Abstract
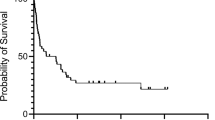
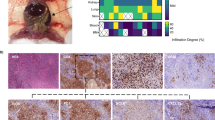
Nodal T follicular helper cell lymphoma (nTFHL) exhibits unique immunophenotypes and somatic alterations, while the prognostic value of these alterations remains unclear. By analyzing 173 nTFHL cases, we identified 36 driver genes, including 4 novel ones (TET3, HLA-C, NRAS, and KLF2). Then, we classified nTFHL cases into four molecular subgroups by major driver alterations. TR-I (+) and TR-I (−) were characterized by TET2 and/or RHOA mutations with and without IDH2 mutations; AC53 by TP53 and/or CDKN2A alterations and aneuploidy; and NSD with no subgroup-defining alterations (namely without any of the above alterations). AC53 exhibited the worst survival, while NSD, particularly those lacking driver alterations, showed the best prognosis. nTFHL had a better prognosis than peripheral T-cell lymphoma, not otherwise specified, when TP53 and/or CDKN2A alterations were absent. Multivariable analyses showed that AC53, the presence of driver alterations, and international prognostic index high-risk were independently associated with worse survival. Finally, we developed a simple prognostic index (mTFHL-PI), which classified patients into three risk categories with a median OS of 181, 67, and 20 months, respectively. Our study identifies novel prognostic factors, namely TP53 and/or CDKN2A alterations and the presence of driver alterations, demonstrating the clinical relevance of molecular classification in nTFHL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data generated in this study are available upon request from the corresponding author.
References
Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, IBdO Araujo, Berti E, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia. 2022;36:1720–48.
Campo E, Jaffe ES, Cook JR, Quintanilla-Martinez L, Swerdlow SH, Anderson KC, et al. The International Consensus Classification of Mature Lymphoid Neoplasms: a report from the Clinical Advisory Committee. Blood. 2022;140:1229–53.
de Leval L, Alizadeh AA, Bergsagel PL, Campo E, Davies A, Dogan A, et al. Genomic profiling for clinical decision making in lymphoid neoplasms. Blood. 2022;140:2193–227.
de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109:4952–63.
Mourad N, Mounier N, Brière J, Raffoux E, Delmer A, Feller A, et al. Clinical, biologic, and pathologic features in 157 patients with angioimmunoblastic T-cell lymphoma treated within the Groupe d’Etude des Lymphomes de l’Adulte (GELA) trials. Blood. 2008;111:4463–70.
Tokunaga T, Shimada K, Yamamoto K, Chihara D, Ichihashi T, Oshima R, et al. Retrospective analysis of prognostic factors for angioimmunoblastic T-cell lymphoma: a multicenter cooperative study in Japan. Blood. 2012;119:2837–43.
Laurence de L, Marie P, Fabien Le B, Jean-Philippe J, Virginie F, Antoine M, et al. Angioimmunoblastic T-cell lymphoma is the most common T-cell lymphoma in two distinct French information data sets. Haematologica. 2015;100:e361–e364.
Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–96.
Lemonnier F, Couronné L, Parrens M, Jaïs JP, Travert M, Lamant L, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–9.
Lewis NE, Petrova-Drus K, Huet S, Epstein-Peterson ZD, Gao Q, Sigler AE, et al. Clonal hematopoiesis in angioimmunoblastic T-cell lymphoma with divergent evolution to myeloid neoplasms. Blood Adv. 2020;4:2261–71.
Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–5.
Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:371–5.
Palomero T, Couronné L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–70.
Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–6.
Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais J-P, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119:1901–3.
Wang C, McKeithan TW, Gong Q, Zhang W, Bouska A, Rosenwald A, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126:1741–52.
Vallois D, Dobay MPD, Morin RD, Lemonnier F, Missiaglia E, Juilland M, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell–derived lymphomas. Blood. 2016;128:1490–502.
Vose J, Armitage J, Weisenburger D. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26:4124–30.
Horwitz S, O’Connor OA, Pro B, Trümper L, Iyer S, Advani R, et al. The ECHELON-2 Trial: 5-year results of a randomized, phase III study of brentuximab vedotin with chemotherapy for CD30-positive peripheral T-cell lymphoma. Ann Oncol. 2022;33:288–98.
Sabattini E, Pizzi M, Tabanelli V, Baldin P, Sacchetti CS, Agostinelli C, et al. CD30 expression in peripheral T-cell lymphomas. Haematologica. 2013;98:e81–82.
Schmitz N, Lenz G, Stelljes M. Allogeneic hematopoietic stem cell transplantation for T-cell lymphomas. Blood. 2018;132:245–53.
Schmitz N, Truemper L, Bouabdallah K, Ziepert M, Leclerc M, Cartron G, et al. A randomized phase 3 trial of autologous vs allogeneic transplantation as part of first-line therapy in poor-risk peripheral T-NHL. Blood. 2021;137:2646–56.
Basha BM, Bryant SC, Rech KL, Feldman AL, Vrana JA, Shi M, et al. Application of a 5 marker panel to the routine diagnosis of peripheral T-cell lymphoma with T-follicular helper phenotype. American J Surgical Pathol. 2019;43:1282–90.
Watatani Y, Sato Y, Miyoshi H, Sakamoto K, Nishida K, Gion Y, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33:2867–83.
Fukuhara S, Oshikawa-Kumade Y, Kogure Y, Shingaki S, Kariyazono H, Kikukawa Y, et al. Feasibility and clinical utility of comprehensive genomic profiling of hematological malignancies. Cancer Sci. 2022;113:2763–77.
Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041.e1021.
Dobay MP, Lemonnier F, Missiaglia E, Bastard C, Vallois D, Jais JP, et al. Integrative clinicopathological and molecular analyses of angioimmunoblastic T-cell lymphoma and other nodal lymphomas of follicular helper T-cell origin. Haematologica. 2017;102:e148–e151.
Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263:6–21.
Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37:551–568.e514.
Bonfiglio F, Bruscaggin A, Guidetti F, Terzi di Bergamo L, Faderl M, Spina V, et al. Genetic and phenotypic attributes of splenic marginal zone lymphoma. Blood. 2022;139:732–47.
Kataoka K, Iwanaga M, Yasunaga J-i, Nagata Y, Kitanaka A, Kameda T, et al. Prognostic relevance of integrated genetic profiling in adult T-cell leukemia/lymphoma. Blood. 2018;131:215–25.
Heavican TB, Bouska A, Yu J, Lone W, Amador C, Gong Q, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. 2019;133:1664–76.
Yoon SE, Cho J, Kim YJ, Ko YH, Park WY, Kim SJ, et al. Comprehensive analysis of clinical, pathological, and genomic characteristics of follicular helper T-cell derived lymphomas. Exp Hematol Oncol. 2021;10:33.
Johnson WT, Ganesan N, Epstein-Peterson ZD, Moskowitz AJ, Stuver RN, Maccaro CR, et al. TP53 mutations identify high-risk events for peripheral T-cell lymphoma treated with CHOP-based chemotherapy. Blood Adv. 2023.
Cortes JR, Ambesi-Impiombato A, Couronné L, Quinn SA, Kim CS, da Silva Almeida AC, et al. RHOA G17V Induces T Follicular Helper Cell Specification and Promotes Lymphomagenesis. Cancer Cell. 2018;33:259–273.e257.
Ng SY, Brown L, Stevenson K, deSouza T, Aster JC, Louissaint A, et al. RhoA G17V is sufficient to induce autoimmunity and promotes T-cell lymphomagenesis in mice. Blood. 2018;132:935–47.
Leca J, Lemonnier F, Meydan C, Foox J, El Ghamrasni S, Mboumba DL, et al. IDH2 and TET2 mutations synergize to modulate T Follicular Helper cell functional interaction with the AITL microenvironment. Cancer Cell. 2023;41:323–339.e310.
O’Connor OA, Horwitz S, Masszi T, Van Hoof A, Brown P, Doorduijn J, et al. Belinostat in patients with relapsed or refractory peripheral t-cell lymphoma: results of the pivotal phase II BELIEF (CLN-19) Study. J Clin Oncol. 2015;33:2492–9.
Shi Y, Dong M, Hong X, Zhang W, Feng J, Zhu J, et al. Results from a multicenter, open-label, pivotal phase II study of chidamide in relapsed or refractory peripheral T-cell lymphoma. Ann Oncol. 2015;26:1766–71.
Tan D, Phipps C, Hwang WY, Tan SY, Yeap CH, Chan YH, et al. Panobinostat in combination with bortezomib in patients with relapsed or refractory peripheral T-cell lymphoma: an open-label, multicentre phase 2 trial. Lancet Haematol. 2015;2:e326–333.
Ghione P, Faruque P, Mehta-Shah N, Seshan V, Ozkaya N, Bhaskar S, et al. T follicular helper phenotype predicts response to histone deacetylase inhibitors in relapsed/refractory peripheral T-cell lymphoma. Blood Adv. 2020;4:4640–7.
Falchi L, Ma H, Klein S, Lue JK, Montanari F, Marchi E, et al. Combined oral 5-azacytidine and romidepsin are highly effective in patients with PTCL: a multicenter phase 2 study. Blood. 2021;137:2161–70.
Ruan J, Moskowitz A, Mehta-Shah N, Sokol L, Chen Z, Kotlov N, et al. Multicenter phase 2 study of oral azacitidine (CC-486) plus CHOP as initial treatment for PTCL. Blood. 2023;141:2194–205.
Rai S, Kim WS, Ando K, Choi I, Izutsu K, Tsukamoto N, et al. Oral HDAC inhibitor tucidinostat in patients with relapsed or refractory peripheral T-cell lymphoma: phase IIb results. Haematologica. 2023;108:811–21.
Camus V, Thieblemont C, Gaulard P, Cheminant M, Casasnovas RO, Ysebaert L, et al. Romidepsin Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone Versus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone in Patients With Previously Untreated Peripheral T-Cell Lymphoma: Final Analysis of the Ro-CHOP Trial. J Clin Oncol. 2024;42:1612–8.
Chen Z, John Wherry E. T cell responses in patients with COVID-19. Nature Rev Immunol. 2020;20:529–36.
Zheng L, Qin S, Si W, Wang A, Xing B, Gao R, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374:abe6474.
Rosetti F, Madera-Salcedo IK, Rodríguez-Rodríguez N, Crispín JC. Regulation of activated T cell survival in rheumatic autoimmune diseases. Nature Rev Rheumatol. 2022;18:232–44.
Young LS, Rickinson AB. Epstein–Barr virus: 40 years on. Nature Rev Cancer. 2004;4:757–68.
Fujisawa M, Nguyen TB, Abe Y, Suehara Y, Fukumoto K, Suma S, et al. Clonal germinal center B cells function as a niche for T-cell lymphoma. Blood. 2022;140:1937–50.
Meeuwes FO, Brink M, van der Poel MWM, Kersten MJ, Wondergem M, Mutsaers P, et al. Impact of rituximab on treatment outcomes of patients with angioimmunoblastic T-cell lymphoma; a population-based analysis. Eur J Cancer. 2022;176:100–9.
Acknowledgements
The authors thank the hematologists and pathologists who were involved in North Japan Hematology Study Group (Supplementary Appendix) for patient care and Y.Hokama, F.Ueki, and Yoshiko Ito for technical assistance. The supercomputing resources were provided by the Human Genome Center, the Institute of Medical Science, The University of Tokyo.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (JP21H05051), Japan Science and Technology Agency Moonshot R&D Program (JPMJMS2022), and Takeda Science Foundation to K.Kataoka, and by JSPS KAKENHI (JP21H02775) to M.N.
Author information
Authors and Affiliations
Contributions
Y.I.: Formal analysis, investigation, data curation, writing – original draft, writing – review and editing, visualization. J.S.: Investigation, data curation, resources, writing – review and editing. K.Kawamoto: Investigation, resources, writing – original draft, writing – review and editing. K.C.H.: Investigation, resources, writing – review and editing. Y.K.: Formal analysis, writing – original draft, writing – review and editing. M.T.: Formal analysis, writing – review and editing. Y.Saito.: Formal analysis, writing – review and editing. K.Mizuno: Formal analysis, writing – review and editing. S.H.: Formal analysis, writing – review and editing. Y.Mizukami: Formal analysis, writing – review and editing. J.K.: Investigation, writing – review and editing. K.Murakami: Investigation, writing – review and editing. T.T.: Investigation, resources, writing – review and editing. Y.H.: Investigation, resources, writing – review and editing. K.C.: Software, writing – review and editing. A.O.: Software, writing – review and editing. Y.Shiraishi: Software, writing – review and editing. H.M.: Investigation, resources, writing – original draft, writing – review and editing. Y.Matsuno: Investigation, resources, writing – review and editing. K.O.: Conceptualization, investigation, resources, writing – original draft, writing – review and editing, project administration. K.Kataoka: Conceptualization, formal analysis, writing – original draft, writing–review and editing, project administration, funding acquisition. M.N.: Conceptualization, resources, data curation, writing – original draft, writing – review and editing, project administration, funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
K.C.H. has received research funding from NEC Corporation, Eli Lilly, and SEKISUI CHEMICAL. Y.K. has received honoraria from Kyowa Kirin, Nippon Shinyaku, Daiichi Sankyo, and Takeda Pharmaceutical. J.K. has received honoraria from TOMY DIGITAL BIOLOGY, Scrum, and Eisai. T.T. has received honoraria from Astellas Pharma, AstraZeneca, Kyowa-Kirin, SymBio Pharmaceuticals, Sumitomo Pharma, Nippon Shinyaku, Novartis, Janssen Pharmaceutical, and Bristol-Myers Squibb; has received research grants from Asahi Kasei Pharma, Eisai, Ono Pharmaceutical, Kyowa-Kirin, Shionogi, Sumitomo Pharma, Chugai Pharmaceutical, and Nippon Shinyaku; and has received research funding from Astellas Pharma, Otsuka Pharmaceutical, LUCA Science, and Priothera SAS. Y.H. has received honoraria from Eli Lilly, Daiichi Sankyo, AstraZeneca, Merck, Novartis, and Merck Sharp & Dohme; has received research funding from NEC Corporation, Eli Lilly, Shionogi, Daiichi Sankyo, and Sysmex; and has received consultancy fee from NEC Corporation. K.Kataoka has received honoraria from Ono Pharmaceutical, Eisai, Astellas Pharma, Novartis, Chugai Pharmaceutical, AstraZeneca, Sumitomo Pharma, Kyowa Kirin, Janssen Pharmaceutical, Takeda Pharmaceutical, Otsuka Pharmaceutical, SymBio Pharmaceuticals, Bristol Myers Squibb, Pfizer, Nippon Shinyaku, Daiichi Sankyo, Alexion Pharmaceuticals, AbbVie, Meiji Seika Pharma, Sanofi, Sysmex, Mundipharma, Incyte Corporation, and Kyorin Pharmaceutical; has received research funding from Asahi Kasei Pharma, Eisai, Otsuka Pharmaceutical, Ono Pharmaceutical, Kyowa Kirin, Shionogi, Takeda Pharmaceutical, Sumitomo Dainippon Pharma, Chugai Pharmaceutical, Teijin Pharma, Japan Blood Products Organization, Mochida Pharmaceutical, JCR Pharmaceuticals, Nippon Shinyaku, Chordia Therapeutics, and Meiji Seika Pharma; holds individual stocks in Asahi Genomics; and has a patent for Genetic alterations as a biomarker in T-cell lymphomas licensed to Kyoto University and PD-L1 abnormalities as a predictive biomarker for immune checkpoint blockade therapy licensed to Kyoto University. M.N. has received honoraria from Takeda Pharmaceutical, Meiji Seika Pharma, AstraZeneca, and Mundipharma; and has received research funding from Takeda Pharmaceutical and AbbVie. All remaining authors have declared no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ito, Y., Shimono, J., Kawamoto, K. et al. TP53 and CDKN2A alterations define a poor prognostic subgroup in patients with nodal T follicular helper cell lymphoma. Leukemia 39, 1723–1734 (2025). https://doi.org/10.1038/s41375-025-02631-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02631-5