Abstract
Acute myeloid leukemia with mutations in TP53 (TP53mut AML) is fatal with a median survival of 6 months. RNA sequencing on purified AML patient samples showed that TP53mut AML had higher expression of mevalonate pathway genes. Using novel, isogenic TP53mut AML cell lines and primary samples, we determined that TP53mut AML resistance to AML chemotherapy cytarabine (AraC) correlated with increased mevalonate pathway activity, a lower induction of reactive oxygen species (ROS), and a mitochondrial response with increased mitochondrial mass and oxidative phosphorylation. Pretreatment with the statin class of mevalonate pathway inhibitors reversed these effects and chemosensitized TP53mut AML. The geranylgeranyl pyrophosphate (GGPP) branch of the mevalonate pathway was required for TP53mut AML chemoresistance. In addition to its role in mitochondria biogenesis, we identified a novel function of GGPP in regulating glutathione for management of AraC-induced ROS. However, statins alone were inadequate to fully reverse chemoresistance in vivo and in a retrospective study of 364 TP53mut AML patients who received chemotherapy concurrently with a statin. Finally, we identified clinical settings and strategies to successfully target the mevalonate pathway, particularly to address the unmet need of TP53mut AML.
Similar content being viewed by others
Introduction
TP53 mutant acute myeloid leukemia (TP53mut AML) is the paramount clinical challenge in the field of leukemia. For TP53mut AML patients, particularly those with bi-allelic alterations, standard therapies, including cytarabine (AraC)-based regimens as well as hypomethylating (HMA) agents with or without the BCL2 inhibitor venetoclax (Ven) all lead to a similar and dismal median overall survival of 6–9 months [1,2,3,4]. Due to this inherent therapy resistance, international guidelines recommend a clinical trial as the first approach to treatment of TP53mut AML [5].
Our group and others have demonstrated that AML requires enhanced mitochondrial oxidative phosphorylation (OXPHOS) to survive chemotherapy, including AraC and Ven [6,7,8,9,10]. The correlation between chemoresistance and increased mitochondrial activity is especially interesting in the context of TP53mut AML as loss of p53 leads to increased glycolysis and OXPHOS in both solid tumors and a recent AML model [11,12,13]. However, the relationship between increased mitochondrial activity and chemoresistance specifically in TP53mut AML has yet to be addressed. Moreover, direct targeting of mitochondrial metabolism to reverse chemoresistance has been limited by toxicity due to essential mitochondrial functions in non-malignant cells [14]. Novel strategies are needed to identify and target pathways that contribute to mitochondrial metabolism primarily and specifically in cancer cells.
Here, we present transcriptional analysis of human AML samples demonstrating that mevalonate pathway genes are overexpressed in TP53mut compared to more chemosensitive TP53 wildtype (TP53WT) AML samples. The mevalonate pathway is a complex metabolic pathway that branches at the level of farnesyl pyrophosphate (FPP), forming isoprenoids like geranylgeranyl pyrophosphate (GGPP) or other cholesterol derivatives. Importantly, there is interdependence between the mevalonate pathway and mitochondrial metabolism [15, 16]. The mevalonate pathway is also necessary for tumorigenesis and metastatic potential in multiple TP53mut solid tumor models [17,18,19,20,21]. In AML, several studies have implicated the mevalonate pathway in therapy resistance [22,23,24,25,26]. Early phase clinical trials investigating the addition of statins to AraC-based induction therapy showed a promising benefit in patients with relapsed/refractory, poor risk AML [27,28,29] but did not lead to statin incorporation into standard therapy. Notably, TP53mut AML patients should be enriched in this population, but sequencing of TP53 was not routinely performed at the time. Altogether, the role of p53 in AML metabolism and chemoresistance remains poorly understood.
We hypothesized that the mevalonate pathway is required for mitochondria-dependent chemoresistance in TP53mut AML. We addressed this hypothesis with multiple orthogonal approaches, including previously unpublished, multi-institutional, retrospective clinical data, novel, isogenic TP53mut AML cell lines and primary TP53mut AML patient samples. Our studies indicate that the mevalonate pathway should be targeted in chemotherapy resistant TP53mut AML.
Methods
Additional methods provided in the supplement.
Cell culture
Human AML cell lines (Supplementary Table 1E) were maintained in minimum essential medium-alpha (Gibco) supplemented with 10% fetal bovine serum (Hyclone). The cultured cells were split every 2-3 days and maintained in an exponential growth phase. Authenticated MOLM14 and HL60 cells were obtained from DSMZ and the liquid nitrogen stock was renewed every 2 years. MOLM13-TP53 isogenic AML cell lines were generously provided by Dr. Stefen Boettcher and generated as previously described [30]. Cell lines were routinely tested for mycoplasma contamination.
Primary AML specimens
Mononuclear cells (MNCs) from patients with AML were obtained from the Stem Cell and Xenograft Core (SCXC, IRB Protocol Number: 703185) at The University of Pennsylvania (UPenn). All samples were acquired through the SCXC following informed consent on the institutional review board-approved protocol. Samples were collected in accordance with federal and institutional guidelines and provided in a pathologically annotated and de-identified fashion. Sample sizes were chosen based on availability.
Statistics
Statistical analysis was performed in Prism10 using unpaired two-sided Student’s T test, ANOVA, and two-sided Fisher’s exact test, as appropriate for each experiment and indicated in the legends. Multiple test correction was implemented using the Benjamini–Hochberg/FDR approach or the Bonferroni’s multiple comparisons test as indicated in the legends. For multiple comparisons, a range of significant adjusted p values, or FDR values, have been provided. The number of independent, biological replicates performed in the laboratory and used for statistical calculations are noted in the legends. The number of replicates for in vitro cell line assays was at least 3 to allow for statistical analysis. Asterisks denote statistical significance as described in figure legends. Error bars represent standard deviations.
Results
Primary TP53 mut AML RNA sequencing show upregulation of a cholesterol synthesis gene signature
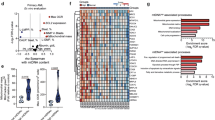
To elucidate biological differences between TP53mut and TP53WT AML, we performed RNA sequencing on untreated primary AML blasts. We obtained 9 bi-allelic TP53mut AML patient samples as well as 21 TP53WT AML patient samples with either a “good” overall survival (OS) (n = 9; median OS not reached) or a “bad” OS (n = 12, median OS 294 days) (Fig. 1A, Supplementary Table 1A). The median OS of the TP53mut AML patients was poor at 65 days (Fig. 1A; log-rank p < 0.0001 versus TP53WT “good” or “bad” outcome patients). Principal component analysis suggested that the TP53mut AML patient samples shared transcriptional similarity whereas the TP53WT AML patient samples were more heterogeneous (Fig. 1B; PC1 48.9% and PC2 15%). Single sample GSEA (ssGSEA) demonstrated that primary TP53mut compared to all TP53WT AML had lower expression of TP53 targets [31] (Fig. 1C, D, Supplementary Table 1B–D). Notably, multiple gene sets involved in cholesterol synthesis were upregulated in TP53mutAML (Fig. 1C, D, Supplementary Table 1C, D). These signatures were recapitulated in a cohort of primary TP53mut (n = 17) versus TP53WT (n = 87) AML samples identified in the BeatAML [32] dataset (Supplementary Fig. 1B–D).
RNA sequencing was performed on highly purified, flow-sorted blasts of primary human AML diagnostic samples from patients with TP53mut (n = 9), TP53WT “bad” outcome AML (n = 12), and TP53WT “good” outcome AML (n = 9). A Kaplan–Meier survival outcomes with statistical analysis by log-rank test. B principal component analysis with principal component 1 (PC1; 48.9%) and PC2 (15%), C GSEA scores from the Hallmark and other denoted gene sets, and D single sample GSEA with calculation of normalized signature scores for “TP53 Targets,” “Hallmark Cholesterol,” and “Maxwell Cholesterol.” Statistical analysis by Student’s T Test. p-values: * = <0.05, ** = <0.01, *** = <0.001.
TP53 mut AML cell lines require the mevalonate pathway to survive AML chemotherapy, AraC
Isogenic TP53mut AML cell line clones with complete loss (M14-Mut1, M14-Mut3) or missense-like mutations (M14-Mut2, M14-Mut4) of the p53 protein were developed using CRISPR/Cas9 editing of the AML cell line, MOLM14, with published guides [33, 34] and single cell cloning via serial dilution. The TP53mut AML clones had consistently higher cell viability in response to AraC than the TP53WT AML clone, M14-WT1, indicating that the TP53mut AML clones were more chemoresistant (Fig. 2A). Furthermore, only the TP53WT clones exhibited a rapid and significant increase in downstream p53 target, p21, at 6 and 24 h following AraC treatment or 6 h after irradiation (Supplementary Fig. 2A, B). To determine the impact of inhibition of the mevalonate pathway on chemoresistance, we pretreated the isogenic clones with the mevalonate pathway inhibitor, rosuvastatin, for 24 h prior to adding AraC. Cell death induced by AraC was augmented by rosuvastatin pre-treatment in all TP53mut and TP53WT AML clones (Fig. 2B, Supplementary Fig. 2C). Importantly, rosuvastatin chemosensitized the TP53mut MOLM14 AML clones to AraC now to the same level as the TP53WT clones (Fig. 2B, Supplementary Fig. 2C). We validated these findings in isogenic TP53mut and TP53WT MOLM13 AML clones [30, 35] (Supplementary Fig. 2D) and the HL60 AML cell line with parental homozygous loss of p53 (Supplementary Fig. 2E). XTT viability assays showed that the combination of AraC and rosuvastatin was synergistic in the isogenic MOLM14 clones using the SynergyFinder tool [36] (Supplementary Fig. 2F). Statin-induced chemosensitization is a class-effect as pitavastatin also chemosensitized the isogenic MOLM14 AML clones to AraC (Supplementary Fig. 2G) at concentrations achievable in patients [22, 37].
A Cell viability of isogenic TP53mut AML clones treated with increasing concentrations of AraC for 24 h and assessed by flow cytometry following staining with AnnexinV and 7AAD (n = 3). B Cell viability of representative isogenic TP53mut and TP53WT AML clones pretreated with 24 h of rosuvastatin (50 μM) followed by an additional 24 h of AraC (1μM) and assessed by flow cytometry following staining with AnnexinV and 7AAD (n = 7). Statistical analysis by one-way ANOVA with false discovery rate (FDR) correction for multiple comparisons. C Schema of the mevalonate pathway with key enzymes highlighted in blue and byproducts in squares. Subunits: FNT = FNTA & FNTB, GGTI = FNTA & PGGT1B, GGTII = RABGGTA & RABGGTB, GGTIII = PTAR1. D Gene expression of select mevalonate pathway genes in isogenic TP53mut and TP53WT AML clones following 24 h of AraC (1 μM) treatment, normalized to GAPDH, and compared to the vehicle for each clone as measured by qRT-PCR (n = 3). E Protein expression of HMGCR normalized to total binding protein (TBP) in isogenic TP53mut and TP53WT AML clones treated with vehicle or 18 h of AraC (1 μM), performed by western blot and quantified by imageJ as presented in relative units (R.U.) of HMGCR to TBP. F CoQ10 as pmol per 2 million cells and G cholesterol as nmol per 2 million cells in the M14-WT1 and M14-Mut1 clones in the same conditions as (B) and quantified by LC-HRMS (n = 5). Statistical analysis for (F, G) by two-way ANOVA with FDR correction for multiple comparisons. Adjusted p-values: * = <0.05, ** = <0.01, *** = <0.001. n is the number of replicates.
Chemoresistant TP53 mut AML cell lines upregulate the mevalonate pathway in response to AraC
To study the mechanism by which statins chemosensitize TP53mut AML, we first addressed the impact of p53 loss on the mevalonate pathway (Fig. 2C) at baseline and in response to AraC. Mevalonate pathway gene expression was assessed by qRT-PCR in representative TP53mut and TP53WT MOLM14 AML clones. AraC induced a time-dependent upregulation of multiple genes in the mevalonate pathway as well as the pathway’s primary transcription factor, SREBF2, in both TP53mut and TP53WT AML clones (Fig. 2D, Supplementary Fig. 2H for time course). The rate-limiting enzyme of the mevalonate pathway and the target of statins, HMGCR, is also regulated at the post-translational level [38, 39]. HMGCR protein expression was significantly increased by AraC in representative clones and highest in those with TP53 mutations (Fig. 2E, Supplementary Fig. 2I). We also detected a significant increase in mevalonate byproducts, CoQ10 and cholesterol, in AraC-treated isogenic clones (Fig. 2F, G, respectively). Rosuvastatin pretreatment abrogated the AraC-induced increase in CoQ10 [Fig. 2F; M14-WT1: F(1,16) = 22, p < 0.001; M14-Mut1: F(1,16) = 26, p < 0.001], while rosuvastatin pretreatment alone or in combination with AraC was associated with a significant increase in cholesterol potentially via uptake (Fig. 2G).
Increased OXPHOS and regulation of ROS correlate with AraC resistance in TP53 mut AML cell lines
Our group and others have demonstrated that AML AraC resistance correlates with increased OXPHOS. However, the impact of TP53 mutations on mitochondrial metabolism in AML is not well understood. We performed a series of functional and structural assays to characterize mitochondria in our isogenic MOLM14 AML clones and TP53mut HL60 cells. In response to AraC, only TP53mut AML cells induced a significant increase in basal and maximum uncoupler-induced oxygen consumption as assessed by Seahorse (Fig. 3A–C, Supplementary Fig. 3A–J). The increase in OXPHOS was associated with a significant increase in mitochondrial mass as determined by measuring the ratio of mitochondrial to nuclear DNA by PCR in isogenic MOLM14 AML clones (Fig. 3D). We next assessed mitochondrial reactive oxygen species (ROS). Treatment of M14-WT1 with AraC was associated with a significant increase in ROS (6-fold increase, p < 0.001) (Fig. 3E). Correlating with chemoresistance, M14-Mut1 and M14-Mut4 exhibited a significantly lower induction of ROS in response to AraC (2-fold increases, p < 0.01 for M14-Mut1 and p = 0.4 for M14-Mut4) (Fig. 3E). Thus, there was a paradoxical improvement in ROS management in TP53mut AML clones despite a significant increase in OXPHOS in response to AraC (Fig. 3A–C, E). Furthermore, AraC-induced ROS and cell viability was at least partially rescued by pretreatment with the antioxidant, reduced glutathione (GSH, Fig. 3E, F), suggesting that AraC-induced ROS directly correlates to induction of cell death.
Oxygen consumption in pmol per minute in A M14-WT1, B M14-Mut1, and C M14-Mut3 treated with vehicle or AraC (1 μM) for 24 h and measured by Seahorse technology (n = 7, n = 8, and n = 8, respectively). AraC versus vehicle fold-changes: Basal OXPHOS of 1.2 (p = 0.05) for M14-WT1, 1.5 (p < 0.001) for M14-Mut1, 2.1 (p = 0.002) for M14-Mut3; Maximum-uncoupler-induced OXPHOS of 1.3 (p = 0.04) for M14-WT1, 2 (p < 0.001) for M14-Mut1, 2.7 (p < 0.001) for M14-Mut3. D Mitochondrial content presented as the ratio of mitochondrial to nuclear DNA measured by PCR in representative isogenic TP53mut and TP53WT AML clones treated with vehicle or AraC (1 μM) for 24 h. E Mitochondrial ROS presented as the ratio to vehicle of mean fluorescence intensity (MFI) of MitoSox in viable cells [determined by Fixable Viability Stain Red (FVS-R)] as measured by flow cytometry in representative isogenic TP53mut and TP53WT AML clones treated with AraC (1 μM, 24 h), glutathione (10 mM, 36 h), the combination, or their respective vehicles (n = 3). F Viability of cells normalized to respective vehicles treated as per (E) and measured by flow cytometry after staining with AnnexinV and 7AAD. G Schema of one-carbon metabolism with bold arrows highlighting the observed pathway increases in AraC-treated cells, particularly in TP53mut AML clones. H Metabolomics in M14-WT1, M14-Mut1, and M14-Mut3 AML cell lines treated with vehicle, 18 h AraC (1 μM), or 24 h AraC (1 μM) (n = 5 per condition) presented as the ratio to respective cell line vehicle. I Relative abundance of dTMP for each condition relative to vehicle (n = 5). Statistical analysis by Student’s T test for (D–F), Z-tests for (H), and two-way ANOVA with Bonferroni’s multiple comparisons test for (I). Adjusted p-values: * = <0.05, ** = <0.01, *** = <0.001. n is the number of replicates.
Metabolomics correlate TP53 mut AML induction of one-carbon metabolism to management of AraC-induced ROS
With the significant differences in AraC-induced OXPHOS and ROS between TP53WT and TP53mut AML cell lines, we next performed metabolomics in representative isogenic clones treated with AraC (Fig. 3G–I, Supplementary Fig. 3K, Supplementary Table 1F). TP53mut AML AraC-treated cells exhibited striking alterations in one-carbon metabolism (Fig. 3G–I, Supplementary Fig. 3K). AraC induced a significant increase in methionine and downstream S-adenosylmethionine (SAM) (Fig. 3H). Only the TP53mut AML clones increased SAM conversion to S-adenosylhomocysteine (SAH), which was associated with an increase in downstream cystathionine, γ-glutamyl-cysteine (γ-Glu-Cys), and GSH (Fig. 3H). GSH is a highly abundant essential cofactor for the enzyme-based glutathione system [40]. Polyamines are also an important ROS scavenger [41,42,43]. AraC was associated with a significant increase in the polyamine precursors arginine and ornithine (Fig. 3H). Only in TP53mut AML clones did AraC induce a significant increase in the SAM-dependent polyamines, spermine and spermidine (Fig. 3H). One of the most significantly increased metabolites in response to AraC was deoxythymidine monophosphate (dTMP; Fig. 3I) and its precursors (Supplementary Fig. 3K), which may reflect an increase in the folic acid cycle. Altogether, this data suggests TP53mut AML-specific induction of one-carbon metabolism, particularly glutathione and polyamines synthesis, for AraC resistance.
AraC-induced alterations in OXPHOS and glutathione synthesis are mevalonate pathway-dependent
Multiple mevalonate pathway byproducts are important for mitochondrial metabolism and redox management. Thus, we next assessed the impact of rosuvastatin on mitochondrial and redox parameters in representative TP53mut and TP53WT MOLM14 AML clones and TP53mut HL60 cells. Rosuvastatin alone decreased basal and max OXPHOS and completely abrogated the AraC-induced increase in OXPHOS (Fig. 4A, Supplementary Fig. 4A–H). We then evaluated intracellular levels of TOMM20, a mitochondria-specific protein that is a surrogate for mitochondrial mass (Fig. 4B), and mitochondrial ROS (Fig. 4C). Cells treated with single agent rosuvastatin trended towards decreased TOMM20 (Fig. 4B) and increased mitochondrial ROS (Fig. 4C). Importantly, rosuvastatin pretreatment in TP53mut MOLM14 AML cells completely abrogated the AraC-induced increase in TOMM20 while significantly increasing mitochondrial ROS to the same degree as M14-WT1 (Fig. 4B, C). We then assessed total glutathione and the ratio of reduced to oxidized glutathione (i.e., GSH/GSSG). In correlation with improved ROS management in TP53mut AML clones, M14-Mut1 and M14-Mut4 responded to AraC with a significant increase in total glutathione, including a specific increase in GSH, which were both abrogated by rosuvastatin pretreatment (Fig. 4D, Supplementary Fig. 4I). This data suggested that a mevalonate pathway-dependent mitochondrial response and ROS management was key for AraC chemoresistance in TP53mut AML.
For (A–D), representative isogenic TP53mut and TP53WT AML clones were treated with rosuvastatin (50 μM) for a total of 48 h with AraC (1uM) added for the last 24 h and the following experiments subsequently performed: A Basal oxygen consumption rate in pmol per minute assessed by Seahorse technology (n = 7), B TOMM20 presented as MFI in viable cells (by FVS-R) assessed by flow cytometry (n = 6, n = 7, n = 4 for respective cell lines), C mitochondrial ROS presented as the ratio to vehicle of MFI of MitoSox in viable cells (by FVS-R) as measured by flow cytometry (n = 3), and D total glutathione presented as μM per 25,000 cells (n = 4, n = 5, n = 3 for respective cell lines). Statistical analysis by two-way ANOVA with FDR correction for multiple comparisons. Adjusted p-values: * = <0.05, ** = <0.01, *** = <0.001. n is the number of replicates.
AraC resistance in TP53 mut AML requires the mevalonate pathway byproduct, GGPP
To determine the specific mevalonate pathway byproducts necessary for mitochondria-dependent chemoresistance, we performed rescue experiments in the representative TP53mut AML clone, M14-Mut1, treated with a rescue agent and rosuvastatin for 48 h with AraC added for the last 24 h (Schema summarizing targets in Fig. 5A). Co-treatment of M14-Mut1 with mevalonate, which is produced by the direct statin target, HMGCR, completely rescued the effect of rosuvastatin on cell viability and OXPHOS (Supplementary Fig. 5A–D). Subsequently, co-treatment of M14-Mut1 with downstream GGPP also rescued the effects of a statin on cell viability (Fig. 5B), OXPHOS (Fig. 5C, Supplementary Fig. 5E, F), TOMM20 as a mitochondria mass surrogate (Fig. 5D), mitochondrial ROS (Fig. 5E), and total and reduced glutathione (Fig. 5F, Supplementary Fig. 5G). GGPP did not rescue the rosuvastatin-induced decrease in coenzyme Q10 (CoQ10) in either M14-Mut1 or M14-WT1 (Supplementary Fig. 5H). These data indicated that rosuvastatin’s activity is specific to the mevalonate pathway, dependent on the GGPP branch of the pathway, and independent of the cholesterol and CoQ10 branches. The presence of GGPP correlated with improved glutathione synthesis and reduced ROS, specifically in the M14-Mut1 clone (M14-WT1 experiments in Supplementary Fig. 6). Finally, we validated that specific geranylgeranyl transferase (GGT) inhibitors, GGTI2417 [44, 45] and GGTI298 [46,47,48], that block the incorporation of GGPP onto targets, but not the farnesyltransferase (FNT) inhibitor, FTI277, recapitulated rosuvastatin’s impact on cell viability alone or in combination with AraC (Fig. 5G, Supplementary Figs. 5I–K, 6M–O).
A Abridged schema of latter half of the mevalonate pathway with essential genes in green and known chemical inhibitors in blue. All experiments in Fig. 6B-I were performed in the M14-Mut1 clone pretreated for 24 h with rosuvastatin (50 μM) and/or either vehicle, GGPP (1 μM) or GGTI2417 (4 μM) followed by an additional 24 h with vehicle or AraC (1 μM) with the subsequent assessment of B cell viability presented as the percentage of annexinV and 7AAD negative cells by flow cytometry with or without GGPP (n = 7), C basal oxygen consumption rate in pmol/minute as assessed by Seahorse technology with or without GGPP (n = 8), D TOMM20 presented as MFI in viable cells (by FVS-R) assessed by flow cytometry with or without GGPP (n = 5), E mitochondrial ROS presented as the MFI of MitoSox in viable cells (by FVS-R) as measured by flow cytometry with or without GGPP (n = 3), F total glutathione presented as μM per 25,000 cells with or without GGPP (n = 5), and G cell viability presented as the percentage of annexinV and 7AAD negative cells by flow cytometry with or without GGTI2417 (n = 3). Statistical analysis by three-way ANOVA with FDR correction for multiple comparisons, including comparison of a treatment with or without the rescue agent or additional inhibitor (i.e., AraC + Statin vs AraC + Statin + GGPP). Adjusted p-values: * = <0.05, ** = <0.01, *** = <0.001. n is the number of replicates.
In vitro statin pretreatment fully abrogates OXPHOS and chemoresistance in primary TP53 mut AML
We then evaluated the efficacy of the drug combination in four primary TP53mut AML samples in vitro (annotations in Supplementary Table 1E). Three of the four samples demonstrated a significant increase in basal and max OXPHOS in response to AraC, which was blocked by rosuvastatin pretreatment (Fig. 6A). Those 3 samples also demonstrated a dose-dependent decrease in colony forming unit ability by the combination of AraC and rosuvastatin (Fig. 6B). Furthermore, the AraC-induced increase in OXPHOS correlated with an increase in mitochondrial mass as measured by flow cytometry after staining for mitochondrial protein TOMM20 in two of the three primary samples (Supplementary Fig. 7A). Of note, these three OXPHOS-inducing primary samples all exhibited bi-allelic TP53 mutations without other AML-driver mutations (Supplementary Table 1E). The fourth TP53mut AML sample, SCXC-7575, had co-mutations in IDH2, ASXL1, KRAS and CEBPA (Supplementary Table 1E) with recent studies correlating IDH2 or ASXL1 co-mutations with improved responses to therapy in TP53mut AML1. SCXC-7575 did not demonstrate a significant increase in OXPHOS in response to AraC (Supplementary Fig. 7B), which correlated with a smaller benefit of combination treatment on CFUs (Supplementary Fig. 7C). These studies suggested that primary human TP53mut AML cells required the mevalonate pathway to survive in vitro AraC treatment and that survival correlated with increased OXPHOS. Furthermore, the absence of an AraC-induced OXPHOS phenotype predicted higher sensitivity to AraC and a decreased benefit of rosuvastatin pretreatment.
A Oxygen consumption rate (pmol/min) assessed by Seahorse technology in previously viably frozen primary TP53mut AML samples that were resuspended in X-Vivo media with 20% BIT serum and 10 ng/mL of cytokines FLT3, SCF, IL3 and IL6, pretreated with rosuvastatin (50 μM) for 24 h followed by AraC (1 μM) treatment for an additional 24 h, and depleted of dead cells via the AnnexinV dead cell removal kit prior to plating for Seahorse analysis. Each sample (SCXC-4708, -6865, -5052) was performed once with 5 technical replicates per condition. AraC vs vehicle fold changes were as follows (basal and max OXPHOS, respectively): SCXC-4708 1.4-fold (p < 0.001) and 1.3-fold (p < 0.001); SCXC-6865 1.9-fold (p = 0.005) and 1.7-fold (p = 0.02); SCXC-5052 1.9-fold (p < 0.001) and 1.7-fold (p < 0.001). B Total number of colony forming units assessed 14 days after plating previously viably frozen primary TP53mut AML samples (SCXC-4708, -6865, -5052) treated on day 0 with rosuvastatin (15 μM or 30 μM) and/or AraC (5 nM or 10 nM) with 3 replicates per condition. Statistical analysis by one-way ANOVA with FDR correction for multiple comparisons. n is the number of replicates.
PDX modeling of TP53 mut AML reveals only partial reversal of AraC resistance by a statin
We established PDX mouse models of TP53mut AML in NOD scid gamma−/− (NSG) mice. Following engraftment of SCXC-5052, mice were treated with vehicle, AraC (50 mg/kg/day intraperitoneally on days 1–5) as routinely performed in the laboratory [6], rosuvastatin (1 mg/kg/day by oral gavage on days 1–7, the maximum dose tolerated in patients [49]) or the combination (schema in Fig. 7A). Mouse bone marrow (BM) and spleen were harvested on days 8–9 and leukemic burden was quantified. Only the drug combination led to a significant though modest decrease in leukemic burden (Fig. 7B). Surviving leukemic cells from AraC-treated mice had a significant increase in mevalonate pathway byproducts, CoQ10 (Fig. 7C) and cholesterol (Fig. 7D), and a significant increase in maximal OXPHOS with a trend towards increased basal OXPHOS (Fig. 7E, Supplementary Fig. 8A). However, rosuvastatin did not fully reverse AraC-induced mevalonate pathway byproduct accumulation or OXPHOS, suggesting inadequate inhibition of the mevalonate pathway by rosuvastatin in vivo (Fig. 7C–E). The experiment was also performed with the primary sample, SCXC-7575. Correlating with the in vitro data, AraC alone significantly reduced leukemic burden in the chemosensitive SCXC-7575 TP53mut IDH2mut ASXL1mut AML PDX mouse model with no additional benefit from rosuvastatin [Supplementary Fig. 8B; F(1,22) = 0.5, p = 0.5]. Furthermore, AraC-persisting leukemic cells did not demonstrate increased basal or max OXPHOS (Supplementary Fig. 8C, D). Altogether, TP53mut AML AraC resistance in vivo correlated with a mevalonate pathway-dependent increase in OXPHOS.
A Schema (created in BioRender) of TP53mut AML PDX model. In brief, primary, human TP53mut AML cells (chemoresistant SCXC-5052) were injected into the tail veins of busulfan-pretreated NSG mice. Engraftment was confirmed on bone marrow aspirates followed by initiation of treatment with vehicle, AraC (50 mg/kg/day IP for D1-5), rosuvastatin (1 mg/kg/day PO for D1-7) or the two drugs in combination. On D8 mice were humanely sacrificed and bilateral femurs/tibias and spleen were harvested (n = 7 mice per condition, early death of 3 mice in AraC condition). B Leukemic burden in millions of hCD45 + hCD33+ cells in the bone marrow and spleen combined for each mouse as quantified with counting beads by flow cytometry. C CoQ10 (ng) and D cholesterol (μg) per 0.5 million magnetic bead-purified hCD45+ leukemic cells from the bone marrow of mice with each circle representing the average of 1 mouse, as assessed by MS (5 replicates per mouse; n = 7, 4, 7, 7 mice for Control, AraC, Statin and AraC + Statin, respectively). E Basal and maximum coupler-induced oxygen consumption in pmol/minute assessed by Seahorse technology in magnetic bead-purified hCD45+ leukemic cells from the bone marrow with each circle representing the average of 1 mouse (n = 4, 3, 2, 2 mice for Control, AraC, Statin and AraC + Statin, respectively). F Kaplan–Meier survival estimates from a retrospective study of newly diagnosed TP53mut AML patients (n = 364) who did or did not receive a concurrent statin during induction therapy (n = 115 and n = 249, respectively). Unless otherwise noted, statistical analysis by two-way ANOVA with FDR correction for multiple comparisons. Adjusted p-values: * = <0.05, ** = <0.01, *** = <0.001. n is the number of replicates. OCR oxygen consumption rate.
In a retrospective analysis of TP53 mut AML patients undergoing induction therapy, co-administration of a statin is safe but does not improve survival
Statins are one of the most prescribed drugs for cardiovascular disease. Thus, a retrospective analysis was performed to determine the survival impact of concomitant statin with induction therapy in patients with newly diagnosed TP53mut AML. Retrospective chart reviews were performed on 364 TP53mut AML patients treated at one of two tertiary centers in the United States between 2013 and 2023 (Supplementary Table 1G). 32% of patients received a statin during induction therapy for TP53mut AML. Statin-treated patients had more indications for a statin, such as a history of coronary artery disease and stroke (Fisher’s exact p < 0.001) but were otherwise well matched. An unadjusted Kaplan–Meier survival estimate suggested a non-significant 1.2-month improvement in median OS in patients who received a statin with induction therapy (Fig. 7F).
Discussion
TP53 mutations confer chemoresistance in AML, rendering standard treatments largely ineffective and leading to dismal patient outcomes [3, 4, 50]. Our work identifies metabolic drivers of AraC resistance in TP53mut AML. We demonstrate for the first time that TP53mut AML requires a mevalonate pathway-dependent adaptive stress response to survive chemotherapy. We provide compelling evidence that the GGPP branch of the pathway is crucial for chemoresistance through its regulation of (1) glutathione to manage chemotherapy-induced ROS and (2) mitochondrial mass and activity. We delve into mechanistic insights and therapeutic implications of targeting the mevalonate pathway in AML.
Our study has furthered our understanding of how cells resist AraC-induced cell death. Using novel isogenic TP53mut MOLM14 AML cell lines, we observe that TP53WT AML cells exhibit a significant increase in mitochondrial ROS in response to AraC. Conversely, TP53mut AML has a higher antioxidant capacity and ability to regulate AraC-induced ROS that correlates with survival. We subsequently show that TP53mut AML cells respond to AraC with significantly increased one-carbon metabolites directed towards synthesis of GSH and polyamines, which both play a role in cellular defense against ROS [40,41,42,43]. Notably, the upregulation of one-carbon metabolism is more robust in TP53mut AML and only the TP53mut AML clones increase levels of the polyamines, spermidine and spermine, in response to AraC. Interestingly, active wildtype p53 is known to inhibit the transcription of genes involved in polyamine synthesis (ARG1, ODC1) and promotes transcription of genes involved in polyamine catabolism (SSAT), which then increases ROS [41]. Altogether, upregulation of one-carbon metabolism may be crucial for TP53mut AML cells to survive AraC, particularly through production of antioxidants and subsequent ROS management.
Survival following AraC treatment is also associated with an increase in mitochondrial mass and OXPHOS, specifically in TP53mut AML cell lines and primary samples in vitro and in vivo. A study evaluating the impact of TP53 missense mutations in solid tumors found that stable endogenous TP53 mutations led to decreased OXPHOS whereas inducible and transient TP53mut models had increased OXPHOS [51]. Our data support these findings in which stable TP53mut AML single cell clones at baseline have decreased OXPHOS (Fig. 3A–C, Supplementary Fig. 3A–J). However, when the cells are stressed with AraC, for example, we observe a significant increase in OXPHOS and mitochondrial mass similar to the inducible/transient TP53mut solid tumor models. Thus, it is crucial to evaluate OXPHOS specifically in the context of stress in TP53mut versus TP53WT cancer models. Furthermore, the critical determinants of chemoresistance may be both the basal level of OXPHOS in AML cells and their ability to dynamically modify OXPHOS in response to stress.
Targeting metabolism, particularly direct mitochondrial inhibition, to reverse chemoresistance remains a challenge in the field [14]. Inhibiting the mevalonate pathway may be an indirect but effective way to overcome toxicity from direct mitochondrial inhibition. However, the optimal mevalonate pathway inhibitor may not be a statin. Combination rosuvastatin and AraC only modestly reduced TP53mut AML leukemic burden in vivo, which correlated with partial abrogation of the mevalonate pathway and OXPHOS (Fig. 7B–E). Furthermore, while our retrospective analysis showed that a broad range of statins at different doses appear to be well tolerated and safe in combination with AML therapy in TP53mut AML patients, there was no survival benefit (Fig. 7F). Statin anti-cancer efficacy in mouse and human models remains limited by (1) statin pharmacokinetics and pharmacodynamics that make it challenging to achieve doses in humans that inhibit geranylgeranylation [52, 53], which is further exacerbated in rodent models [54], (2) rescue of mevalonate pathway inhibition by dietary presence of mevalonate byproducts [55], and (3) highly conserved feedback loops that induce mevalonate pathway upregulation following inhibition [38]. Future preclinical studies should focus on robust mevalonate pathway inhibition in chemoresistant AML.
To optimize mevalonate pathway inhibition for anti-cancer benefit, it is essential to understand the specific role(s) of the mevalonate pathway in chemoresistance. We determined that GGPP is the essential byproduct required for AraC resistance. Other groups have validated the importance of this byproduct in multiple tumor models [21,22,23, 25, 56, 57]. However, the specific function of GGPP has remained elusive. GGPP may have multiple roles, such as regulation of GTPases that (1) play a role in mitochondrial biogenesis and (2) control glutathione levels. Notably, GGPP-dependency for maintaining glutathione has only been previously described in adipose tissue [58]. Future studies will define the specific mechanisms by which GGPP regulates mitochondrial biogenesis and glutathione synthesis and determine the therapeutic benefit of direct targeting of GGPP in TP53mut AML. We anticipate that novel and specific GGT inhibitors, such as GGTI2418 which has shown significant promise in clinical trials for therapy-refractory peripheral T-cell lymphomas [59], will also be efficacious in TP53mut AML.
Data availability
Sequencing data available: GSE295181. Code available at https://github.com/bowmanr/Skuli_TP53_mevalonate .
References
Badar T, Nanaa A, Atallah E, Shallis RM, Craver EC, Li Z, et al. Prognostic impact of ‘multi-hit’ versus ‘single hit’ TP53 alteration in patients with acute myeloid leukemia: results from the Consortium on Myeloid Malignancies and Neoplastic Diseases. Haematologica. 2024;109:3533–42.
Grob T, Al Hinai ASA, Sanders MA, Kavelaars FG, Rijken M, Gradowska PL, et al. Molecular characterization of mutant TP53acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood. 2022;139:2347–54.
Bewersdorf JP, Shallis RM, Gowda L, Wei W, Hager K, Isufi I, et al. Clinical outcomes and characteristics of patients with TP53-mutated acute myeloid leukemia or myelodysplastic syndromes: a single center experience*. Leuk Lymphoma. 2020;61:2180–90.
Pasca S, Haldar SD, Ambinder A, Webster JA, Jain T, Dalton WB, et al. Outcome heterogeneity of TP53-mutated myeloid neoplasms and the role of allogeneic hematopoietic cell transplantation. Haematologica. 2024;109:948–52.
Pollyea DA, Altman JK, Chair V, Assi R, Bachiashvili K, Raj Bhatt V, et al. NCCN guidelines version 2.2025 acute myeloid leukemia (age ≥18 years) continue NCCN guidelines panel disclosures ξ Bone marrow transplantation ‡ Hematology/Hematology oncology Þ Internal medicine † Medical oncology ≠ Pathology * Discussion Section Writing Committee Member. 2025. https://www.nccn.org/home/member-.
Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7:716–35.
Scotland S, Saland E, Skuli N, De Toni F, Boutzen H, Micklow E, et al. Mitochondrial energetic and AKT status mediate metabolic effects and apoptosis of metformin in human leukemic cells. Leukemia. 2013;27:2129–38.
Bosc C, Saland E, Bousard A, Gadaud N, Sabatier M, Cognet G, et al. Mitochondrial inhibitors circumvent adaptive resistance to venetoclax and cytarabine combination therapy in acute myeloid leukemia. Nat Cancer. 2021;2:1204–23.
Jones CL, Stevens BM, D'Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell. 2018;34:724–740.e4.
Stevens BM, Jones CL, Pollyea DA, Culp-Hill R, D’Alessandro A, Winters A, et al. Fatty acid metabolism underlies venetoclax resistance in acute myeloid leukemia stem cells. Nat Cancer. 2020;1:1176–87.
Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D'Alessandro A, et al. The TP53 apoptotic network is a primary mediator of resistance to BCL2 inhibition in AML cells. Cancer Discov. 2019;9:910–25.
Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:2935
Basu S, Gnanapradeepan K, Barnoud T, Kung CP, Tavecchio M, Scott J, et al. Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1α. Genes Dev. 2018;32:230–43.
Yap TA, Daver N, Mahendra M, Zhang J, Kamiya-Matsuoka C, Meric-Bernstam F, et al. Complex I inhibitor of oxidative phosphorylation in advanced solid tumors and acute myeloid leukemia: phase I trials. Nat Med. 2023;29:115–26.
Tricarico P, Crovella S, Celsi F. Mevalonate pathway blockade, mitochondrial dysfunction and autophagy: a possible link. Int J Mol Sci. 2015;16:16067–84.
Ishihara N, Otera H, Oka T, Mihara K. Regulation and physiologic functions of GTPases in mitochondrial fusion and fission in Mammals. Antioxid Redox Signal. 2013;19:389–99.
Freed-Pastor WA, Mizuno H, Zhao X, Langerød A, Moon SH, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–58.
Moon SH, Huang CH, Houlihan SL, Regunath K, Freed-Pastor WA, Morris JP, et al. p53 represses the mevalonate pathway to mediate tumor suppression. Cell. 2019;176:564–580.e19.
Oni TE, Biffi G, Baker LA, Hao Y, Tonelli C, Somerville TDD, et al. SOAT1 promotes mevalonate pathway dependency in pancreatic cancer. J Exper Med. 2020;217.
Kaymak I, Maier CR, Schmitz W, Campbell AD, Dankworth B, Ade CP, et al. Mevalonate pathway provides ubiquinone to maintain pyrimidine synthesis and survival in p189-203-deficient cancer cells exposed to metabolic stress. Cancer Res. 2020;80.
Guo C, Wan R, He Y, Lin SH, Cao J, Qiu Y, et al. Therapeutic targeting of the mevalonate–geranylgeranyl diphosphate pathway with statins overcomes chemotherapy resistance in small cell lung cancer. Nat Cancer. 2022;3:614–28.
Lee JS, Roberts A, Juarez D, Vo TTT, Bhatt S, Herzog L, et al. Statins enhance efficacy of venetoclax in blood cancers. Sci Transl Med. 2018;10.
Zhou C, Li J, Du J, Jiang X, Xu X, Liu Y, et al. HMGCS1 drives drug-resistance in acute myeloid leukemia through endoplasmic reticulum-UPR-mitochondria axis. Biomed Pharmacother. 2021;137:111378.
Pandyra A, Mullen PJ, Kalkat M, Yu R, Pong JT, Li Z, et al. Immediate utility of two approved agents to target both the metabolic mevalonate pathway and its restorative feedback loop. Cancer Res. 2014;74:4772–82.
Xia Z, Tan M, Wei-Lynn Wong W, Dimitroulakos J, Minden M, Penn L. Blocking protein geranylgeranylation is essential for lovastatin-induced apoptosis of human acute myeloid leukemia cells. Leukemia. 2001;15:1398–407.
Mueller J, Schimmer RR, Koch C, Schneiter F, Fullin J, Lysenko V, et al. Targeting the mevalonate or Wnt pathways to overcome CAR T-cell resistance in TP53-mutant AML cells. EMBO Mol Med. 2024;16:445–74.
Kornblau SM, Banker DE, Stirewalt D, Shen D, Lemker E, Verstovsek S, et al. Blockade of adaptive defensive changes in cholesterol uptake and synthesis in AML by the addition of pravastatin to idarubicin + high-dose Ara-C: a phase 1 study. Blood. 2007;109:2999–3006.
Advani AS, Mcdonough S, Copelan E, Willman C, Mulford DA, List AF, et al. SWOG0919: a Phase 2 study of idarubicin and cytarabine in combination with pravastatin for relapsed acute myeloid leukaemia. Br J Haematol. 2014;167:233–7.
Advani AS, Li H, Michaelis LC, Medeiros BC, Liedtke M, List AF, et al. Report of the relapsed/refractory cohort of SWOG S0919: a phase 2 study of idarubicin and cytarabine in combination with pravastatin for acute myelogenous leukemia (AML). Leuk Res. 2018;67:17–20.
Boettcher S, Miller PG, Sharma R, Mcconkey M, Leventhal M, Krivtsov AV, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science. 2019;365:599–604.
Schavolt KL, Pietenpol JA. p53 and Δnp63α differentially bind and regulate target genes involved in cell cycle arrest, DNA repair and apoptosis. Oncogene. 2007;26:6125–32.
Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–31.
Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat Genet. 2018;50:1381–7.
Guernet A, Mungamuri SK, Cartier D, Sachidanandam R, Jayaprakash A, Adriouch S, et al. CRISPR-barcoding for intratumor genetic heterogeneity modeling and functional analysis of oncogenic driver mutations. Mol Cell. 2016;63:526–38.
Schimmer RR, Kovtonyuk LV, Klemm N, Fullin J, Stolz SM, Mueller J, et al. TP53 mutations confer resistance to hypomethylating agents and BCL-2 inhibition in myeloid neoplasms. Blood Adv. 2022;6:3201–6.
Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48:488
Brem EA, O'Brien SM, Becker PS, Jeyakumar D, Pinter-Brown LC, Kirk C, et al. A phase 1 study of adding pitavastatin to venetoclax-based therapy in AML and CLL/SLL. Blood. 2023;142:5940
DeBose-Boyd RA. Feedback regulation of cholesterol synthesis: Sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008;18:609–21.
Lu XY, Shi XJ, Hu A, Wang JQ, Ding Y, Jiang W, et al. Feeding induces cholesterol biosynthesis via the mTORC1–USP20–HMGCR axis. Nature. 2020;588:479–84.
Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta Gen Subj. 2013;1830:3217–66.
Li J, Meng Y, Wu X, Sun Y. Polyamines and related signaling pathways in cancer. Cancer Cell Int. 2020;20:539.
Ha HC, Sirisoma NS, Kuppusamy P, Zweier JL, Woster PM, Casero RA. The natural polyamine spermine functions directly as a free radical scavenger. Proc Natl Acad Sci USA. 1998;95:11140–5.
Zahedi K, Barone S, Soleimani M. Polyamines and their metabolism: from the maintenance of physiological homeostasis to the mediation of disease. Med Sci. 2022;10.
Falsetti SC, Wang D, Peng H, Carrico D, Cox AD, Der CJ, et al. Geranylgeranyltransferase I inhibitors target RalB to inhibit anchorage-dependent growth and induce apoptosis and RalA to inhibit anchorage-independent growth. Mol Cell Biol. 2007;27:8003–14.
Kazi A, Carie A, Blaskovich MA, Bucher C, Thai V, Moulder S, et al. Blockade of protein geranylgeranylation inhibits Cdk2-dependent p27 Kip1 phosphorylation on Thr187 and accumulates p27 Kip1 in the nucleus: implications for breast cancer therapy. Mol Cell Biol. 2009;29:2254–63.
Vogt A, Sun J, Qian Y, Hamilton AD, Sebti M. The geranylgeranyltransferase-I inhibitor GGTI-298 arrests human tumor cells in G0/G1 and induces p21(WAF1/CIP1/SD11) in a p53-independent manner. J Biol Chem. 1997;272:27224–9.
Sun J, Qian Y, Chen Z, Marfurt J, Hamilton AD, Sebti M. The geranylgeranyltransferase I inhibitor GGTI-298 induces hypophosphorylation of retinoblastoma and partner switching of cyclin- dependent kinase inhibitors: a potential mechanism for GGTI-298 antitumor activity. J Biol Chem. 1999;274:6930–4.
Dan HC, Jiang K, Coppola D, Hamilton A, Nicosia SV, Sebti SM, et al. Phosphatidylinositol-3-OH kinase/AKT and survivin pathways as critical targets for geranylgeranyltransferase I inhibitor-induced apoptosis. Oncogene. 2004;23:706–15.
Goss GD, Jonker DJ, Laurie SA, Weberpals JI, Oza AM, Spaans JN, et al. A phase I study of high-dose rosuvastatin with standard dose erlotinib in patients with advanced solid malignancies. J Transl Med. 2016;14:83.
Daver NG, Iqbal S, Renard C, Chan RJ, Hasegawa K, Hu H, et al. Treatment outcomes for newly diagnosed, treatment-naïve TP53-mutated acute myeloid leukemia: a systematic review and meta-analysis. J Hematol Oncol. 2023;16:19.
Eriksson M, Ambroise G, Ouchida AT, Lima Queiroz A, Smith D, Gimenez-Cassina A, et al. Effect of mutant p53 proteins on glycolysis and mitochondrial metabolism. Mol Cell Biol. 2017;37.
Abdullah MI, de Wolf E, Jawad MJ, Richardson A. The poor design of clinical trials of statins in oncology may explain their failure – lessons for drug repurposing. Cancer Treat Rev. 2018;69:84–89.
Juarez D, Fruman DA. Targeting the mevalonate pathway in cancer. Trends Cancer. 2021;7:525–40.
Schonewille M, Freark de Boer J, Mele L, Wolters H, Bloks VW, Wolters JC, et al. Statins increase hepatic cholesterol synthesis and stimulate fecal cholesterol elimination in mice. J Lipid Res. 2016;57:1455–64.
De Wolf E, Abdullah MI, Jones SM, Menezes K, Moss DM, Drijfhout FP, et al. Dietary geranylgeraniol can limit the activity of pitavastatin as a potential treatment for drug-resistant ovarian cancer. Sci Rep. 2017;7:5410
Juarez D, Buono R, Matulis SM, Gupta VA, Duong M, Yudiono J, et al. Statin-induced mitochondrial priming sensitizes multiple myeloma cells to BCL2 and MCL-1 inhibitors. Cancer Res Commun. 2023;3:2497–509.
Jiao Z, Cai H, Long Y, Sirka OK, Padmanaban V, Ewald AJ, et al. Statin-induced GGPP depletion blocks macropinocytosis and starves cells with oncogenic defects. Proc Natl Acad Sci USA. 2020;117:4158–68.
Shu X, Wu J, Zhang T, Ma X, Du Z, Xu J, et al. Statin-induced geranylgeranyl pyrophosphate depletion promotes ferroptosis-related senescence in adipose tissue. Nutrients. 2022;14.
Chew T, Yannakou CK, Ganju V, Grant S, Sebti S, Prince HM. Phase 1 pharmacodynamic and pharmacokinetic study of the geranylgeranyltransferase I inhibitor PTX-100 (GGTI-2418) in patients with advanced malignancies. Blood. 2023;142:1705
Acknowledgements
We thank the UPenn Stem Cell and Xenograft Core, UPenn Cytomics and Cell Sorting Shared Resource Laboratory (RRID:SCR_022376; NIH P3016520), and Mass Spectrometry Shared Resource of the University of Colorado (NIH P30CA046934). Research support provided by: the UPenn Hematology Clinical Research Training Program (T32HL0439, SJS), UPenn Clinical and Translational Research Award KL2 Program (KL2TR001879, SJS), American Society of Hematology (SJS, RLB), American Society of Clinical Oncology (SJS), the NIH (R35CA197731, SMS; P30ES013508, CM; R35CA220483, MCS; K08CA230190, PJS; P30CA16056, PJS, LNM-G; R00CA248460 RLB; U54CA283759, MC), Department of Defense (W81XWH-20-1-0867, DAF), Veterans Merit Award (I01BX000918, MC), Laboratoire d’Excellence Toulouse Cancer (ANR11-LABEX, TOUCAN and TOUCAN2.0, J-ES), INCA (HRHG-MP22-036, IRONAML53, J-ES), Ligue Nationale de Lutte Contre le Cancer (J-ES), Association GAEL (J-ES), Leukemia Research Foundation (RLB), Swiss Cancer League (KFS-4885-08-2019, SB) and Helmut Horten Foundation (SB). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Author information
Authors and Affiliations
Contributions
Supervision: KNM, JES, DAF, BK, MCS, PJS, RLB, AM, MC. Funding acquisition: SJS, KNM, JES, DAF, MC. Conceptualization: SJS, KNM, JES, DAF, BK, MCS, NS, RLB, AM, MC. Methodology/Provision of models/reagents: SJS, BM, GBS, DD, JAR, ES, SB, SMS, AD, CM, GW. Systems Development: BM, GW. Data collection: SJS, AB, MK, PP, AK, OR, GBS, DD, LNMG, CCA, RB, MP, ES, FL, AD, SW, JPX, CM, PJS. Data analysis: SJS, AB, MK, PP, AK, OR, CCA, RB, MP, FL, EN, CM. Bioinformatics: MB, RH, RLB, AM. Statistical analysis: SJS, MB, CCA, EN, RH, CM, RLB, AM. Figure preparation: SJS, CCA, RLB, AM. Interpretation of data: SJS, JAR, ES, CL, KNM, JES, DAF, AD, CM, BK, MCS, GW, NS, RLB, AM, MC. Writing – original draft: SJS. Critical review of the manuscript & writing – review and editing prior to finalization: All authors.
Corresponding author
Ethics declarations
Competing interests
SMS is co-inventor of GGTI-2418/17 and is Scientific Founder of Prescient Therapeutics, the company that licensed GGTI-2418/17 and sponsoring current clinical trials. Competing interests for unrelated studies include the following: CL is a consultant for Bristol Myers Squibb (BMS), AbbVie and Astellas; CL is on advisory boards for Servier, Daiichi, AbbVie, Rigel and Genentech; CL receives research funding from JAZZ and BMS; DAF is a consultant for AbbVie. Other authors declare no relevant competing interests.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. All animal-related experiments received ethical approval from the UPenn Institutional Animal Care and Use Committee (IACUC Protocol #803506). The use of deidentified human samples and data collected from the electronic health record received ethical approval from the UPenn and Roswell Park Comprehensive Cancer Center Institutional Review Boards (IRB Protocols #703185 and #BDR173423, respectively). Informed consent was obtained from all participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Skuli, S.J., Bakayoko, A., Kruidenier, M. et al. Chemoresistance of TP53 mutant acute myeloid leukemia requires the mevalonate byproduct, geranylgeranyl pyrophosphate, for induction of an adaptive stress response. Leukemia 39, 2087–2098 (2025). https://doi.org/10.1038/s41375-025-02668-6
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02668-6