Abstract
Synovial sarcoma is characterized by variable epithelial differentiation and specific SS18-SSX gene fusions. The diagnosis is primarily based on phenotype, but fusion gene detection is increasingly being considered indispensable, with SS18 break-apart fluorescence in situ hybridization (FISH) being favored in many laboratories. However, SS18 FISH assay produces negative or atypical results in a minority of cases, leaving uncertainties in diagnosis and management. Here, we analyzed this challenging subset of SS18 FISH-negative/atypical synovial sarcoma using RNA sequencing and monoclonal antibodies that recognize SS18-SSX and the SSX C-terminus. Among 99 synovial sarcoma cases that were previously subjected to SS18 break-apart FISH, eight cases were reported as negative and three cases were indeterminate, owing to atypical signal patterns. Three of these 11 tumors (two monophasic and one biphasic) harbored novel EWSR1-SSX1 fusions, were negative for SS18-SSX staining, and were positive for SSX C-terminus staining. One monophasic tumor harbored a novel MN1-SSX1 fusion, and showed negative SS18-SSX expression and positive SSX C-terminus staining. Another monophasic tumor carried an SS18L1-SSX1 fusion, and was weakly positive for SS18-SSX, while SMARCB1 expression was reduced. The presence of these novel and/or rare fusions was confirmed using RT-PCR and Sanger sequencing. EWSR1-SSX1 was further validated by EWSR1 FISH assay. The remaining six tumors (five monophasic and one biphasic) showed strong SS18-SSX expression, and RNA sequencing successfully performed in three cases identified canonical SS18-SSX2 fusions. Based on a DNA methylation-based unsupervised clustering, the tumors with EWSR1-SSX1 and SS18L1-SSX1 clustered with synovial sarcoma, while the MN1-SSX1-positive tumor was not co-clustered despite classic histology and immunoprofile. In summary, we discovered novel and rare SSX1 fusions to non-SS18 genes in synovial sarcoma. The expanded genetic landscape carries significant diagnostic implications and advances our understanding of the oncogenic mechanism.
Similar content being viewed by others
Introduction
Synovial sarcoma (SS) accounts for up to 10% of soft tissue sarcomas, and often affects the deep soft tissues of young adults. The monophasic spindle cell subtype consists of dense fascicles of uniform spindle cells, whereas epithelial glands and nests are present in the biphasic subtype. A poorly differentiated pattern is defined by the presence of mitotically active, highly atypical round or spindle cells. Immunohistochemically, SS often shows scattered cytokeratin and more widespread epithelial membrane antigen (EMA) expression, along with intense diffuse TLE1 expression in >90% of cases1, while CD34 expression is typically negative2. Occasional BCOR expression is a confusing finding in the differential diagnosis of sarcoma with BCOR genetic abnormalities3. SS is genetically characterized by SS18 gene fusions including SS18-SSX1, SS18-SSX2, and SS18-SSX4. The alternative fusions SS18L1-SSX1 and SS18-NEDD4 have been reported in two and one cases, respectively3,4,5. SS18-SSX fusion oncoprotein binds to the BAF (SWI/SNF) complex and causes displacement of native SS18 and eviction of SMARCB1 (also known as INI1 and BAF47)6. Reduced immunohistochemical expression of SMARCB1 is accordingly observed in up to 90% of SS samples7,8.
The diagnosis of SS is primarily based on histomorphology and immunophenotype. However, molecular genetic detection of SS18-SSX fusion is increasingly considered indispensable. The fusion can be detected by various methods, including reverse transcription polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH). SS18 break-apart FISH is favored by most laboratories, as it is readily applicable to a single thin FFPE section with good morphological control. As a result, SS18 break-apart FISH is often considered the gold standard of SS diagnosis. However, FISH is not entirely sensitive. In previous studies using >20 cases and a commercially available SS18 break-apart FISH probe, the sensitivity of the SS18 FISH assay was 82–97%9,10,11,12,13, with most studies reporting a rate less than 90%10,12,13, which is generally lower than that of RT-PCR assays9,10,12,14. Even taking into account an uncommon but accepted positive pattern of isolated SS18 5′ signal predominance9,10,15, a significant minority of SS shows negative or atypical/uninterpretable signals. In our experience, we have occasionally observed sarcomas that phenotypically match with SS, yet fail to demonstrate evidence of SS18 rearrangement by FISH. While some of these cases received definitive diagnosis based on classic phenotypes including TLE1 positivity and reduced SMARCB1 expression, other cases were only suspected, leaving uncertainties in diagnosis and clinical management.
The objective of this study was to further characterize this challenging subset of SS18 FISH-negative/atypical SS using tools that have recently become available, including next-generation sequencing and two monoclonal antibodies that recognize SS18-SSX and the SSX carboxy-terminal (C-term) antigen. The latter reagents identify the most common SS18-SSX fusion breakpoint and the C-term portion of SSX, respectively. Previous large studies16,17,18 collectively reported 87–95% sensitivity and 100% specificity for SS18-SSX antibody, and 92–100% sensitivity and 93–96% specificity for SSX (C-term) antibody. Our investigation led to the discovery of novel SSX1 fusions to non-SS18 genes in a small subset of SS.
Materials and methods
Cases
After approval by the institutional review board (Approval No. 2014-089), we retrieved cases that were diagnosed as SS, or as sarcoma most consistent with SS, from the pathology archive of the National Cancer Center Hospital (Tokyo, Japan) from January 2008 to March 2021. All cases were histologically reviewed with available reports by a bone/soft tissue pathologist (AY). After excluding several cases that were found to represent different diseases, the diagnoses of 158 SS cases were confirmed. The patients consisted of 83 males and 75 females, with a median age of 42.5 years (range, 5–87 years). The histological subtype was monophasic in 131 cases and biphasic in 27 cases, and a poorly differentiated pattern was recognized in 40 cases (37 in monophasic; three in biphasic). One or more ancillary tests were previously performed for all cases. Among all cases, 99 were previously subjected to SS18 break-apart FISH assay. Of these, 88 cases were reported as positive (including seven cases with an isolated 5′ pattern), eight were negative, and three were indeterminate owing to atypical signal patterns. Atypical patterns include small isolated 3′ signals with equivocally (narrowly) split 5′/3′ (n = 2) and equivocally (narrowly) split 5′/3′ (n = 1). The FISH-negative/atypical cohort (n = 11, representing 11% of SS tested by FISH) formed the basis of this study. Among the 59 tumors that were not subjected to FISH assays, 12 tested positive for SS18-SSX fusion by RT-PCR and/or sequencing. The remaining 47 cases did not receive molecular testing, among which 36 were tested for SMARCB1 immunohistochemistry and all showed reduced SMARCB1 expression. The remaining 11 (recent) cases were diagnosed based on positive SS18-SSX immunohistochemical staining after the introduction of this recently developed antibody.
Immunohistochemistry
Immunohistochemistry was performed on whole tissue sections. The primary antibodies used were SS18-SSX (E9X9V, 1:2000, Cell Signaling Technology, Danvers, MA, USA) and SSX (C-term) (E5A2C, 1:2000, Cell Signaling Technology). Antigen retrieval was performed using Target Retrieval Solution pH 9 (Dako, Glostrup, Denmark) in water bath. The slides were incubated with the primary antibody for 1 h and subsequently labeled using the EnVision system (Dako). Other immunohistochemical results were retrieved from the pathology reports or were performed afresh when needed, using the following primary antibodies: cytokeratin (AE1/AE3, 1:100; Dako), EMA (E29, 1:100; Dako), TLE1 (M-101, 1:200; Santa Cruz Biotechnology, Dallas, TX, USA), BCOR (C-10, 1:200; Santa Cruz), CD34 (QBEnd10, 1:100; Dako), SMARCB1 (25/BAF47, 1:100; BD Biosciences, Franklin Lakes, NJ, USA), and MUC4 (8G7, 1:400; Santa Cruz).
FISH
SS18 FISH assay was previously performed using the SS18 Split Dual Color FISH Probe (Abbott Molecular, Abbott Park, IL, USA). EWSR1 Split Dual Color FISH Probe (Abbott Molecular) and MN1 Split Dual Color FISH Probe (Cytotest, Rockville, MD, USA) were used in this study when needed. FISH images were captured using the Metafer Slide Scanning Platform (Metasystem, Altlussheim, Germany) and at least 60 non-overlapping tumor cells were evaluated. Tumors in which >20% of the tumor cells showed split 5′/3′ or isolated 5′ signals were considered positive for rearrangement.
Sequencing
Total RNA was extracted from 4 μm-thick FFPE tumor sections using an RNeasy FFPE kit (Qiagen, Hilden, Germany). An RNA sequencing library was prepared using a TruSight RNA Pan-Cancer library kit (Illumina, San Diego, CA, USA). The library was subjected to paired-end sequencing on a MiSeq DNA sequencer (Illumina, cases 1, 2, 3, 4, 6, and 7) or NextSeq sequencer (Illumina, cases 5 and 9). For case 1, the paired-end reads were aligned to known RNA sequences in the RefSeq, Ensembl, and LincRNA databases using the BWA-MEM program. For cases 2, 3, 4, 6, and 7, fusion genes were identified using the RNA-Seq Alignment application (Illumina). For cases 5 and 9, fusion genes were searched using the FusionCatcher ver 1.00 algorithm. Novel and/or rare fusions (EWSR1-SSX1, MN1-SSX1, and SS18L1-SSX1) were confirmed using RT-PCR and Sanger sequencing with the primers listed in Supplementary Table 1.
DNA methylation-based classification
Adequate materials were available for methylation analysis for case 1 (EWSR1-SSX1), case 4 (MN1-SSX1), and case 5 (SS18L1-SSX1). One SS with SS18-SSX1 was also analyzed as a positive control. Genomic DNA was extracted from FFPE sections using a DNeasy Blood & Tissue Kit (Qiagen). Bisulfite modification was performed using 500 ng genomic DNA and an EZ DNA Methylation Kit (Zymo Research Corporation, Irvine, CA, USA). The bisulfite-modified DNA was retrieved using an Infinium HD Assay Kit FFPE Restore Kit (Illumina). The samples were subjected to methylation analysis using Infinium MethylationEPIC BeadChips (EPIC, Illumina).
The methylation profiling classifier developed by the German Cancer Research Center (DKFZ) was used (DKFZ classifier, https://www.molecularneuropathology.org/msp/) to assign subtype scores for each tumor. All computational analyses were performed in R version 4.0.4. Raw signal intensities were obtained from IDAT files of 4 SS samples using the minfi Bioconductor package version 1.34.0. Unprocessed IDAT files from 1077 samples were downloaded from the NCBI Gene Expression Omnibus (GEO) under accession number GSE140686 and used as a reference19. Datasets of 1081 Illumina EPIC and 450k samples were merged using the combineArrays function in minfi. A correction for the type of material tissue (FFPE/frozen) and array (450k/EPIC) was performed with the removeBatchEffect function in the limma package (version 3.44.3). The methylated and unmethylated signals were individually corrected and the beta values were calculated using an offset of 100. After probe-filtering criteria were applied according to the GitHub repository (https://github.com/mwsill/mnp_training), 428,230 probes were retained for subsequent analysis.
To perform unsupervised non-linear dimension reduction, the 10,000 most variable probes (according to standard deviation) were selected among 1077 reference samples. The t-distributed stochastic neighbor embedding (t-SNE)20 plots for the 4 SS and 1077 reference samples were made using the Rtsne package (version 0.15) using 3000 iterations and a perplexity value of 30.
RNA differential expression analysis
Total RNA was extracted from 10 μm FFPE sections using the RNeasy FFPE kit (Qiagen). RNA quality was assessed using DV200 values, and cases with DV200 more than 30% were included for library preparation. An input of 75 ng of total FFPE RNA was used with the TruSeq RNA Access Library Prep Kit, which captures the coding transcriptome/RNA exome (Illumina). Libraries were quantified by real-time PCR using KAPA Library Quantification kits (Kapa Biosystems, Wilmington, MA, USA) on ABI StepOne thermocycler, and sequenced on a Miseq (Illumina) using a 76 cycle, paired-end protocol providing approximately 10–12 million reads per sample. All RNA-seq reads were aligned to the human reference genome (GRCh19) using RNA-Seq Alignment (version 2.0.2) and RNA-Seq Differential Expression (version 1.0.1) (Illumina).
Results
Summary of SS18 FISH-negative/atypical synovial sarcoma
The results are summarized in Table 1. Briefly, 11 SS18 FISH-negative/atypical synovial sarcomas occurred in three men and eight women with a median age of 47 years (range, 5–71 years). Six cases were primary to the somatic soft tissue, while five arose in the internal trunk. Nine cases were monophasic and two were biphasic, with three cases at least focally showing a poorly differentiated pattern. AE1/AE3 and/or EMA was expressed, at least focally, in all ten tumors that were tested. TLE1 was expressed in all seven tumors tested. SMARCB1 expression was reduced in 6 of 11 tumors. The SS18 FISH pattern was negative in eight tumors and atypical/uninterpretable in three tumors. All 11 tumors were tested using SS18-SSX immunohistochemistry, and SSX (C-term) staining was performed in five tumors. Eight tumors were successfully analyzed by targeted RNA sequencing.
Recurrent EWSR1-SSX1 fusions in synovial sarcomas
Cases 1–3 were immunohistochemically negative for SS18-SSX and diffusely positive for SSX (C-term), while SMARCB1 expression was not reduced. RNA sequencing showed that case 1 harbored an in-frame EWSR1 (exon 10, NM_001163285.2)-SSX1 (exon 4, NM_005635.4) fusion, and cases 2 and 3 harbored identical in-frame EWSR1 (exon 10, NM_001163285.2)-SSX1 (exon 5, NM_005635.4) fusions. The EWSR1-SSX1 fusions were confirmed using RT-PCR, Sanger sequencing, and EWSR1 break-apart FISH assays in all three cases. Their clinicopathological data are provided below and are summarized in Tables 1 and 2. The pathological findings are illustrated in Figs. 1 and 2.
A–C Case 1 showed monophasic histology composed of swirling fascicles of uniform spindle cells (A, B). A poorly differentiated round cell component was focally present (C). D–F Case 2 showed papillary and glandular architecture (D). The tumor was biphasic, including fascicles of uniform spindle cells (E) and complex glandular formations of epithelial cells (F). G–I Case 3 displayed long fascicular (G) to whorled (H, necrosis is present in the bottom) spindle cell proliferation. The monomorphic tumor cells harbored a high nuclear/cytoplasmic ratio (I).
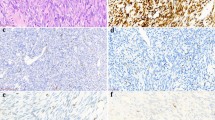
Two tumors each were variably positive for cytokeratin (A case 1) and EMA (B case 3). All were diffusely positive for TLE1 (C case 1). SMARCB1 immunoreactivity was not reduced (D case 3). SS18-SSX immunostaining was negative (E case 2), while SSX (C-term) immunostaining showed diffuse strong reactivity (F case 2, notice both epithelial and spindle cell components are positively stained in this biphasic tumor). Targeted RNA sequencing identified in-frame EWSR1-SSX1 fusions in all three cases. The presence of the fusion genes was confirmed using EWSR1 break-apart FISH assay (G case 3; split green and orange signals indicate gene rearrangements) and RT-PCR/Sanger sequencing (H case 1) for all three cases.
Case 1 was a 45-year-old woman who presented with a 5 cm intramuscular mass in the right thigh. After neoadjuvant chemotherapy, the patient received wide en bloc tumor resection and adjuvant chemotherapy. The tumor locally recurred three times as 1.5–3.5 cm masses over 8 years. Each recurrence was surgically resected and treated with chemotherapy. The tumor ultimately recurred in the inguinal and paraaortic lymph nodes, and pleura, and the patient is alive with disease 10.5 years after the initial surgery. The examined specimen was a resection of the third local recurrence, in which the tumor showed intersecting dense fascicles and whorls of spindle cells with a mitotic count of 15/10 high power fields (HPF; 0.237 mm2). No necrosis was observed. A poorly differentiated round cell pattern (30%), which was focally associated with sclerosis, and myxoid change (<5%) were also observed.
Case 2 involved a 63-year-old woman who presented with dyspnea and cough. A workup identified a 7 cm focally calcified mass in the left lung and mediastinum, pleural dissemination, and pleural fluid. The tumor was partially excised, and the patient is alive on chemotherapy 4 months after presentation. The tumor displayed a biphasic histology consisting of papillary to glandular epithelial structure and intervening fascicles of spindle cell proliferation. The tumor cells were monomorphic with mitotic activity (5/10 HPF). Necrosis and focal metaplastic ossification were present.
Case 3 was a 53-year-old man who had a 5 cm heart mass with pericardial fluid that was identified in a health check-up. The tumor was resected with an initial diagnosis elsewhere of probable benign spindle cell tumor. The tumor locally recurred in 2 years and was excised again. The patient is alive with disease 2.5 years after the initial presentation. The recurrent tumor showed poorly differentiated monophasic histology consisting of fascicular proliferation of monomorphic spindle cells with focal whirling and necrosis. Mitosis was brisk (>20/10 HPF).
Discovery of an MN1-SSX1 fusion
Case 4 showed negative SS18-SSX protein expression, diffuse SSX (C-term) positivity, and intact SMARCB1 expression. RNA sequencing identified an in-frame MN1 (exon 1, NM_002430.3)-SSX1 (exon 5, NM_005635.4) fusion. The presence of the fusion was confirmed using RT-PCR and Sanger sequencing. MN1 break-apart FISH assay did not identify a positive pattern of MN1 rearrangements, despite the sequencing results.
The patient was a 47-year-old woman who presented with a 15-cm mass in the retroperitoneum. The tumor was resected with a segment of inferior vena cava. Multiple lung metastases were identified subsequently, several of which were resected. The patient is currently alive with additional small lung metastases at 14 months after initial surgery. The tumor showed alternating loose and dense areas. The latter areas were composed of intersecting and swirling fascicles of uniform spindle cells with a mitotic count of 3/10 HPF. Nuclear palisading was occasionally prominent. The loose areas displayed myxoid stroma with mast cells. Other findings include hemangiopericytomatous vessels and wiry collagen associated with scattered calcification. No necrosis or poorly differentiated component was observed. The metastatic tumors in the lung showed similar histology to the primary. Additional clinicopathological data are provided in Tables 1 and 2, and the pathological findings are illustrated in Fig. 3.
The tumor showed alternating loose and dense areas (A), the latter composed of intersecting and swirling fascicles of spindle cells (B). Nuclear palisading was occasionally observed (C). The tumor cells were monomorphic and possessed a high nuclear/cytoplasmic ratio (D). Immunohistochemically, the tumor showed focal strong cytokeratin expression (E), diffuse weak EMA expression (F), and diffuse strong TLE1 (G), and BCOR (H) expressions. SS18-SSX immunostaining was negative (I), while SSX (C-term) immunostaining showed diffuse strong reactivity (J). Targeted RNA sequencing identified in-frame MN1-SSX1 fusion transcripts, which were confirmed using RT-PCR/Sanger sequencing (K).
Identification of an SS18L1-SSX1 fusion
Case 5 was weakly positive for SS18-SSX protein and strongly positive for SSX C-term. SMARCB1 expression was reduced. RNA sequencing identified an in-frame SS18L1 (exon 10, NM_198935.3)-SSX1 (exon 6, NM_005635.4) fusion, which was confirmed using RT-PCR and Sanger sequencing.
The patient was a 26-year-old woman with a 7.1 cm intramuscular mass in the right thigh. After neoadjuvant chemotherapy, the patient received wide en bloc tumor resection with negative margins and adjuvant treatment. The patient is alive with no evidence of disease 8.5 years after the surgery. The resected tumor showed intersecting dense fascicles of spindle cells with a mitotic count of 2/10 HPF. Myxoid change (10%) and calcification/ossification (<5%) were present, but a poorly differentiated component was not observed. The tumor was within the skeletal muscle, and there was no clinical and histological evidence of intraneural tumor location despite extensive sampling (15 tumor sections). Additional clinicopathological data are provided in Tables 1 and 2. The pathological findings are illustrated in Fig. 4.
The tumor consisted of fascicular proliferation of uniform spindle cells (A, B). Focal stromal calcification/ossification was observed (C). SMARCB1 immunoreactivity was reduced (D notice strongly stained endothelial cells and immune cells as internal positive controls). SS18-SSX immunostaining was weakly positive (E compare with the intensity observed in an SS18-SSX2-positive tumor, depicted in Fig. 5C), while SSX (C-term) immunostaining showed diffuse strong reactivity (F). Targeted RNA sequencing identified SS18L1-SSX1 fusion transcripts, which were confirmed using RT-PCR/Sanger sequencing (G).
SS18-SSX2 fusions in synovial sarcoma with negative/atypical SS18 FISH
The remaining six cases (cases 6–11) showed strong diffuse SS18-SSX expression. RNA sequencing was successfully performed in cases 6, 7, and 9, and canonical SS18 (exon 10, NM_001007559.3)-SSX2 (exon 6, NM_003147.5) fusion transcripts were identified. Additional clinicopathological data are provided in Table 1 and the pathological findings are illustrated in Fig. 5. The atypical FISH patterns observed in cases 9–11 are shown in Supplementary Fig. 1.
The tumor showed classic monophasic synovial sarcoma histology (A). However, SS18 FISH assay demonstrated no signals indicating gene rearrangement (B all tumor cell nuclei harbored 2 yellow or fused green/orange signals). SS18-SSX immunohistochemistry showed diffuse strong expression of the fusion oncoprotein (C). Targeted RNA sequencing identified an SS18-SSX2 fusion.
DNA methylation-based classification
The tumor in case 5 (SS18L1-SSX1) and one control tumor with canonical SS18-SSX1, matched with “methylation class synovial sarcoma” by the DKFZ classifier (score = 0.99). The two remaining samples (case 1 with EWSR1-SSX1 and case 4 with MN1-SSX1) showed no matching methylation classes (calibrated score ≥0.3). However, t-SNE analysis showed that both cases 1 (EWSR1-SSX1) and 5 (SS18L1-SSX1), along with a control SS18-SSX1-positive tumor, clustered within the SS cluster. The remaining 1 sample (case 4 with MN1-SSX1) was located in the vicinity of, but not within, the Ewing sarcoma cluster (Fig. 6).
The reference cohort (n = 1077) and our synovial sarcoma samples (n = 4; one control with SS18-SSX1, case 1 with EWSR1-SSX1, case 4 with MN1-SSX1, and case 5 with SS18L1-SSX1) were plotted using t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction. Our synovial sarcomas are indicated as red squares. Tumor name abbreviations and the color code of the 62 reference tumors and three control DNA methylation classes were the same as in Koelsche et al.19. Cases 1 and 5 are plotted within the synovial sarcoma cluster along with a positive control, while case 4 is plotted in the vicinity of Ewing sarcoma.
RNA differential expression analysis
As case 4 (MN1-SSX1) was not co-clustered with other synovial sarcomas by DNA methylation profiling, we attempted to probe for distinct mRNA expression patterns for this case. When case 4 (MN1-SSX1) was compared with a group of cases 2, 3, 6, and 7 (tumors with EWSR1-SSX1 and SS18-SSX2), approximately 200 significantly upregulated or downregulated genes were identified with an adjusted p-value <0.05. However, the gene list did not include those that encode relevant immunohistochemical markers that are currently used in sarcoma diagnosis (Supplementary Table 2). We noted two genes, KRT17 and PRAME, expression of which was upregulated and downregulated, respectively, in case 4. Their encoded proteins are occasionally assessed in diagnostic pathology; CK17 and PRAME immunostaining in case 4 was negative. In contrast, there was no significant expression difference in genes whose products can be used for SS diagnosis (TLE1, BCL2, MUC1, KRT19, CD99, and BCOR).
Validation of SS18-SSX immunohistochemistry
We validated the SS18-SSX immunohistochemistry sensitivity by staining 67 randomly selected SS samples that were not included in the above 11 tumors. All were previously tested positive for SS18 FISH, SS18-SSX by RT-PCR/sequencing, and/or reduced SMARCB1 expression. The staining was positive for 66 tumors, with diffuse strong reactivity in 61 cases. These included a single case (25 years/male, chest wall, biphasic) with an SS18-SSX4 fusion. Five tumors displayed weak to moderate intensity, all of which were positive for SS18 FISH. The only tumor (38 years/male, abdominal wall, monophasic) that was negative for SS18-SSX staining was positive for SS18 FISH. This tumor showed conventional histology and reduced SMARCB1 expression.
Discussion
This study highlights an uncommon but recurrent challenge in the use of SS18 FISH for SS diagnosis. We found that 11% of SS that were tested by SS18 FISH assays produced negative or atypical/uninterpretable results, which was consistent with previous reports9,10. Because these events create problems in diagnostic practice, we performed an in-depth clinicopathological and molecular analysis of 11 such cases collected over a 13-year period, taking advantage of newly available RNA sequencing technology and monoclonal antibodies that recognize SS18-SSX and SSX (C-term).
The most remarkable finding of our study was the discovery of novel SSX1 fusions in four cases: EWSR1-SSX1 (n = 3) and MN1-SSX1 (n = 1). These tumors occurred in the extremity (n = 1) and internal trunk (n = 3) in middle-aged adults. Histology of all four tumors fits well within the known spectrum of SS and comprised intersecting fascicles of uniform spindle cells with a high nuclear/cytoplasmic ratio. One tumor had a biphasic epithelial structure and two displayed a poorly differentiated pattern. Additional characteristics observed that are common in SS included an alternating dense and loose pattern (cases 1, 2, and 4), swirling growth (cases 1, 3, and 4), hemangiopericytomatous vessels (case 4), scattered mast cells (cases 1 and 4), nuclear palisading (case 4), wiry collagen deposition (cases 2–4), and calcification/ossification (cases 2 and 4). The immunoprofiles were also typical, including frequent expression of CK, EMA, TLE1, and bcl-2, while CD34 expression was negative. As expected, SS18-SSX staining was negative and SSX (C-term) staining was diffusely positive, but SMARCB1 was not reduced. The incidence of these novel fusions in SS is estimated to be up to 2.5% (4/158), provided that all the SS cases in the initial cohort that did not receive SS18 FISH showed SS18-SSX by sequencing, SS18-SSX immunoreactivity, and/or SMARCB1 reduction.
The histological classification of these cases was partly supported using a DNA methylation-based unsupervised clustering, in which the cases with EWSR1-SSX1, SS18L1-SSX1, and SS18-SSX1 belonged to the known SS cluster. Interestingly, however, the case with the MN1-SSX1 fusion was plotted away from the SS group, and in the vicinity of, albeit not within, the Ewing sarcoma cluster. This was a perplexing finding as the tumor showed a classic SS histology and immunoprofile without any round cells, and the Ewing sarcoma markers CD99 and NKX2.221 were entirely negative. Subsequent RNA differential expression analysis showed no significant expression differences in genes, whose encoded proteins are immunohistochemically assessed widely in the contemporary sarcoma diagnosis. DNA methylation-based sarcoma classification is an emerging research tool that has a high concordance with histological diagnosis, but is associated with occasional misleading or failed prediction22. Whether an MN1-SSX1 sarcoma belongs to SS, as supported by its phenotype, or might represent an altogether new disease, requires future studies with a larger number of cases.
Although all three cases of SS with EWSR1-SSX1 in this cohort showed fascicular spindle cell proliferation, a poorly differentiated round cell pattern, such as that focally observed in case 1, could mimic Ewing sarcoma when this pattern predominates in a sample. This represents a potential diagnostic pitfall if a panel-FISH approach with EWSR1 and SS18 break-apart probes is used. Careful phenotypic characterization including markers such as NKX2.2 and SSX (C-term) would be helpful in diagnosing difficult cases.
EWSR1 (22q12.2) encodes an RNA-binding protein that is a member of the FET family along with FUS and TAF15. EWSR1 protein possesses an N-terminal transcriptional activation domain and a C-terminal RNA-binding domain. In oncology, EWSR1 is best known for its fusion with a number of 3ʹ partner genes, such as EWSR1-FLI1 in Ewing sarcoma. As with other fusion events, the putative EWSR1-SSX1 fusion protein retains the N-terminal transcriptional activation domain (Fig. 7). However, SSX1 does not have a DNA-binding domain, unlike many other EWSR1 fusion partners. MN1 (22q12.1) encodes a transcriptional coactivator, with no homology to other proteins. The MN1 protein has an N-terminal nuclear localization signal, proline-rich sequences, and two polyglutamine stretches, but lacks a DNA-binding domain23. MN1 interacts with several transcription factors including PBX1, PKNOX1, and the E3 ubiquitin ligase RING124. Although originally described as being disrupted in meningioma23, the oncogenic role of MN1 is best known in acute myeloid leukemia (AML), a rare subset of which harbors an MN1-ETV6 fusion25. MN1 overexpression has also been associated with poor prognosis in AML with normal karyotype26. More recently, brain tumors with MN1 alterations, most commonly MN1-BEND2 fusions, were identified to form a distinct DNA methylation class with variable histology (such as astroblastoma), immunophenotype, and clinical outcome27,28. MN1 C-terminal truncating mutations define a distinct syndrome with craniofacial and brain abnormality24,29.
The existence of SS with fusions that lack involvement of SS18 or its paralog SS18L1 sheds new light on SS pathogenesis. The mechanisms of action exerted by the SS18-SSX fusion oncoprotein include disruption of epigenetic control, mainly by disturbing the balance between the polycomb repressive complex (PRC) and the BAF chromatin remodeling complex. In one model, SS18-SSX recruits PRC2 by bridging its member TLE1 and activating transcriptional factor 2 (ATF2), thereby repressing ATF2 targets such as the tumor suppressor genes CDKN2A and EGR130. More recent studies emphasize crucial roles of SS18-SSX-containing “hijacked” BAF complexes, which target and reactivate genes bivalently marked with H3K4me3 and H3K27me36. These BAF complexes also interact with KDM2B, a member of PRC1.1, and bring together BAF and PRC1.1 on unmethylated CpG islands to aberrantly reactivate the expression of developmentally regulated genes31. Detailed functional studies are clearly needed to understand the role of EWSR1-SSX1 and MN1-SSX1 in SS. However, the recurrent presence of EWSR1-SSX1 in three cases, shared 3′ SSX1 to canonical fusions, and the abundant SSX (C-term) immunoreactivity in tumor nuclei suggest that these SSX1 fusions play driver roles. Because neither EWSR1 nor MN1 is a component of the BAF complex, EWSR1-SSX and MN1-SSX1 fusion oncoproteins are not expected to be incorporated into the BAF complex in the same way as SS18-SSX. This hypothesis is partly supported by intact (not reduced) SMARCB1 immunoexpression in all four tumors harboring these new fusions.
However, it is conceivable that the downstream effect of these fusions may converge into the same molecular perturbation incurred by SS18-SSX. It is proposed that the EWSR1 fusion protein mediates liquid–liquid phase separation owing to the prion-like intrinsically disordered region (IDR) found in the EWSR1 N-terminal low-complexity sequence, which is conserved in the fusion product. EWSR1 fusion protein binds with BAF complex without altering the core protein composition likely in these condensates, thereby altering genome-wide transcription patterns32,33. In Ewing sarcoma, EWSR1-FLI1 recruits the BAF complex via the IDR to GGAA microsatellite repeat enhancers to activate tumor-specific target genes32. Similarly, a substantial portion of MN1 consists of an IDR24, and MN1 has recently been shown, in an abstract paper34, to interact with the BAF complex via the IDR in MN1-driven AML. The EWSR1-SSX1 and MN1-SSX1 fusions that we identified conserve IDRs, and thus, it is tempting to speculate that the abnormal pattern of BAF complex recruitment promoted by these fusions might overlap with that induced by SS18-SSX-containing BAF complex, resulting in a shared transcriptional program leading to SS6,35.
In this connection, emerging data suggest an interchangeable role of SS18, EWSR1, and MN1 in fusion-driven oncogenesis. Recent reports documented two soft tissue tumors with SS18-POU5F1 fusions with a myoepithelial phenotype36,37, and one of these tumors was clustered, by RNA expression profiling, with myoepithelial tumors containing EWSR1-POU5F136. Similarly, one case of astroblastoma harboring an EWSR1-BEND2 fusion was reported38, instead of the more common MN1-BEND2 fusions27,39, as was an MN1-PATZ1-positive pediatric intracranial sarcoma with expression and phenotypes resembling EWSR1-PATZ1-positive sarcoma40.
Very recently, Antonescu et al. reported two unusual sarcomas with EWSR1 (exon 7)-SSX1 (exon 5) fusions41. These tumors, which affected the extremities of young adults, showed nests and cords of epithelioid cells in a sclerotic background and were diffusely immunoreactive to cytokeratin, EMA, TLE1, and BCOR, while focally positive for MUC4. These tumors seemed distinct from SS that we describe herein, as they entirely lacked a fascicular spindle cell component. Keratin expression pattern (focal vs. diffuse) and the EWSR1 fusion breakpoint (exon 10 vs. exon 7) were also different. However, further study is required to determine the relationship, as our case 1 focally exhibited sclerosis in a poorly differentiated round cell component (Supplementary Fig. 2), although sclerosis in this specimen could be related to previous excision. We conducted MUC4 staining for cases 1 and 4, and found that case 1 exhibited weak focal expression, which is a known phenomenon in a subset of SS42.
One tumor harbored a known SS18L1-SSX1 fusion. This is the 3rd such case in literature with an identical SS18L1 (exon 10) to SSX1 (exon 6) combination to that reported previously3,4. The tumor occurred in the lower extremity in a young adult, similar to the two previous cases, but unlike them, it was not associated with a peripheral nerve. The tumor displayed a classic monophasic histology and immunoprofile. Notably, SS18-SSX immunohistochemistry was weakly positive, rather than a typical strong intensity. SS18L1 and SS18 are ~60% homologous4,43, and eight of the nine last amino acids of SS18 (QQRPYGYDQ) and SS18L1 (QQRPYGYEQ) before the breakpoint are identical, with the remaining amino acid being similar (Asp vs. Glu), suggesting close antigenicity and cross-reactivity to the SS18-SSX antibody. This adds further value to the SS18-SSX antibody, and weak SS18-SSX immunoexpression along with negative SS18 FISH may predict an SS18L1-SSX1 fusion. Interestingly, SMARCB1 staining was reduced in this case, similar to the tumors with SS18-SSX fusions. The oncogenic mechanism of SS18L1-SSX1 remains unclear, but SS18L1 (also known as CREST) is a component of the BAF complex, and is physiologically involved in the neuron-specific BAF, where SS18L1 replaces SS18 and plays an important role in dendrite development43,44. SS18L1-SSX1 chimeric protein may enter the BAF complex and cause downstream effects similar to SS18-SSX, including BAF47 eviction.
Lastly, 6 of 11 cases tested showed evidence of canonical SS18-SSX fusions despite the negative/atypical FISH results. All six cases showed strongly positive staining for SS18-SSX. Three cases tested by RNA sequencing harbored SS18-SSX2 fusions. All these cases showed classic histology, with five displaying reduced SMARCB1 expression. The reason these tumors demonstrated negative/atypical FISH signals may be cryptic or complex rearrangement that escaped the resolution of FISH, as speculated for other sarcoma types45.
In conclusion, we investigated a small subset of SS that was negative or atypical in the SS18 FISH assay and discovered novel and rare SSX1 fusions to non-SS18 genes. The expanded genetic landscape of SS carries significant diagnostic implications and advances our understanding of the oncogenic mechanism. Our data also underscore the value of next-generation sequencing and recently developed antibodies to SS18-SSX and SSX (C-term) for SS diagnosis.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request and IRB approval.
References
Terry, J. et al. TLE1 as a diagnostic immunohistochemical marker for synovial sarcoma emerging from gene expression profiling studies. Am. J. Surg. Pathol. 31, 240–246 (2007).
Pelmus, M. et al. Monophasic fibrous and poorly differentiated synovial sarcoma: immunohistochemical reassessment of 60 t(X;18)(SYT-SSX)-positive cases. Am. J. Surg. Pathol. 26, 1434–1440 (2002).
Kao, Y. C. et al. BCOR upregulation in a poorly differentiated synovial sarcoma with SS18L1-SSX1 fusion-A pathologic and molecular pitfall. Genes Chromosomes Cancer 56, 296–302 (2017).
Storlazzi, C. T. et al. A novel fusion gene, SS18L1/SSX1, in synovial sarcoma. Genes Chromosomes Cancer 37, 195–200 (2003).
Argani, P. et al. Novel SS18-NEDD4 gene fusion in a primary renal synovial sarcoma. Genes Chromosomes Cancer 59, 203–208 (2020).
McBride, M. J. et al. The SS18-SSX fusion oncoprotein hijacks BAF complex targeting and function to drive synovial sarcoma. Cancer Cell 33, 1128–1141 e1127 (2018).
Arnold, M. A. et al. A unique pattern of INI1 immunohistochemistry distinguishes synovial sarcoma from its histologic mimics. Hum. Pathol. 44, 881–887 (2013).
Ito, J., Asano, N., Kawai, A. & Yoshida, A. The diagnostic utility of reduced immunohistochemical expression of SMARCB1 in synovial sarcomas: a validation study. Hum. Pathol. 47, 32–37 (2016).
Amary, M. F. et al. Detection of SS18-SSX fusion transcripts in formalin-fixed paraffin-embedded neoplasms: analysis of conventional RT-PCR, qRT-PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod. Pathol. 20, 482–496 (2007).
Chuang, H. C. et al. Reappraisal of TLE-1 immunohistochemical staining and molecular detection of SS18-SSX fusion transcripts for synovial sarcoma. Pathol. Int. 63, 573–580 (2013).
Sun, B. et al. The diagnostic value of SYT-SSX detected by reverse transcriptase-polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH) for synovial sarcoma: a review and prospective study of 255 cases. Cancer Sci. 99, 1355–1361 (2008).
Ten Heuvel, S. E., Hoekstra, H. J. & Suurmeijer, A. J. Diagnostic accuracy of FISH and RT-PCR in 50 routinely processed synovial sarcomas. Appl. Immunohistochem. Mol. Morphol. 16, 246–250 (2008).
Terry, J. et al. Fluorescence in situ hybridization for the detection of t(X;18)(p11.2;q11.2) in a synovial sarcoma tissue microarray using a breakapart-style probe. Diagn. Mol. Pathol. 14, 77–82 (2005).
Guillou, L. et al. Detection of the synovial sarcoma translocation t(X;18) (SYT;SSX) in paraffin-embedded tissues using reverse transcriptase-polymerase chain reaction: a reliable and powerful diagnostic tool for pathologists. A molecular analysis of 221 mesenchymal tumors fixed in different fixatives. Hum. Pathol. 32, 105–112 (2001).
Jiang, D. et al. Synovial sarcoma showing loss of a green signal in SS18 fluorescence in situ hybridization: a clinicopathological and molecular study of 12 cases. Virchows Arch. 471, 799–807 (2017).
Baranov, E. et al. A novel SS18-SSX fusion-specific antibody for the diagnosis of synovial sarcoma. Am. J. Surg. Pathol. 44, 922–933 (2020).
Perret, R., Velasco, V., Le Guellec, S., Coindre, J. M. & Le Loarer, F. The SS18-SSX antibody has perfect specificity for the SS18-SSX fusion protein: a validation study of 609 neoplasms including 2 unclassified tumors with SS18-Non-SSX fusions. Am. J. Surg. Pathol. 45, 582–584 (2021).
Zaborowski, M. et al. When used together SS18-SSX fusion-specific and SSX C-terminus immunohistochemistry are highly specific and sensitive for the diagnosis of synovial sarcoma and can replace FISH or molecular testing in most cases. Histopathology 77, 588–600 (2020).
Koelsche, C. et al. Sarcoma classification by DNA methylation profiling. Nat. Commun. 12, 498 (2021).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Yoshida, A. et al. NKX2.2 is a useful immunohistochemical marker for Ewing sarcoma. Am. J. Surg. Pathol. 36, 993–999 (2012).
Lyskjaer, I. et al. DNA methylation-based profiling of bone and soft tissue tumours: a validation study of the ‘DKFZ Sarcoma Classifier’. J. Pathol. Clin. Res. https://doi.org/10.1002/cjp2.215 (2021).
Lekanne Deprez, R. H. et al. Cloning and characterization of MN1, a gene from chromosome 22q11, which is disrupted by a balanced translocation in a meningioma. Oncogene 10, 1521–1528 (1995).
Miyake, N. et al. Gain-of-function MN1 truncation variants cause a recognizable syndrome with craniofacial and brain abnormalities. Am. J. Hum. Genet. 106, 13–25 (2020).
Buijs, A. et al. Translocation (12;22) (p13;q11) in myeloproliferative disorders results in fusion of the ETS-like TEL gene on 12p13 to the MN1 gene on 22q11. Oncogene 10, 1511–1519 (1995).
Heuser, M. et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood 108, 3898–3905 (2006).
Sturm, D. et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164, 1060–1072 (2016).
Tauziede-Espariat, A. et al. Pediatric methylation class HGNET-MN1: unresolved issues with terminology and grading. Acta Neuropathol. Commun. 7, 176 (2019).
Mak, C. C. Y. et al. MN1 C-terminal truncation syndrome is a novel neurodevelopmental and craniofacial disorder with partial rhombencephalosynapsis. Brain 143, 55–68 (2020).
Su, L. et al. Deconstruction of the SS18-SSX fusion oncoprotein complex: insights into disease etiology and therapeutics. Cancer Cell 21, 333–347 (2012).
Banito, A. et al. The SS18-SSX oncoprotein hijacks KDM2B-PRC1.1 to drive synovial sarcoma. Cancer Cell 33, 527–541 e528 (2018).
Boulay, G. et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171, 163–178 e119 (2017).
Linden, M. et al. FET family fusion oncoproteins target the SWI/SNF chromatin remodeling complex. EMBO Rep. 20, e45766 (2019).
Riedel, S. S. et al. SWI/SNF dysregulation through a prion-like domain causes AML. Blood 134, 2512 (2019).
Nacev, B. A. et al. The epigenomics of sarcoma. Nat. Rev. Cancer 20, 608–623 (2020).
Antonescu, C. R., Agaram, N. P., Sung, Y. S., Zhang, L. & Dickson, B. C. Undifferentiated round cell sarcomas with novel SS18-POU5F1 fusions. Genes Chromosomes Cancer 59, 620–626 (2020).
Shenoy, A. et al. Malignant round cell tumor with SS18-POU5F1 fusion: is it a myoepithelial neoplasm, a synovial sarcoma or a new entity? Histopathology 77, 681–684 (2020).
Yamasaki, K. et al. Spinal cord astroblastoma with an EWSR1-BEND2 fusion classified as a high-grade neuroepithelial tumour with MN1 alteration. Neuropathol. Appl. Neurobiol. 46, 190–193 (2020).
Hirose, T. et al. Astroblastoma: a distinct tumor entity characterized by alterations of the X chromosome and MN1 rearrangement. Brain Pathol. 28, 684–694 (2018).
Burel-Vandenbos, F. et al. A polyphenotypic malignant paediatric brain tumour presenting a MN1-PATZ1 fusion, no epigenetic similarities with CNS high-grade neuroepithelial tumour with MN1 alteration (CNS HGNET-MN1) and related to PATZ1-fused sarcomas. Neuropathol. Appl. Neurobiol. 46, 506–509 (2020).
Antonescu, C. R. et al. Sarcomas with sclerotic epithelioid phenotype harboring novel EWSR1-SSX1 fusions. Genes Chromosomes Cancer https://doi.org/10.1002/gcc.22970 (2021 Epub).
Doyle, L. A. et al. MUC4 is a highly sensitive and specific marker for low-grade fibromyxoid sarcoma. Am. J. Surg. Pathol. 35, 733–741 (2011).
Aizawa, H. et al. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303, 197–202 (2004).
Staahl, B. T. et al. Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J. Neurosci. 33, 10348–10361 (2013).
Yoshida, A. et al. CIC break-apart fluorescence in-situ hybridization misses a subset of CIC-DUX4 sarcomas: a clinicopathological and molecular study. Histopathology 71, 461–469 (2017).
Acknowledgements
The authors thank Sachiko Miura, Toshiko Sakaguchi, Chizu Kina, Susumu Wakai, Hiroshi Chigira, Kaori Yamaguchi, and Hiroki Kakishima for superb technical assistance.
Funding
This work was supported in part by JSPS KAKENHI (Grant Number JP21K06919, A.Y.) and the National Cancer Center Rare Cancer Grant (Award Number G007, A.Y.).
Author information
Authors and Affiliations
Contributions
A.Y. designed the project, performed pathology review, and conducted IHC and FISH analyses. Y.A., T.K., E.R., N.H., T.S., H.I. and T.M. performed sequencing. K.Sa., Y.M. and K.I. performed DNA methylation analysis. K.Su., M.K., and Y.Y. and A.K. provided samples and clinical data. A.Y. wrote the original draft, which was reviewed and approved by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The study was approved by the institutional review board at the National Cancer Center Hospital, Tokyo, Japan (No. 2014-089).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yoshida, A., Arai, Y., Satomi, K. et al. Identification of novel SSX1 fusions in synovial sarcoma. Mod Pathol 35, 228–239 (2022). https://doi.org/10.1038/s41379-021-00910-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41379-021-00910-x
This article is cited by
-
Molecular and epigenetic oncogenesis in synovial sarcoma: implications for cancer biology, diagnosis and treatment
Oncogene (2025)
-
Biphasic synovial sarcoma with myoepithelial features: a distinctive variant with a predilection for the foot
Virchows Archiv (2024)
-
Molecularly defined sinonasal malignancies: an overview with focus on the current WHO classification and recently described provisional entities
Virchows Archiv (2024)
-
Novel characteristics for immunophenotype, FISH pattern and molecular cytogenetics in synovial sarcoma
Scientific Reports (2023)
-
Aberrant gene activation in synovial sarcoma relies on SSX specificity and increased PRC1.1 stability
Nature Structural & Molecular Biology (2023)