction of cells in S-phase (Fig. 4C) and minimal effects upon cell viability (Fig. 4D). Taken together, DYRK1A inhibition in KMT2A-R ALL appears to affect cell proliferation specifically by increasing the number of cells in G1/G0 and limiting the number of cells in S-phase.
A Molecular structure of EHT161040. BKMT2A::AFF1 (dark blue; SEM and RS4;11), KMT2A::MLLT1 (light blue; HB11;19 and KOPN8), and non-KMT2A-R ALL cell lines (red; MN60 and Tanoue) were treated with increasing concentrations of EHT161040. Viability/cell proliferation was determined via an XTT assay after 72 h. C Cell cycle analysis of the indicated KMT2A-R ALL cell lines after treatment with 5 µM EHT1610 for 72 h. On the left are representative examples of the flow cytometric analysis (n = 3), and on the right is the summary of all three experiments. D Cell viability and apoptosis were tested in control- and DYRK1A inhibitor-treated (EHT1610; 5 µM; 72 h) KMT2A-R ALL. On the left are representative examples of the flow cytometric analysis (n = 3) and on the right is the summary of all three experiments. Data are represented as individual values with mean ± SEM bars. *P < 0.05; **P < 0.01; ***P < 0.001 by t-test.
Dual inhibition of DYRK1A and MYC increases apoptosis in certain KMT2A-R ALL cell lines but does not consistently enhance the therapeutic efficacy of DYRK1A inhibition
The role of MYC is highly complex, functioning as a well-known oncogene that drives cell proliferation and growth, while also paradoxically inducing apoptosis. In leukemia and certain solid tumors, MYC increases the expression of BIM, a proapoptotic protein, which can lead to cell death under specific conditions [48, 49]. Furthermore, elevated MYC levels can activate p53 [50], resulting in cell cycle arrest or apoptosis. Given the upregulation of MYC induced by DYRK1A inhibition, we investigated whether MYC upregulation would confer resistance to, or sensitize, KMT2A-R ALL cells to DYRK1A inhibition. Our flow cytometry (7-AAD/Annexin V) data indicate that dual inhibition of MYC (via the menin inhibitor VTP50469) and DYRK1A (EHT1610) significantly reduced viability in only one cell line (HB11;19) and had a moderate effect on KOPN8 cells, while other tested cell lines did not show increased cell death following dual DYRK1A and menin inhibition (Supplementary Fig. 2D). To confirm these findings, we conducted additional experiments using an MYC inhibitor (MYCi975) that prevents the dimerization of MYC and MAX, thereby blocking the expression of MYC target genes, including MYC itself [51, 52]. Western blot analysis confirmed the efficacy of MYCi975 in reducing MYC protein expression (Supplementary Fig. 2E). Additionally, we performed an XTT assay, which demonstrated that the response to dual treatment (MYC inhibitor + DYRK1A inhibitor) was consistent with the menin inhibitor data. Specifically, HB11;19 and KOPN8 were the only cell lines affected by the dual treatment (Supplementary Fig. 2F). These findings suggest that while MYC modulation can influence the response to DYRK1A inhibition in specific KMT2A-R ALL cell lines, its effects are not broadly consistent across all tested models, highlighting the complexity of MYC’s role in ALL.
DYRK1A represses ERK signaling and protects KMT2A-R ALL cells from cell cycle arrest
It has been demonstrated that DYRK1A overexpression results in increased ERK signaling output in brain cells and synovial tissues of rheumatoid arthritis patients [53,54,55]. Based on these findings, we hypothesized that the induction of cell cycle arrest in KMT2A-R ALL cells after pharmacologic inhibition may be via DYRK1A inhibitor-mediated downregulation of ERK signaling. To assess the effect of DYRK1A inhibition on ERK signaling, we treated different KMT2A-R ALL cell lines with EHT1610 and collected protein samples at four different time points. Surprisingly, we instead observed that DYRK1A inhibition resulted in hyperphosphorylation of ERK (Fig. 5A). To determine if this was a specific effect in KMT2A-R leukemias or if DYRK1A also mediates hyperphosphorylation in other ALL subtypes, we performed a Western blot to measure and compare ERK phosphorylation levels in vehicle- or EHT1610-treated KMT2A-R ALL and non-KMT2A-R ALL cell lines and in KMT2A-R AML cell lines. Interestingly, DYRK1A inhibitor-induced ERK hyperphosphorylation was restricted to KMT2A-R ALL samples (Fig. 5B). It has been demonstrated that hyperactivation of B cell receptor signaling molecules can result in negative selection of B cells [56] and that specifically the RAS/MEK/ERK signaling pathway is involved in this process [57]. Based on these findings, we hypothesized that DYRK1A inhibitor-mediated ERK hyperphosphorylation may result in negative selection and cell cycle arrest in KMT2A-R ALL cells. To test this hypothesis, we first treated 4 different KMT2A-R ALL cell lines either with vehicle control, EHT1610, the MEK1/2 inhibitor trametinib, or a combination of both drugs (Fig. 5C). As expected, DYRK1A inhibition resulted in potent ERK hyperphosphorylation in all tested KMT2A-R ALL cell lines with activated ERK signaling at a steady state, while trametinib inhibited ERK signaling. Most importantly, trametinib co-exposure effectively abrogated the EHT1610-mediated hyperphosphorylation of ERK. To test if trametinib would rescue KMT2A-R ALL cell proliferation, we performed a synergy study using increasing concentrations of EHT1610 and trametinib. Strikingly, our results demonstrate that trametinib rescued KMT2A-R ALL cells from DYRK1A inhibition-induced cell cycle arrest (Fig. 5D). Taken together, our results demonstrate an unexpected involvement of DYRK1A in RAS/MEK/ERK signaling regulation in KMT2A-R ALL cells and illustrate that DYRK1A inhibitor-mediated cell cycle arrest is induced via ERK hyperphosphorylation.
A Protein expression levels of the indicated proteins were determined via Western blot using three KMT2A-R ALL cell lines treated with 5 µM EHT1610 for the indicated time points. B KMT2A-R (SEM and HB11;19) and Ph-like (MUTZ5 and MHH-CALL4) ALL cell lines as well as KMT2A-R (MOLM14, MV4;11) AML cell lines were treated either with control or 5 µM EHT1610. After 72 h pERK, ERK, and β-ACTIN protein levels were determined via Western blot. C The indicated KMT2A-R ALL cell lines were treated either with control, 5 µM EHT1610, 20 nM trametinib, or both drugs. After 72 h, the expression levels of the indicated proteins were determined via Western blotting. D The combinatorial effect of EHT1610 and trametinib was determined via synergy analyses using Synergy Finder software.
DYRK1A inhibition induces the expression of BIM and reduces BCL-XL
The proapoptotic factor BIM is regulated via multiple regulatory mechanisms. One of these regulators is MYC, which has been demonstrated to directly bind to the BIM promoter region to activate BIM-mediated apoptosis [49]. ERK has also been studied as a molecule that can either increase the proapoptotic activity via phosphorylation of BIM at S44, T56, and S58 residues (exon 3) [58] or decrease its protein stability and induce proteasome-dependent degradation via phosphorylation of S69 [59, 60]. To ascertain if DYRK1A inhibition and consequent activation of MYC and ERK signaling results in increased BIM expression and activity in KMT2A-R ALL cells, we treated four cell lines with EHT1610 and performed a Western blot analysis of BIM and BCL2. The proapoptotic activity of BIM is neutralized when BCL2 binds to BIM [61]. Strikingly, DYRK1A inhibition increased BIM expression in three of the four KMT2A-R ALL cell lines, while the BCL2 levels remained mostly unchanged (Fig. 6A). We validated that BIM activation starts as early as 24 h (Supplementary Fig. 4C). Furthermore, DYRK1A inhibition resulted in the reduction of the antiapoptotic factor BCL-XL, further indicating that DYRK1A inhibition renders cells sensitive to apoptosis (Fig. 6B). Taken together, these data indicate that DYRK1A inhibition may induce apoptosis via upregulation of BIM and downregulation of BCL-XL.
A Western blot analysis of the indicated KMT2A-R ALL cell lines treated either with vehicle control or 5 µM EHT1610. After 72 h, the expression levels of the indicated proteins were determined. B Western blot analysis of KMT2A-R ALL cell lines treated with control or 5 µM EHT1610. After 72 h, the expression levels of BCL-XL and β-actin were determined. C SEM and KOPN8 cells were treated with increasing concentrations of EHT1610 and venetoclax. The synergistic effect of both drugs was determined via Synergy Finder software as in Fig. 5. D Flow cytometric analysis of 4 KMT2A-R ALL cell lines treated with 5 µM EHT1610 and 20 nM venetoclax for 72 h. Shown is the statistical analysis (n = 3). E Kaplan–Meier analysis of the overall survival of mice injected with 1 million KMT2A-AFF1 rearranged PDX cells (150MD) and treated either with control, GNF2133 (50 µM), venetoclax (50 µM), or a combination of both drugs. The treatment was started after ~5% CD19+/CD45+ALL cells were detected in the blood and continued until ~80% CD19+/CD45+ ALL replacements were detected in the blood of the mice. Significance was calculated via a log-rank test in Prism. F Kaplan–Meier survival analysis of KMT2A::AFF1 ALL PDX model (ALL26MD) mice treated for 20 days with vehicle control, GNF2133 (50 µM), venetoclax (50 µM), or a combination of both drugs (n = 5 mice/cohort). Significance was calculated via a log-rank test in Prism. Data are represented as individual values with mean ± SEM bars. *P < 0.05; **P < 0.01; ***P < 0.001 by t-test or Log-rank (Mantel–Cox) test.
DYRK1A inhibition renders cells sensitive to venetoclax and increases overall survival of mice in preclinical in vivo studies
To determine if DYRK1A inhibition sensitizes KMT2A-R ALL cells to BCL2 inhibition, we treated KMT2A-R ALL cells in vitro with increasing concentrations of EHT1610 and the BCL2 inhibitor venetoclax and observed synergistic killing (Fig. 6C) and decreased cell viability, as assessed by flow cytometry (Fig. 6D and Supplementary Fig. 4B).
Given our observations of significant EHT1610-associated toxicity in mice, we synthesized the recently described DYRK1A inhibitor GNF2133 (Supplementary Fig. 5A), which has been shown to be well tolerated in vivo in mice and rats in preclinical studies [62]. First, we compared GNF2133 to EHT1610 and their ability in vitro to induce ERK hyperphosphorylation in KMT2A-R ALL cells (Supplementary Fig. 5B) and confirmed that GNF2133 exposure decreased the percentage of cells in S-phase compared to control treatment (Supplementary Fig. 5C). We further observed synergistic killing of KMT2A-R ALL cell lines when co-treated with GNF2133 and venetoclax (Supplementary Fig. 5D, E) and GNF2133-mediated ERK hyperphosphorylation in KMT2A-R PDX cases (Supplementary Fig. 5F).
We then confirmed in a pilot feasibility study that optimized GNF2133 doses of up to 50 mg/kg daily ×14 days in vivo did not induce untoward toxicity in NSG mice (data not shown). KMT2A-R ALL PDX model (ALL150MD) mice were then treated with vehicle control, GNF2133 (50 mg/kg), venetoclax (50 mg/kg), or both inhibitors. Single-agent GNF2133 or venetoclax each reduced leukemia burden, but when GNF2133 and venetoclax were given in combination, the leukemic burden was further reduced (Supplementary Fig. 6A) and prolonged animal survival (Fig. 6E). Dual inhibitor therapy again appeared tolerable in vivo given weight stability of treated mice over time (Supplementary Fig. 6B). The criteria for mice to reach the study endpoint included either >80% CD19+/CD45+ cells in the blood, 20% weight loss, and/or reduced physical mobility due to leukemia progression or potential drug toxicity as described [28, 29, 63]. Once any of these criteria were met, the mice were sacrificed. Supplementary Fig. 6C shows that, at the time of sacrifice, the spleens were equally enlarged across all treatment groups. Results were validated in a second KMT2A-R ALL PDX model (ALL26MD) with the greatest inhibition of leukemia proliferation (Supplementary Fig. 6D), improved survival (Fig. 6F), and tolerability (Supplementary Fig. 6E) observed with dual GNF2133 and venetoclax treatment. Furthermore, spleen enlargement was equal among the different treatment groups (Supplementary Fig. 6F).
To test whether DYRK1A inhibition affects healthy hematopoietic cells, we treated C57BL/6 mice for two weeks with either vehicle control or GNF2133 (50 mg/kg) and subsequently performed a complete blood count (CBC). The results show no significant changes in hematopoietic cell populations between the control and GNF2133-treated mice, except for a reduction in eosinophils and basophils following GNF2133 treatment (Supplementary Fig. 7A, B). During the two-week treatment period, none of the mice experienced significant weight loss (Supplementary Fig. 7C).
Taken together, these data credential the combined inhibition of DYRK1A and BCL2 as an additional effective and potentially clinically translatable therapeutic strategy for KMT2A-R ALL.
Discussion
To identify new targets in KMT2A-R ALL for precision medicine approaches, we performed a kinome-wide CRISPR screen and identified multiple kinases, including DYRK1A, as required for leukemia cell survival. We selected to further study the importance of DYRK1A for KMT2A-R ALL as it met the following three criteria: (1) Growth inhibition upon DYRK1A targeting was stronger in KMT2A-R leukemic cells as in non-KMT2A-R ALL cells, (2) DYRK1A is not a common essential gene assessed via the Cancer Dependency MAP, and (3) DYRK1A has not been studied specifically in KMT2A-R ALL. Previous studies have suggested the involvement of DYRK1A in Down syndrome-associated megakaryoblastic leukemia [23] and normal hematopoiesis [24, 64]. More recently, additional studies support a critical role of DYRK1A in DS-ALL [25] and the closely related CRLF2-R Ph-like ALL subtype [25]. While higher DYRK1A expression levels are expected in ALL cells from patients with Down syndrome, given that DYRK1A is located on chromosome 21, it is not known how DYRK1A is transcriptionally regulated. Here we demonstrate a direct transcriptional regulation of DYRK1A via KMT2A fusions in ALL. In addition, we report that pharmacologic inhibition of menin, an essential transcription factor involved in the regulation of KMT2A-R target genes [65,66,67], and direct DYRK1A inhibition results in cell cycle arrest of KMT2A-R ALL. Given that DYRK1A is not the only target gene regulated by KMT2A, we specifically focused on studying the importance of DYRK1A for KMT2A-R ALL survival using DYRK1A inhibitors. We specifically tested two DYRK1A inhibitors (EHT1610 and GNF2133) for our studies, which are both specifically inhibiting DYRK1A with very little off-target effects. However, in our in vivo studies EHT1610 treatments were toxic to the recipient mice resulting in a premature ending of the study. We attribute the toxicity observed with EHT1610 in part to incomplete solubilization as evidenced by peritoneal crystallization of the compound resulting in toxicity and likely decreased bioavailability. Batch variability and long-term storage of lyophilized EHT1610 may contribute to inconsistent solubilization; a recurrent obstacle in developing DYRK1A-selective ATP-competitive inhibitors has been poor aqueous solubility [68]. Using the DYRK1A inhibitor GNF2133 was significantly better tolerated by the mice based on our data as well as published data [62]. In previous experiments, we also tested harmine, a widely used DYRK1A inhibitor, to examine the combinatorial effect of dual DYRK1A and BCL2 inhibition. Similar to GNF2133, harmine treatment in combination with venetoclax led to a significant reduction in KMT2A-R ALL in mice, thereby validating our findings and underscoring DYRK1A’s critical role in KMT2A-R ALL (data not shown). However, harmine’s application in humans is limited due to associated toxicities, particularly neurotoxicity. Thus, the in vivo efficacy of GNF2133 becomes particularly valuable and highlights the need for further development of pharmacologic agents targeting DYRK1A.
Our results demonstrate that KMT2A-R ALL requires DYRK1A for normal cell proliferation and dual inhibition of DYRK1A and BCL2 decreases leukemia progression in KMT2A-R ALL PDX models.
ALL cells are bone marrow-derived B cell precursors that have lost the ability to differentiate but retain some properties of B cells. In B cells, it has been shown that the PI3K and ERK signaling pathways are regulated via the B cell receptor, and depending on the signaling strength, may mediate negative selection to prevent autoimmunity. Previous results in normal B cells have suggested that DYRK1A regulates B cell differentiation [24]. Additionally, it has been demonstrated that DYRK1A regulates B cell survival and B cell autoimmunity via activation of NF-κB [64]. Our results demonstrate that DYRK1A inhibition in KMT2A-R ALL prevented cell proliferation while interestingly resulting in hyperphosphorylation of ERK, in contrast to prior reports in other cell types [53, 55]. Dual inhibition of DYRK1A and the ERK regulating kinases MEK1/2 prevented ERK hyperphosphorylation and consequently rescued cell proliferation. demonstrating that DYRK1A-mediated hyperphosphorylation of ERK is specifically negatively affecting cell proliferation and survival. Given that KMT2A-R ALL cells do not express a (pre) B cell receptor on the surface, we propose that the mechanism of negative selection is induced independently of the B cell receptor in these cells and that DYRK1A downregulates the negative selection of KMT2A-R B-ALL, thereby facilitating rapid leukemia cell proliferation. Upon DYRK1A inhibition, ERK hyperphosphorylation may stimulate a type of negative selection that makes cells vulnerable to inhibition of BH3-based cell survival mechanisms.
Another important observation is that DYRK1A and MYC negatively regulate each other in a feedback loop. First, we identified that pharmacologic DYRK1A inhibition resulted in increased MYC levels via decreased phosphorylation of Thr58 and Ser62 on MYC, which is concordant with a recent publication demonstrating that viral overexpression of DYRK1A in non-KMT2A-R AML results in phosphorylation of Thr58 and Ser62 on MYC and consequently in MYC degradation [38]. A critical regulator of apoptosis is BIM, which is directly regulated via MYC. Per our results, DYRK1A inhibition results in upregulation of MYC and consequently in higher BIM levels, while the BIM negative regulator BCL2 is not affected. Furthermore, we demonstrate that upon DYRK1A inhibition, the expression levels of the antiapoptotic factor BCL-XL are reduced, which consequently renders KMT2A-R ALL cells sensitive to dual DYRK1A and venetoclax inhibition and significantly extends the survival of mice transplanted with KMT2A-R ALL, indicating that a DYRK1A inhibitor-based therapy is a novel approach for the treatment of KMT2A-R ALL.
Despite significant improvements in the treatment of patients with acute leukemias, including improved access to relevant targeted inhibitors and antibody-based or cellular immunotherapies, children and adults with KMT2A-R ALL continue to experience inferior clinical outcomes. Our preclinical data suggest that co-targeting of DYRK1A and menin or BCL2 has particular activity against this high-risk leukemia subtype that may warrant clinical investigation in patients in the future to address issues of chemoresistance and relapse risk.
Data availability
All data used in this study are publicly available and can be accessed from the Gene Expression Omnibus: GSE83671, GSE38403, GSE84116, GSE127508, GSE26408, GSE36354, and GSE120246.
References
Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700.
Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–707.
Winters AC, Bernt KM. MLL-rearranged leukemias-an update on science and clinical approaches. Front Pediatr. 2017;5:4.
Schultz KR, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children’s oncology group study. J Clin Oncol. 2009;27:5175–81.
Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slayton WB, et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group study AALL0031. Leukemia. 2014;28:1467–71.
Byrd JC, Brown JR, O’Brien S, Barrientos JC, Kay NE, Reddy NM, et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med. 2014;371:213–23.
Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood. 2005;105:812–20.
Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–25.
Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65.
Krivtsov AV, Evans K, Gadrey JY, Eschle BK, Hatton C, Uckelmann HJ, et al. A Menin-MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell. 2019;36:660–673.e611.
Issa GC, Aldoss I, DiPersio J, Cuglievan B, Stone R, Arellano M, et al. The menin inhibitor revumenib in KMT2A-rearranged or NPM1-mutant leukaemia. Nature. 2023;615:920–4.
Brown PA, Kairalla JA, Hilden JM, Dreyer ZE, Carroll AJ, Heerema NA, et al. FLT3 inhibitor lestaurtinib plus chemotherapy for newly diagnosed KMT2A-rearranged infant acute lymphoblastic leukemia: Children’s Oncology Group trial AALL0631. Leukemia. 2021;35:1279–90.
Stein EM, Garcia-Manero G, Rizzieri DA, Tibes R, Berdeja JG, Savona MR, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood. 2018;131:2661–9.
Campbell CT, Haladyna JN, Drubin DA, Thomson TM, Maria MJ, Yamauchi T, et al. Mechanisms of pinometostat (EPZ-5676) treatment-emergent resistance in MLL-rearranged leukemia. Mol Cancer Ther. 2017;16:1669–79.
Perner F, Stein EM, Wenge DV, Singh S, Kim J, Apazidis A, et al. MEN1 mutations mediate clinical resistance to menin inhibition. Nature. 2023;615:913–9.
Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med. 2019;381:1728–40.
Erba HP, Montesinos P, Kim HJ, Patkowska E, Vrhovac R, Žák P, et al. Quizartinib plus chemotherapy in newly diagnosed patients with FLT3-internal-tandem-duplication-positive acute myeloid leukaemia (QuANTUM-First): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2023;401:1571–83.
Pollard JA, Alonzo TA, Gerbing R, Brown P, Fox E, Choi J, et al. Sorafenib in combination with standard chemotherapy for children with high allelic ratio FLT3/ITD+ acute myeloid leukemia: a report from the Children’s Oncology Group Protocol AAML1031. J Clin Oncol. 2022;40:2023–35.
DiNardo CD, Jonas BA, Pullarkat V, Thirman MJ, Garcia JS, Wei AH, et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med. 2020;383:617–29.
Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130:722–31.
Becker W, Weber Y, Wetzel K, Eirmbter K, Tejedor FJ, Joost HG. Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J Biol Chem. 1998;273:25893–902.
Arbones ML, Thomazeau A, Nakano-Kobayashi A, Hagiwara M, Delabar JM. DYRK1A and cognition: a lifelong relationship. Pharm Ther. 2019;194:199–221.
Malinge S, Bliss-Moreau M, Kirsammer G, Diebold L, Chlon T, Gurbuxani S, et al. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J Clin Invest. 2012;122:948–62.
Thompson BJ, Bhansali R, Diebold L, Cook DE, Stolzenburg L, Casagrande AS, et al. DYRK1A controls the transition from proliferation to quiescence during lymphoid development by destabilizing Cyclin D3. J Exp Med. 2015;212:953–70.
Bhansali RS, Rammohan M, Lee P, Laurent AP, Wen Q, Suraneni P, et al. DYRK1A regulates B cell acute lymphoblastic leukemia through phosphorylation of FOXO1 and STAT3. J Clin Invest. 2021;131:1–18.
Deshmukh V, O’Green AL, Bossard C, Seo T, Lamangan L, Ibanez M, et al. Modulation of the Wnt pathway through inhibition of CLK2 and DYRK1A by lorecivivint as a novel, potentially disease-modifying approach for knee osteoarthritis treatment. Osteoarthr Cartil. 2019;27:1347–60.
Kim S, Ko E, Choi HG, Kim D, Luchi M, Khor B, et al. FRTX-02, a selective and potent inhibitor of DYRK1A, modulates inflammatory pathways in mouse models of psoriasis and atopic dermatitis. J Transl Autoimmun. 2023;6:100185.
Kruth KA, Fang M, Shelton DN, Abu-Halawa O, Mahling R, Yang H, et al. Suppression of B-cell development genes is key to glucocorticoid efficacy in treatment of acute lymphoblastic leukemia. Blood. 2017;129:3000–8.
Tasian SK, Teachey DT, Li Y, Shen F, Harvey RC, Chen IM, et al. Potent efficacy of combined PI3K/mTOR and JAK or ABL inhibition in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2017;129:177–87.
Gotesman M, Vo TT, Herzog LO, Tea T, Mallya S, Tasian SK, et al. mTOR inhibition enhances efficacy of dasatinib in ABL-rearranged Ph-like B-ALL. Oncotarget. 2018;9:6562–71.
Surrey LF, MacFarland SP, Chang F, Cao K, Rathi KS, Akgumus GT, et al. Clinical utility of custom-designed NGS panel testing in pediatric tumors. Genome Med. 2019;11:32.
Joseph PL, Anella Y, Patrick AB, Lisa MN, Asen B, Min W, et al. Combinatorial efficacy of entospletinib and chemotherapy in patient-derived xenograft models of infant acute lymphoblastic leukemia. Haematologica. 2020;106:1067–78.
Hurtz C, Wertheim GB, Loftus JP, Blumenthal D, Lehman A, Li Y, et al. Oncogene-independent BCR-like signaling adaptation confers drug resistance in Ph-like ALL. J Clin Invest. 2020;130:3637–53.
Kerry J, Godfrey L, Repapi E, Tapia M, Blackledge NP, Ma H, et al. MLL-AF4 spreading identifies binding sites that are distinct from super-enhancers and that govern sensitivity to DOT1L inhibition in leukemia. Cell Rep. 2017;18:482–95.
Geng H, Brennan S, Milne TA, Chen WY, Li Y, Hurtz C, et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012;2:1004–23.
Lin S, Luo RT, Ptasinska A, Kerry J, Assi SA, Wunderlich M, et al. Instructive role of MLL-fusion proteins revealed by a model of t(4;11) pro-B acute lymphoblastic leukemia. Cancer Cell. 2016;30:737–49.
Loftus JP, Yahiaoui A, Brown PA, Niswander LM, Bagashev A, Wang M, et al. Combinatorial efficacy of entospletinib and chemotherapy in patient-derived xenograft models of infant acute lymphoblastic leukemia. Haematologica. 2021;106:1067–78.
Liu Q, Liu N, Zang S, Liu H, Wang P, Ji C, et al. Tumor suppressor DYRK1A effects on proliferation and chemoresistance of AML cells by downregulating c-Myc. PLoS ONE. 2014;9:e98853–e98853.
Chaikuad A, Diharce J, Schröder M, Foucourt A, Leblond B, Casagrande A-S, et al. An unusual binding model of the methyl 9-anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) confers high selectivity for dual-specificity tyrosine phosphorylation-regulated kinases. J Med Chem. 2016;59:10315–21.
Green MR, Monti S, Dalla-Favera R, Pasqualucci L, Walsh NC, Schmidt-Supprian M, et al. Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc Natl Acad Sci USA. 2011;108:2873–8.
Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212.
Hurtz C, Chan LN, Geng H, Ballabio E, Xiao G, Deb G, et al. Rationale for targeting BCL6 in MLL-rearranged acute lymphoblastic leukemia. Genes Dev. 2019;33:1265–79.
Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67.
Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med. 2013;19:1014–22.
de Alboran IM, O’Hagan RC, Gärtner F, Malynn B, Davidson L, Rickert R, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55.
Sabò A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–92.
Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167:353–71.
Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA. 2004;101:6164–9.
Muthalagu N, Junttila MR, Wiese KE, Wolf E, Morton J, Bauer B, et al. BIM is the primary mediator of MYC-induced apoptosis in multiple solid tissues. Cell Rep. 2014;8:1347–53.
Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–33.
Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, et al. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell. 2019;36:483–497.e415.
Truica MI, Burns MC, Han H, Abdulkadir SA. Turning up the heat on MYC: progress in small-molecule inhibitors. Cancer Res. 2021;81:248–53.
Abekhoukh S, Planque C, Ripoll C, Urbaniak P, Paul JL, Delabar JM, et al. Dyrk1A, a serine/threonine kinase, is involved in ERK and Akt activation in the brain of hyperhomocysteinemic mice. Mol Neurobiol. 2013;47:105–16.
Kelly PA, Rahmani Z. DYRK1A enhances the mitogen-activated protein kinase cascade in PC12 cells by forming a complex with Ras, B-Raf, and MEK1. Mol Biol cell. 2005;16:3562–73.
Guo X, Zhang D, Zhang X, Jiang J, Xue P, Wu C, et al. Dyrk1A promotes the proliferation, migration and invasion of fibroblast-like synoviocytes in rheumatoid arthritis via down-regulating Spry2 and activating the ERK MAPK pathway. Tissue Cell. 2018;55:63–70.
Keenan RA, De Riva A, Corleis B, Hepburn L, Licence S, Winkler TH, et al. Censoring of autoreactive B cell development by the pre-B cell receptor. Science. 2008;321:696–9.
Limnander A, Weiss A. Ca-dependent Ras/Erk signaling mediates negative selection of autoreactive B cells. Small GTPases. 2011;2:282–8.
Stang SL, Lopez-Campistrous A, Song X, Dower NA, Blumberg PM, Wender PA, et al. A proapoptotic signaling pathway involving RasGRP, Erk, and Bim in B cells. Exp Hematol. 2009;37:122–134.e122.
Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G, et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene. 2003;22:6785–93.
Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem. 2003;278:18811–6.
Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403.
Liu YA, Jin Q, Zou Y, Ding Q, Yan S, Wang Z, et al. Selective DYRK1A inhibitor for the treatment of type 1 diabetes: discovery of 6-Azaindole derivative GNF2133. J Med Chem. 2020;63:2958–73.
Hurtz C, Tasian SK, Wertheim GB, Ruggeri B, Stubbs MC, Perl AE, et al. Redundant JAK, SRC and PI3 kinase signaling pathways regulate cell survival in human Ph-like ALL cell lines and primary cells. Blood. 2017;130:717–717.
Li Y, Xie X, Jie Z, Zhu L, Yang JY, Ko CJ, et al. DYRK1a mediates BAFF-induced noncanonical NF-κB activation to promote autoimmunity and B-cell leukemogenesis. Blood. 2021;138:2360–71.
Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–18.
Chen YX, Yan J, Keeshan K, Tubbs AT, Wang H, Silva A, et al. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Natl Acad Sci USA. 2006;103:1018–23.
Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67:7275–83.
Meine R, Becker W, Falke H, Preu L, Loaëc N, Meijer L, et al. Indole-3-carbonitriles as DYRK1A inhibitors by fragment-based drug design. Molecules. 2018;23:1–23.
Acknowledgements
We thank Drs. Thanh-Trang Vo and David Fruman at the University of California, Irvine for sharing TVA-1 cells, Dr. Liling Wang at the University of Pennsylvania for helpful scientific discussions, and George Morton at Temple University for providing us with GNF2133.
Funding
These studies were supported by the National Institutes of Health/National Cancer Institute K22CA251649 (CH), 1U01CA232486 (SKT), and 1U01CA243072 (SKT) awards, the Rally Foundation for Childhood Cancer (CH, SKT), an American Society of Hematology Restart research grant (CH), the Department of Defense Translational Team Science Award CA180683P1 (SKT), the Leukemia Research Foundation (CH), the V Foundation for Cancer Research (CH, SKT), the SchylerStrong Foundation for Childhood Cancer Research (SKT), Alex’s Lemonade Stand Foundation (CH), and a Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program CURE award (MC, SKT). SS was supported by Scholar Awards from the American Society of Hematology and the St. Baldrick’s Foundation, and grants from the Andrew McDonough B+ Foundation, Leukemia Research Foundation, and the Leukemia Lymphoma Society Translational Research Program (6624-21). RSB is a Damon Runyon Physician-Scientist supported in part by the Damon Runyon Cancer Research Foundation (PST-45-24), the Hematopoiesis Training Program grant at the University of Pennsylvania (NIH/NIDDK T32 DK07780), and the University of Pennsylvania Measey Scholars in Molecular Medicine Program. TB acknowledges the University of Rouen Normandie, INSA Rouen Normandie, CNRS (Centre National de la Recherche Scientifique), and Region Normandie for multiform support. TAM is supported by the MRC Molecular Haematology Unit grant MC_UU_00029/6. SKT is a Scholar of the Leukemia and Lymphoma Society and holds the Joshua Kahan Endowed Chair in Pediatric Leukemia Research at the Children’s Hospital of Philadelphia. Open access funding provided by SCELC, Statewide California Electronic Library Consortium.
Author information
Authors and Affiliations
Contributions
VSSAA performed experiments and analyzed data; GW and JS assisted with the design and execution of the CRISPR screen; SG performed CRISPR-Cas9 experiments; JAC and JPL performed preclinical in vivo animal studies and provided ALL cell lines and PDX model cells for in vitro studies; SJL, AK, SS performed experiments and discussed results; RSB and JDC provided consultation on DYRK1A biology, selection, and use of DYRK1A inhibitors and assisted with the preparation of the manuscript: WC synthesized GNF2133; HG and TM provided and analyzed ChIP-Seq data; XH and KMB provided the Menin inhibitors MI-503 and VTP50469: TB synthesized the DYRK1A inhibitor EHT1610. MC and SKT, assisted with experimental design, interpreted data, provided research funding and experimental materials, and edited the manuscript. CH designed and directed the study, performed experiments, analyzed, and interpreted data, wrote the manuscript, and provided research funding; All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
JDC is on the SAB for Alethiomics and receives research funding from Syndax. TAM is a paid consultant for and shareholder in Dark Blue Therapeutics Ltd. SKT receives research funding from Incyte Corporation and Kura Oncology, serves/d on scientific advisory boards for Aleta Biotherapeutics, AstraZeneca, Jazz Pharmaceuticals, Kura Oncology, Syndax Pharmaceuticals, and Wugen, Inc., and has received travel support from Amgen and Jazz Pharmaceuticals (all for unrelated studies).
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. We have complied with all the relevant ethical regulations for animal testing and research. All animal-related experiments have received ethical approval from Loma Linda University Institutional Animal Care and Use Committee (IACUC-23-029).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ayyadevara, V.S.S.A., Wertheim, G., Gaur, S. et al. DYRK1A inhibition results in MYC and ERK activation rendering KMT2A-R acute lymphoblastic leukemia cells sensitive to BCL2 inhibition. Leukemia 39, 1078–1089 (2025). https://doi.org/10.1038/s41375-025-02575-w
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02575-w