Abstract
Due to methodological reasons, the X-chromosome has not been featured in the major genome-wide association studies on Alzheimer’s Disease (AD). To address this and better characterize the genetic landscape of AD, we performed an in-depth X-Chromosome-Wide Association Study (XWAS) in 115,841 AD cases or AD proxy cases, including 52,214 clinically-diagnosed AD cases, and 613,671 controls. We considered three approaches to account for the different X-chromosome inactivation (XCI) states in females, i.e. random XCI, skewed XCI, and escape XCI. We did not detect any genome-wide significant signals (P ≤ 5 × 10−8) but identified seven X-chromosome-wide significant loci (P ≤ 1.6 × 10−6). The index variants were common for the Xp22.32, FRMPD4, DMD and Xq25 loci, and rare for the WNK3, PJA1, and DACH2 loci. Overall, this well-powered XWAS found no genetic risk factors for AD on the non-pseudoautosomal region of the X-chromosome, but it identified suggestive signals warranting further investigations.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease and the most common cause of dementia among the elderly. AD is caused by a combination of modifiable and non-modifiable risk factors, including genetics. Currently, more than 80 genetic loci are associated with AD risk, highlighting several underlying biological mechanisms for AD, including APP metabolism, Tau-mediated toxicity, lipid metabolism or immune-related processes [1,2,3,4,5,6]. Greater understanding of the genetics of AD is essential to improve the characterization of the pathophysiological processes involved in the disease. However, although the genetic landscape of AD has been extensively studied on the autosomes, little is known about the association of the X-chromosome gene variants with AD risk. To date, large-scale genome-wide association studies (GWAS) did not include the X-chromosome due to the need of specific analyses to account for its features.
While women carry two copies of the X-chromosome, men are hemizygous, meaning they have one X and one Y chromosome. To maintain balance around allelic dosage between the sexes, X-chromosome inactivation (XCI) occurs in females. This process is where one X chromosome is transcriptionally silenced during female development [7, 8]. The choice of the silenced copy is most often random (random XCI or r-XCI), but inactivation can also be skewed toward a specific copy (skewed XCI or s-XCI). Such XCI ‘skewness’ can be subsequently acquired during life and has been described to increase with age in adults [9,10,11,12]. Importantly, up to one‐third of X‐chromosome genes ‘escape’ inactivation and are expressed from both X‐chromosomes in female cells (escape XCI or e-XCI). However, these tend to be expressed less from the inactive X-chromosome. Notably, all the genes in the pseudoautosomal region (PAR) 1 of the X-chromosome have Y-chromosome homologues and escape inactivation. Additionally, some genes variably escape inactivation: their expression from the inactive X-chromosome differs between individuals or between cells and tissues within an individual [7, 13]. The inactivation process and the distinction between the PAR and non-PAR regions are thus important considerations when performing an X-chromosome-wide association study (XWAS). For all these reasons, the X-chromosome needs to be treated separately from the autosomes in the quality control (QC), the imputation process and the analysis [14, 15], and has usually been excluded from GWAS, including for the large-scale AD ones. Yet, the X-chromosome represents about 5% of the genome in terms of size and number of genes (UCSC Genome Browser, https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&chromInfoPage=), and thus the study of AD genetics remains incomplete.
Several X-chromosome genes have been associated with brain imaging phenotypes [16, 17]. Furthermore, the X-chromosome carries, disproportionately for the whole genome, more than 15% of the known genes related to intellectual disabilities [18]. While genes related to intellectual disabilities are considered to modulate early neurodevelopmental stages well before neurodegenerative processes start, they might impact on the development of cognitive abilities and, potentially, on the establishment of cognitive reserve and brain resilience [19]. Additionally, XCI escape or skewness might contribute to observed sex differences reported in AD [7, 20]. Women have a higher risk of developing dementia than men: in the 65–69 and 85–89 age groups, the prevalence is 1.5% and 24.9% respectively for women, compared with 1.1% and 16.3% for men [21, 22]. However, conclusions regarding differences of incidence across men and women are more mixed [23,24,25,26,27]. Indeed, the sex difference in prevalence can be largely explained by a greater longevity of women, although other factors may also be involved, such as a selective survival bias in men, socio-environmental factors, or different AD-related biological mechanisms between sexes [28]. For example, women have a greater tau burden than men [29,30,31,32,33,34,35,36] and the impact of APOE variants on the disease risk or on Tau concentration differs between males and females [37, 38]. Additionally, in the general population, women have better memory performance [39,40,41,42,43]; some studies reported a faster decline of the global cognition in women but results were mixed in the literature [39, 41, 43, 44]. However, among people with mild cognitive impairment, cognitive decline is reportedly faster in women than in men [43, 45, 46]. Additionally, women live longer with AD compared to men [47, 48]. Consistent with this, in AD mouse models, having two X-chromosomes was associated with reduced mortality [47]. This advantage conferred by a second X-chromosome could partly relate to the KDM6A gene, which escapes inactivation. A variant of the human version of this gene was associated with an increase in this gene’s expression in the brain, and with less cognitive decline in aging and preclinical AD [47]. Finally, in humans, expression/level of other X-linked genes or proteins are reportedly associated with cognitive change or tau pathology in a sex-specific manner [49, 50].
To investigate the impact of X-chromosome genetic variants on AD risk, we conducted an in-depth XWAS on 115,841 AD cases or AD proxy cases and 613,671 controls from the IGAP (International Genomics of Alzheimer’s Project), EADB (European Alzheimer & Dementia Biobank), UK Biobank (UKB) and FinnGen studies (Supplementary Table S1). We considered three approaches to account for the different inactivation states in females, i.e. r-XCI, s-XCI, and e-XCI [15]. In the r-XCI model, males were considered as homozygous females, while they were coded hemizygous in the e-XCI model. In the s-XCI model, a dominance effect was added to the r-XCI model to account for non-random inactivation in females.
Method
Samples
The XWAS is based on 115,841 AD or AD-proxy cases (58% females) and 613,671 controls (55% females) of European ancestry from 35 case-control studies, 2 family studies (LOAD and FHS), and 2 biobanks (UKB and FinnGen) (Supplementary Material and Supplementary Table S1). 55,868 of the 115,841 cases were AD-proxy cases from the UKB. Females were considered as AD-proxy cases if they indicated having at least one parent with dementia [51]. For males, only the mother’s status was used to define the proxy status (Supplementary Material). In a sensitivity analysis including only the diagnosed AD cases, a total of 63,838 AD-cases (59% females) and 806,335 controls (55% females) was considered (Supplementary Table S1).
Additionally, we also analyzed levels of the two cerebro-spinal fluid biomarkers Aβ42 and phosphorylated tau (pTau) in 5522 and 5415 EADB-core samples, respectively [52] (Supplementary Table S2). We also considered 2661 samples with cognitive impairment from three population-based longitudinal studies: AgeCode [53], SNAC-K [54] (both included in EADB-core) and 3C [55] (included in EADI) (Supplementary Table S3). Definitions of cognitive impairment are described in the respective references. These individuals are at increased risk for dementia, but their speed of progression is heterogeneous. For each sample, a Mini-Mental State Examination (MMSE) score was obtained at baseline and in 1 to 5 follow-up sessions.
In addition to the classical autosomal QC, an X-chromosome specific QC was performed prior to imputation for each study (Supplementary Material and Supplementary Table S4). We did not analyze the PAR regions due to a lack of variants on most genotyping chips. Related individuals were excluded from UKB samples but were kept in FinnGen, where related individuals’ exclusion accounts for about 40% of the sample size [56].
Thirty-four studies were imputed with the TOPMed [57] panel (N = 112,690) and three studies were imputed with the 1000 Genomes [58] panel (March 2012) (FHS, CHS and RS, N = 10,102, Supplementary Table S4). The FinnGen was imputed with a Finnish reference panel and the UKB with a combination of 1000 Genomes, HRC [59] and UK10K [60] panels.
Main analyses
Association tests
Since random X-chromosome inactivation is the most frequent case, we considered the r-XCI approach for our main analysis and the s-XCI and e-XCI approaches for secondary analyses. The approaches are described briefly below, while additional details are provided in the Supplementary Material. An overview of the study design is represented in Fig. 1. For all the models, the analyses were adjusted on the principal components (PCs) and/or the genotyping center if necessary (Supplementary Table S4). Dosage or genotype probabilities were used for all studies but FinnGen, where best guessed genotypes were considered (Supplementary Material).
a Main analyses and b sensitivity analyses. Box colors indicate the approach: purple, green, orange and blue represent r-XCI, s-XCI, e-XCI and sex-stratified approaches, respectively. Boxes circled in red are the main r-XCI, s-XCI and e-XCI analyses. *Fixed effect meta-analysis with an inverse-variance weighted approach as implemented in METAL [64]. **Sex-stratified models were adjusted on 1) principal components (PCs) and/or the genotyping center; 2) PCs, center and age; 3) PCs, center, age and APOE.
r-XCI approach
The r-XCI approach is equivalent to an additive genetic model, where males are considered as homozygous females. Males’ and females’ genotypes were thus coded: genotype (G) = {0, 2} and G = {0, 1, 2} respectively. The association test was performed for each study in men and women jointly using an additive logistic regression model for case-control studies, a generalized estimating equation (GEE) model for family studies and a logistic mixed model for biobanks. To account for differences in genotypic variance between sexes, we considered a robust estimate of the variance (or Huber-White Sandwich estimator, accounting for heterogeneity of variance within a regression model, Supplementary Material) for case-control studies [61, 62] and an adjustment on sex for family studies and biobanks (Supplementary Table S5). The association test on proxy status in UKB was performed separately for males and females, and a correction factor of 2 was applied to the association statistics (effect sizes and standard errors) of the female-only model (Supplementary Material) [51, 63]. The results were then combined across studies in a fixed effect meta-analysis with an inverse-variance weighted approach with METAL [64].
e-XCI approach
Under the e-XCI hypothesis, males’ and females’ genotypes were coded G = {0, 1} and G = {0, 1, 2} respectively. Variant effects were estimated separately in females and in males, except in FinnGen, where the variant effects were estimated directly in both males and females combined with an adjustment on sex (Supplementary Table S5). Results were then combined across studies, males and females with a fixed effect meta-analysis, inverse variance weighted approach using METAL. We did not include AD-proxy in the e-XCI meta-analysis. As males and females are related in family studies, only female results from LOAD and FHS were included in the meta-analysis. The sex-stratified models were adjusted on PCs and/or the genotyping center only, except for two ADGC studies (PFIZER and TGEN2) and the CHARGE studies (FHS, RS and CHS), where models were additionally adjusted on age (Supplementary Table S6 and Supplementary Material).
s-XCI approach
For the s-XCI approach, males’ and females’ genotypes were coded G = {0, 2} and G = {0, 1, 2} respectively. A general genotypic model, including both an additive and a dominance variable, was estimated in females from case-control studies to account for non-random inactivation through the dominance variable, which equals 1 in female heterozygotes, and 0 otherwise. The χ2 test of the dominance effect was then added to the χ2 test of the additive effect estimated under r-XCI, which results in a two degree of freedom (df) test of the association of the variant with AD risk including its potential skewedness [62, 65] (Supplementary Table S5). We did not include family studies and biobanks in the s-XCI approach.
While analyses and QC of the results (see below) were performed with the coding scheme described above, odds-ratio and confidence intervals are provided on the real XCI scale, i.e. G = {0, 1} for males and G = {0, 0.5, 1} for females under r-XCI and s-XCI, but G = {0, 1} for males and G = {0, 1, 2} for females under e-XCI (Supplementary Table S5).
Sex-stratified analyses
As the XCI mechanism induces variability across females, one might expect stronger effects in males compared to females; we therefore performed an additional sex-stratified analysis and compared the variant effect sizes in males and females. Proxy cases were not included in this analysis. We combined the results across studies in males and females separately with a fixed effect meta-analysis and inverse-variance weighted approach using METAL [43, 64]. The variant effect sizes of males and females were then compared with a Wald test (Supplementary Material).
Quality control of the results and definition of associated loci
A QC of the results was carried out for all the studies. We filtered out variants with at least one missing datum (on effect, standard error, or p-value), an absolute effect size greater than 5, or an imputation quality less than 0.3. We also filtered out the variants whose effective allele count (product of the imputation quality and the expected minimum minor allele count between the cases and the controls) was less than 5, and less than 10 for LOAD [66]. For datasets imputed with 1000 G and the UKB, we excluded variants for which the conversion of position or alleles from GRCh37 to GRCh38 was not possible or problematic, and variants with a difference in frequency >0.5 compared with the reference panels TOPMed or 1000 G.
After the meta-analysis, we filtered the variants analyzed in less than 40% of AD cases (considering the effective sample size of females UKB-proxy, which is the raw sample size divided by four [51]), variants with heterogeneity p-value < 5 × 10−8 and variants where the difference between the maximum frequency and the minimum frequency across studies was higher than 0.4.
Inflation of the test statistics was checked in each study and in the meta-analysis by computing a genomic inflation factor λ with the median approach implemented in the GenABEL 1.8-0 R package [67], on common variants in low LD (r2 < 0.2) (Supplementary Material). A signal was considered genome-wide or X-chromosome-wide significant in either approach if associated with AD risk with P ≤ 5 × 10−8 or P ≤ 1.6 × 10−6. This X-chromosome wide threshold is based on R = 3.12%, the relative number of tests performed on the X-chromosome (n = 888,213, r-XCI approach) versus on the autosomes (n = 27,549,394) in the EADB-core study, the largest dataset imputed with the TOPMed reference panel. As the genome-wide threshold of 5 × 10−8 corresponds to the Bonferroni correction for one million tests, we computed the corresponding threshold for the X-chromosome as 0.05 / (R*1,000,000) = 1.6 × 10−6.
Sensitivity analyses
To account for potential results that we may have missed because of false negatives related to proxy cases or biobanks, we performed sensitivity analyses on the whole X-chromosome excluding these samples for the r-XCI approach and excluding biobank samples for the e-XCI approach (in the first place, proxy cases were not included in the e-XCI analysis and samples from biobanks, including proxy cases, were not included in the s-XCI analysis).
Additionally, several sensitivity analyses of the identified signals were performed. As the robust r-XCI model can generate false positives in the case of differences of frequency between males and females, we performed a sensitivity analysis by adjusting it on sex rather than using a robust variance for the r-XCI signals. The results were obtained by meta-analyzing the sex-stratified models for all case-control studies and UKB, and a sex-combined model adjusted on sex for FinnGen, with males coded as homozygous females for all models (family studies were excluded) (Supplementary Material, Supplementary Table S5). Sensitivity analyses including an adjustment on age and the number of APOEε4 and APOEε2 alleles were also performed for all signals. Results were obtained from the meta-analysis of adjusted sex-stratified models with the adequate coding of males and excluding family studies. Finally, a sensitivity analysis was performed using a stricter imputation quality filter (r2 > 0.8).
Biomarker and cognitive decline analyses
We tested the association of Aβ42 and pTau with the genotypes of each common index variant in the EADB-core samples. The analyses were performed separately in 11 batches from 7 European countries (Supplementary Table S2). Following the protocol used by Jansen et al. 2022 [52], we applied a linear regression of the normalized log-transformed levels of Aβ42 and pTau, adjusted for sex, age, assay type (if applicable), and ten PCs, using SNPTEST ‘expected’ method [68]. Genotype probabilities were used for all batches. Additionally to the filters on variants with missing datum, variants with minor allele frequency or MAF < 1% were filtered out, and for batches with less than 250 samples, variants with MAF < 5% were also excluded. We applied this model to both the r-XCI and e-XCI approaches, using a meta-analysis of sex-combined models with an r-XCI coding of the genotypes and a meta-analysis of sex-stratified models with an e-XCI coding of the genotypes, respectively.
Evaluating the link of AD-related X-chromosomal variants with cognitive decline may inform about their contribution to the trajectory of the disease. An association study of cognitive decline was thus performed for the common index variants of the X-chromosome wide significant signals using samples with cognitive impairment from longitudinal studies. Both the r-XCI and e-XCI approaches were considered. The association tests were performed in each study in males and females separately using a linear mixed model. The models tested the association between the normalized MMSE score [69] and the index variant dosage. To account for the random effect between the individuals, we included a “Time” variable, representing the time between the baseline and each follow-up session for each individual. Females were coded G = {0,0.5,1} and G = {0,1,2} for the r-XCI and e-XCI approaches, respectively, and males were coded G = {0,1} in both approaches. Each model was adjusted for age at baseline and four PCs. We also performed sex-combined quadratic models -including the squared effect of “Time”- adjusted on sex, for both r-XCI and e-XCI approaches, as sensitivity analyses [70]. The sex-combined quadratic models were only computed in 3C and AgeCode (the quadratic models did not converge in SNAC-K due to low sample size). We then meta-analyzed together the results of the male-only and female-only linear models of all three studies, and meta-analyzed together the linear and quadratic effects of the sex-combined quadratic model for both approaches.
The significance threshold used for the Aβ42, pTau and cognitive decline association test is 4.17 × 10−3, which corresponds to the Bonferroni correction for the 4 independent common variants analyzed for 3 phenotypes.
Colocalization with brain tissue eQTL and pQTL
We performed a genetic colocalization of our hits with brain tissue pQTL and eQTL (protein and expression Quantitative Trait Loci, respectively) for all protein-coding genes within 500 kb of each common index variant, using the “coloc.abf” function from the coloc R-package (version 5.2.3). The brain tissue QTL data were extracted from Wingo et al. 2023 [71], GTex (11 brain tissues) and CommonMind; the last two were processed by the eQTL Catalog (https://www.ebi.ac.uk/eqtl/Data_access/). From the GTex data, we identified 20 genes within 500 kb of the common index variants. We performed the colocalization analyses using the results of the r-XCI or e-XCI analysis where each AD association signal was identified (either the meta-analyses including AD-proxy cases, the diagnosed AD cases meta-analysis or the meta-analysis excluding biobanks). A colocalization between a brain tissue eQTL or pQTL of a gene and an AD-association signal was considered significant when PP4 > 0.75 (posterior probability that both traits are associated and share a single causal variant) [2].
Differential expression and methylation analyses
To evaluate the biological significance of the genes in the identified loci, we examined differential expression and methylation data in studies comparing AD vs control brains and assessing amyloid plaque burden. We considered 19 genes located at +/- 500 kb of the index variants.
To explore differential expression of our associated loci, we analyzed postmortem brain pathology expression data from individuals of European ancestry using RNA-seq data obtained from temporal lobe and pre-frontal cortex (GEO: GSE44772, GSE33000, GSE118553). Logistic regressions were performed in males and females separately and were adjusted on age at death (age at last visit for clinical AD diagnosis), postmortem interval, RNA integrity, APOE ε4 status, and first 3 genomic principal components. A significance threshold of 1.32 × 10−3 was considered, corresponding to a Bonferroni correction for 19 genes analyzed in two subgroups, that being 38 tests.
Differential methylation of associated loci was assessed by using the DNA Methylation in Aging and Methylation in AD (MIAMI-AD) database (miami-ad.org) [72] which collates results from epigenome-wide association studies in aging and AD. Studies in the database meet two main criteria: 1) having more than 100 total subjects and 2) conducting a genome-wide study of more than 100k CpGs. Details of individual studies can be found on the MIAMI-AD website. Here we queried genes within the associated loci for association with AD neuropathology or dementia, in males and females separately, but also in the combined sample. A significance threshold of 4.39 × 10−4 was considered, corresponding to a Bonferroni correction for 19 genes analyzed for 2 phenotypes in 3 subgroups, hence a total 114 tests.
X-linked intellectual disability (XLID) enrichment analysis
We also tested the enrichment of AD association signals in XLID genes in each approach. We first performed gene-based analyses for each approach (Supplementary Material) and then compared the enrichment of AD association signals in the 156 XLID genes included in the gene-based analysis (out of 164 XLID genes [18]) with the rest of the X-chromosome genes, using MAGMA v1.08 [73].
Results
XWAS overview
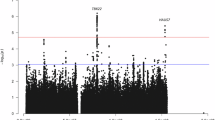
A total of 666,264, 442,001 and 438,420 variants - including 288,320, 276,902 and 263,169 common variants (MAF ≥ 1%) - were analyzed in the r-XCI, e-XCI and s-XCI approaches, respectively. We observed a minor deviation from expected p-values in the r-XCI and e-XCI models (median genomic inflation factor λ = 1.074 and 1.087, respectively) and a deflation in the s-XCI model (median λ = 0.735) (Supplementary Material, Supplementary Figs. S1–S3 and Supplementary Table S7). We did not identify any genome-wide significant signals (P ≤ 5 × 10−8) in any of the models (Figs. 2–4). However, five loci exhibited signals that were X-chromosome-wide significant (P ≤ 1.6 × 10−6) in the r-XCI approach; the index variants were common for the Xp22.32, FRMPD4 and Xq25 loci, and rare for the PJA1 and TMEM187-G6PD/IKBKG loci (Fig. 2, Table 1 and Supplementary Table S8). Additionally, a rare variant in the WNK3 gene was X-chromosome-wide significant in the e-XCI analysis (Fig. 3). No X-chromosome-wide significant signal was found in the s-XCI analysis (Fig. 4). As expected, we observed correlated results between the r-XCI and e-XCI meta-analysis results (Supplementary Table S9).
Association results of a the meta-analysis including AD-proxy cases, b the diagnosed AD cases meta-analysis and c the meta-analysis excluding biobanks. The red and blue lines represent the genome-wide significant threshold (5 × 10−8) and the X-chromosome-wide significant threshold (1.6 × 10−6), respectively. The labels show the closest protein-coding gene (according to GENCODE release 45, https://www.gencodegenes.org/human/releases.html) to the index variant of each X-chromosome-wide significant locus.
In the sensitivity models including only diagnosed AD-cases or excluding biobank samples, we did not identify any genome-wide significant signals among X-chromosome variants either (Figs. 2 and 3 and Supplementary Figs. S1 and S2). However, we identified an X-chromosome-wide significant signal at a common index variant in the DMD locus in the r-XCI meta-analysis excluding biobanks, and at two rare index variants in the WNK3 and DACH2 genes in the r-XCI meta-analysis excluding AD-proxy cases (Table 1 and Fig. 2).
In either the male-only or female-only meta-analyses, we did not identify any genome-wide nor X-chromosome-wide significant signals (Supplementary Fig. S4). We also did not observe any genome-wide nor X-chromosome-wide significant difference of effect between males and females for any X-chromosome variants (Supplementary Fig. S4).
Detailed description of the X-chromosome-wide significant loci
In more details, rs4364769 (MAF = 0.12, OR = 1.079 [1.048–1.110], P = 2.55 × 10−7) was identified as the index variant of the Xp22.32 locus in the r-XCI meta-analysis (Table 1 and Supplementary Fig. S5). The odds-ratio estimate of rs4364769 shows some variability across sensitivity analyses but confidence intervals overlap (Supplementary Table S8). The index variant of the Xp22.32 signal is located more than 300 kb from the closest protein coding gene, NLGN4X (Neuroligin 4 X-Linked).
The index variant in the FRMPD4 (FERM and PDZ Domain Containing 4) locus was rs5933929 (MAF = 0.38, OR = 0.952 [0.935–0.970], P = 1.98 × 10−7) in the r-XCI meta-analysis (Table 1 and Supplementary Fig. S6). This variant is located in an intron within some transcripts of FRMPD4. The odds-ratio of rs5933929 was consistent across sensitivity analyses (Supplementary Table S8).
The common variant rs5972406, located in an intron of the DMD dystrophin gene, was X-chromosome-wide significant only in the r-XCI meta-analysis excluding biobanks (MAF = 0.075, OR = 1.143 [1.083–1.207], P = 1.16 × 10−6, Table 1 and Supplementary Fig. S7). The odds-ratio estimate was lower in the analysis including AD-proxy cases (OR = 1.075 [1.037–1.113], P = 6.75 × 10−5), but confidence intervals overlap (Supplementary Table S8).
rs191195705 was the index variant in the Xq25 signal in the r-XCI meta-analysis (MAF = 0.11, OR = 0.925 [0.896–0.954], P = 7.09 × 10−7, Table 1 and Supplementary Fig. S8). Here the males and the UKB-proxy males carried a large part of the observed effect, leading to a lower signal in the sensitivity analyses excluding proxy or biobank cases, or in the female-only compared to the male-only meta-analyses (Supplementary Table S8 and Supplementary Fig. S8). However, the difference of effect between males and females was not significant (P = 0.51, Supplementary Table S8). rs191195705 is over 500 kb from the closest protein coding gene, GRIA3 (Glutamate Ionotropic Receptor AMPA Type Subunit 3).
Other signals had rare index variants which were not analyzed in the ADGC and CHARGE studies, due to their smaller sample sizes (Supplementary Figs. S9–S12). rs189139822, located in an intron of WNK3, was identified in the r-XCI (MAF = 9.70 × 10−3, OR = 1.481 [1.263–1.735], P = 1.29 × 10−6) and e-XCI (MAF = 9.10 × 10−3, OR = 1.343 [1.192–1.513], P = 1.20 × 10−6, Supplementary Table S8) meta-analyses excluding AD-proxy cases (Table 1 and Supplementary Fig. S8). The odds-ratio of rs189139822 was consistent across studies and sensitivity analyses (Supplementary Table S8). However, the imputation quality for the variant was low (r2 < 0.6) in many studies (Supplementary Fig. S8).
rs771148434, the index variant of the PJA1 (Praja Ring Finger Ubiquitin Ligase 1) signal, was very rare (MAF = 8.00 × 10−4) and analyzed only in the EADB-core and UKB-proxy studies. Detected in the r-XCI meta-analysis (OR = 3.107 [1.967–4.910], P = 1.18 × 10−6, Table 1 and Supplementary Fig. S10), this variant was excluded from most of the sensitivity analyses due to its rarity (Supplementary Table S8).
The rs1326297223 index variant is located in an intron of DACH2. It was identified in the r-XCI meta-analysis excluding AD-proxy cases (MAF = 2.10 × 10−3, OR = 2.281 [1.629–3.192], P = 1.56 × 10−6, Table 1 and Supplementary Fig. S11), and its odds-ratio was consistent across studies and sensitivity analyses (Supplementary Table S8).
Three rare variants, all in high LD (r2 > 0.8), were associated at the X-chromosome-wide significance threshold with AD-risk in the TMEM187-G6PD/IKBKG (Transmembrane Protein 187, Glucose-6-Phosphate Dehydrogenase and Inhibitor Of Nuclear Factor Kappa B Kinase Regulatory Subunit Gamma) locus (Supplementary Table S10). They were only analyzed in the EADB-core and UKB-proxy studies, but the signal was heterogeneous across studies (I2 = 56.5 for the index variant rs782044000, Supplementary Fig. S12), and carried mainly by EADB-core. In this study, most carriers were from Italy and Greece, and the signal disappeared when excluding Greek samples (P = 0.43 for rs782044000), or when meta-analyzing the per-country results in EADB-core (P = 0.12 for rs782044000, Supplementary Material). One of the three index variants, rs5030868, is the G6PD Mediterranean mutation, which is much more frequent in the Mediterranean region than in the rest of Europe [74, 75]. We thus considered this signal to be falsely inflated in EADB-core due to this population structure.
Biomarker and cognitive decline associations
We further investigated the association of the common index variants of the suggestive signals with CSF biomarkers and cognitive decline. We found no significant association (P < 4.17 × 10−3) with Aβ42 or pTau with any common index variant, whatever the approach (Supplementary Table S11), but the FRMPD4 index variant rs5933929 was significantly associated with cognitive decline in both the r-XCI and e-XCI approaches (P = 2.75 × 10−3 and 3.30 × 10−3 respectively), and the direction of effect of the cognitive decline and AD-risk associations were consistent (Supplementary Table S12).
Colocalization with brain tissue eQTL and pQTL
We did not find any significant colocalization of our AD association signals with any brain tissue eQTL or pQTL signal (Supplementary Table S13).
Differential expression and methylation
We also examined expression and methylation data of the genes within the suggestive loci.
The genes FRMPD4 (in males), GRIA3 (Xq25, in females), TSR2 (in the WNK3 locus), and EDA (in the PJA1 locus) showed significant differential expression (P < 1.32 × 10−3) in temporal lobe tissue (Supplementary Table S14). Expression of EDA was also significantly associated with AD status in female pre-frontal cortex tissue. Amyloid plaque burden was found to be significantly associated (P < 4.39 × 10−4) with methylation changes in FRMPD4, ARHGAP6 (in the FRMPD4 locus), and DMD.
XLID gene enrichment
All the suggestive loci contain XLID putative causal genes, except DACH2. However, we did not detect an enrichment of the AD association signals in the XLID genes in any of the analyses (P = 0.37, 0.33 and 0.53 in r-XCI, e-XCI and s-XCI approaches, respectively).
Discussion
We conducted the most comprehensive XWAS on AD to date, including 115,841 AD or AD-proxy cases and 613,671 controls and using three complementary models to account for the complexity related to the X-chromosome. Importantly, 52,214 clinically diagnosed AD cases were included, allowing to assess the impact of proxy-AD or biobank cases on the results. Despite not detecting any genome-wide significant signals regardless of the approach used, seven X-chromosome-wide significant loci passed our post-analysis QC. Index variants were common in four loci; the signal in the FRMPD4 locus was consistent across the sensitivity analyses, showing strong robustness, while the other signals in Xp22.32 (NLGN4X), Xq25 (GRIA3) and DMD showed some variability. Robustness of the results was more difficult to assess for the rare index variants of the WNK3, PJA1, and DACH2 loci.
FRMPD4 (FERM and PDZ domain containing 4) is mostly expressed in brain tissues (GTex Portal, https://gtexportal.org/), and showed differential expression in male temporal lobe between AD cases and controls. Through its interaction with other proteins, the FRMPD4 protein is involved in the regulation of the morphogenesis and density of dendritic spines, and in the maintenance of excitatory synaptic transmission [76]. FRMPD4 is an X-linked intellectual disability gene [77] and is associated with low educational attainment [78]. The association of the index variant of the FRMPD4 locus with cognitive decline could thus be linked to a lower cognitive reserve. The associated variant is in an intron within some transcripts of FRMPD4 but is also close to the MSL3 gene, which interacts with KAT8, a reported genetic risk factor for AD [2, 79, 80]. In addition, FRMPD4 is an inactivated gene in females, while MSL3 escapes inactivation [13].
The signal at the intronic variant within the DMD dystrophin gene decreased when including proxy or biobank cases; further analyses are necessary to determine whether this is due to a falsely inflated signal in the clinically diagnosed samples, or to a less specific diagnosis in the proxy and biobank samples. DMD is inactivated in females [13], and mutations in the gene can cause Duchenne muscular dystrophy. Some patients suffering from this disease can exhibit cognitive impairment, and a shift towards amyloidogenesis in memory-specific brain regions was found in mice mutated in the DMD gene (mdx mouse) compared to wild-type mice [81]. Additionally, the DMD rs5927116 variant was reportedly associated with the volume of entorhinal cortex in a small sample (N = 792); however, this signal is 1.4 Mb away from our AD signal and the variants are independent (LD measured by r2 = 1.65 × 10−4) [82].
The rare variant signals in the WNK3 and PJA1 loci are characterized by a low imputation quality or a limited number of clinically diagnosed AD cases analyzed, and further analyses in sequencing data would be necessary to validate those signals. Inhibition of WNK3 is reportedly neuroprotective in stroke [83] and intracerebral hemorrhage [84], but has a deleterious effect on neurons after traumatic brain injury [85]. In the PJA1 locus, the index variant is located in an enhancer between the PJA1 and NALF2 (NALCN Channel Auxiliary Factor 2) genes. Variants in this locus are associated with educational attainment [78], and PJA1 is expressed in the brain. Another rare variant signal was identified in the DACH2 gene, which is associated with brain shape (segment 7) [86] and edge-level brain connectivity measures [87].
Identifying putative causal genes in the two other loci, Xp22.32 and Xq25, is more challenging, as the index variants are located more than 300 kb away from the closest protein coding gene, NLGN4X and GRIA3, respectively. Additionally, those variants are not eQTL/sQTL for any gene according to GTeX Portal. Expression of the GRIA3 gene in temporal lobe is associated with AD risk in females, and its expression in the dorsolateral prefrontal cortex is reportedly associated with cognitive change in women during aging and AD [47]. However, the rs191195705 index variant of the Xq25 signal is associated with AD risk mainly in males in our analyses (Supplementary Table S8). Regarding the Xp22.32 locus, the rs5916169 variant, located at 127 kb from our index variant, is associated with functional connectivity [16]. However, this variant is not in LD (r2 = 0.005) with the AD index variant.
Different X-chromosome-wide significant loci were detected in the main analysis—considering AD-proxy cases— and in the sensitivity ones including only diagnosed AD-cases or excluding biobank samples. A loss of significance was expected in the sensitivity analyses compared to the main analysis due to lower power; similar odds-ratio and overlapping confidence intervals should however be observed for signals mainly driven by AD rather than non-AD dementia. This was the case for the FRMPD4 locus, and to a lesser extent for the Xp22.32 and Xq25 loci. The loss of a signal in the sensitivity analyses might also be due to purely analytical reasons; for example, the rare index variant of the PJA1 locus did not pass the filtering criteria in the sensitivity analyses. The identification of the DMD, WNK3 and DACH2 loci in the sensitivity analyses but not in the main analysis, despite its higher power, might be due to sampling variation, a dilution of the signal in the main analysis linked to the expected higher proportion of non-AD dementia cases among AD-proxy cases or a falsely inflated signal in the sensitivity analyses. Additionally, the correction factor used in the UKB proxy analysis was designed for common variants with low to moderate effect. It might be less appropriate for rare variants with larger effects, such as the index variants of the WNK3 and DACH2 loci. Further studies in larger samples will help to delineate the real impact of those loci on AD risk.
Although this study represents a powerful XWAS for AD, we did not find any genome-wide-significant genetic association with AD risk among X-chromosome variants. A recent XWAS on AD identified only one genome-wide significant association in the SLC9A7 locus [88]. We do not replicate this result at the X-chromosome-wide significance level (OR = 1.023 [1.005–1.042], P = 1.36 × 10−2 for the index variant rs2142791, and minimum P in the locus of 5.2 × 10−5, in the r-XCI meta-analysis including AD-proxy cases, Supplementary Table S15). The lack of signal overlap between the two studies may be explained in part by a different definition of AD and AD-proxy status, leading to an expected higher proportion of non-AD dementia cases in the other study (Supplementary Material).
Technical or analytical reasons can partly explain the absence of genome-wide significant signals on the X-chromosome, such as: 1) overall lower variant density, 2) lower coverage by genotyping platforms, 3) lower call rate of variants, 4) lower imputation quality, or 5) a lower effective sample size in males on the X-chromosome compared to the autosomes [89]. However, it is also possible that fewer genome-wide significant associations of X-chromosome loci with AD risk exist than on autosomes due to a lower density of functional variants on the X-chromosome. Indeed, Gorlov et al. 2023 [89] found a lower density of variants in both exonic and intronic regions on the X-chromosome compared to autosomes, which they link to a stronger selection against X-chromosome mutations.
In conclusion, this XWAS found no common genetic risk factor for AD on the non-pseudoautosomal region of the X-chromosome but identified suggestive signals with moderate impact on AD risk, which warrant further investigations. In particular, future analyses of sequencing data will help to address some of the technical issues described above, and will allow to study the impact of X-chromosome rare variants or structural variants on AD risk. Additionally, extending XWAS to AD-related phenotypes, such as cognitive decline, AD pathology or AD biomarkers, would further delineate the impact of X-chromosome genetic variations on the processes leading to AD. Lastly, insights into the contribution of the X-chromosome to AD or AD-related phenotypes will be provided by additional studies of the impact of X-chromosome biology beyond genetic variations, for example gene expression or epigenetic alterations, including parental imprinting [49, 90, 91].
Data availability
Summary statistics are available through the European Bioinformatics Institute GWAS Catalog (https://www.ebi.ac.uk/gwas/) under study accessions GCST90449045 to GCST90449052.
Code availability
We used publicly available software for all analyses, which are listed in the Supplementary Materials with their appropriate citations. We also provide command lines in the Supplementary Materials appendix, and some specific scripts on Zenodo (https://doi.org/10.5281/zenodo.14001011).
References
Lambert JC, Ramirez A, Grenier-Boley B, Bellenguez C. Step by step: towards a better understanding of the genetic architecture of Alzheimer’s disease. Mol Psychiatry. 2023;28:2716–27.
Bellenguez C, Küçükali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet. 2022;54:412–36.
Holstege H, Hulsman M, Charbonnier C, Grenier-Boley B, Quenez O, Grozeva D, et al. Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer’s disease. Nat Genet. 2022;54:1786–94
Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30.
Wightman DP, Jansen IE, Savage JE, Shadrin AA, Bahrami S, Holland D, et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat Genet. 2021;53:1276–82.
Sims R, Van Der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. 2017;49:1373–84.
Carrel L, Brown CJ. When the lyon(Ized chromosome) roars: ongoing expression from an inactive X chromosome. Philos Trans R Soc B Biol Sci. 2017;372:20160355.
Lyon MF. Gene action in the X-chromosom (Mus musculus L.). Nature. 1961;190:372–3.
Amos-Landgraf JM, Cottle A, Plenge RM, Friez M, Schwartz CE, Longshore J, et al. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am J Hum Genet. 2006;79:493–9.
Shvetsova E, Sofronova A, Monajemi R, Gagalova K, Draisma HHM, White SJ, et al. Skewed X-inactivation is common in the general female population. Eur J Hum Genet. 2019;27:455–65.
Zito A, Davies MN, Tsai PC, Roberts S, Andres-Ejarque R, Nardone S, et al. Heritability of skewed X-inactivation in female twins is tissue-specific and associated with age. Nat Commun. 2019;10:1–11.
Busque L, Mio R, Mattioli J, Brais E, Biais N, Lalonde Y, et al. Nonrandom X-inactivation patterns in normal females: Lyonization ratios vary with age. Blood. 1996;88:59–65.
Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–8.
Sun L, Wang Z, Lu T, Manolio TA, Paterson AD. eXclusionarY: 10 years later, where are the sex chromosomes in GWASs? Am J Hum Genet. 2023;110:903–12.
Chen B, Craiu RV, Strug LJ, Sun L. The X factor: a robust and powerful approach to X-chromosome-inclusive whole-genome association studies. Genet Epidemiol. 2021;45:694–709.
Smith SM, Douaud G, Chen W, Hanayik T, Alfaro-Almagro F, Sharp K, et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci. 2021;24:737–45.
Mallard TT, Liu S, Seidlitz J, Ma Z, Moraczewski D, Thomas A, et al. X-chromosome influences on neuroanatomical variation in humans. Nat Neurosci. 2021;24:1216–24.
Neri G, Schwartz CE, Lubs HA, Stevenson RE. XLID update 2017. Am J Med Genet A. 2018;176:1375.
Hickman RA, O’Shea SA, Mehler MF, Chung WK. Neurogenetic disorders across the lifespan: from aberrant development to degeneration. Nat Rev Neurol. 2022;18:117–24.
Ferretti MT, Santuccione Chadha A. The missing X factor in Alzheimer disease. Nat Rev Neurol. 2021;17:727–8.
Alzheimer Europe. Dementia in Europe yearbook 2019: estimating the prevalence of dementia in Europe. Alzheimer Europe; 2019. pp. 108.
Nichols E, Steinmetz JD, Vollset SE, Fukutaki K, Chalek J, Abd-Allah F, et al. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: an analysis for the Global Burden of Disease Study 2019. Lancet Public Health. 2022;7:e105–25.
Corrada MM, Brookmeyer R, Paganini-Hill A, Berlau D, Kawas CH. Dementia incidence continues to increase with age in the oldest old the 90+ study. Ann Neurol. 2010;67:114–21.
Gilsanz P, Corrada MM, Kawas CH, Mayeda ER, Glymour MM, Quesenberry CP, et al. Incidence of dementia after age 90 in a multiracial cohort. Alzheimers Dement. 2019;15:497–505.
Mayeda ER, Glymour MM, Quesenberry CP, Whitmer RA. Inequalities in dementia incidence between six racial and ethnic groups over 14 years. Physiol Behav. 2016;176:100–6.
Matthews FE, Stephan BCM, Robinson L, Jagger C, Barnes LE, Arthur A, et al. A two decade dementia incidence comparison from the Cognitive Function and Ageing Studies I and II. Nat Commun. 2016;7:11398.
Wolters FJ, Chibnik LB, Waziry R, Anderson R, Berr C, Beiser A, et al. Twenty-seven-year time trends in dementia incidence in Europe and the United States: the Alzheimer Cohorts Consortium. Neurology. 2020;95:E519–31.
Shaw C, Hayes-Larson E, Glymour MM, Dufouil C, Hohman TJ, Whitmer RA, et al. Evaluation of selective survival and sex/gender differences in dementia incidence using a simulation model. JAMA Netw Open. 2021;4:e211001.
Barnes LL, Wilson RS, Bienias JL, Schneider JA, Evans DA, Bennett DA. Sex differences in the clinical manifestations of Alzheimer disease pathology. Arch Gen Psychiatry. 2005;62:685–91.
Liesinger AM, Graff-Radford NR, Duara R, Carter RE, Hanna Al-Shaikh FS, Koga S, et al. Sex and age interact to determine clinicopathologic differences in Alzheimer’s disease. Acta Neuropathol. 2018;136:873–85.
Oveisgharan S, Arvanitakis Z, Yu L, Farfel J, Schneider JA, Bennett DA. Sex differences in Alzheimer’s disease and common neuropathologies of aging. Acta Neuropathol. 2018;136:887–900.
Buckley RF, Mormino EC, Rabin JS, Hohman TJ, Landau S, Hanseeuw BJ, et al. Sex differences in the association of global amyloid and regional tau deposition measured by positron emission tomography in clinically normal older adults. JAMA Neurol. 2019;76:542–51.
Wisch JK, Meeker KL, Gordon BA, Flores S, Dincer A, Grant EA, et al. Sex-related differences in tau positron emission tomography (PET) and the effects of hormone therapy (HT). Alzheimer Dis Assoc Disord. 2021;35:164–8.
Pereira JB, Harrison TM, La Joie R, Baker SL, Jagust WJ. Spatial patterns of tau deposition are associated with amyloid, ApoE, sex, and cognitive decline in older adults. Eur J Nucl Med Mol Imaging. 2020;47:2155–64.
Edwards L, La Joie R, Iaccarino L, Strom A, Baker SL, Casaletto KB, et al. Multimodal neuroimaging of sex differences in cognitively impaired patients on the Alzheimer’s continuum: greater tau-PET retention in females. Neurobiol Aging. 2021;105:86–98.
Filon JR, Intorcia AJ, Sue LI, Vazquez Arreola E, Wilson J, Davis KJ, et al. Gender differences in Alzheimer disease: brain atrophy, histopathology burden, and cognition. J Neuropathol Exp Neurol. 2016;75:748–54.
Babapour Mofrad R, Tijms BM, Scheltens P, Barkhof F, Van Der Flier WM, Sikkes SAM, et al. Sex differences in CSF biomarkers vary by Alzheimer disease stage and APOE ϵ4 genotype. Neurology. 2020;95:e2378–88.
Hohman TJ, Dumitrescu L, Barnes LL, Thambisetty M, Beecham G, Kunkle B, et al. Sex-specific association of apolipoprotein E with cerebrospinal fluid levels of tau. JAMA Neurol. 2018;75:989–98.
Levine DA, Gross AL, Briceño EM, Tilton N, Giordani BJ, Sussman JB, et al. Sex differences in cognitive decline among US adults. JAMA Netw Open. 2021;4:1–13.
Bloomberg M, Dugravot A, Dumurgier J, Kivimaki M, Fayosse A, Steptoe A, et al. Sex differences and the role of education in cognitive ageing: analysis of two UK-based prospective cohort studies. Lancet Public Health. 2021;6:e106–15.
Lipnicki DM, Crawford JD, Dutta R, Thalamuthu A, Kochan NA, Andrews G, et al. Age-related cognitive decline and associations with sex, education and apolipoprotein E genotype across ethnocultural groups and geographic regions: a collaborative cohort study. PLoS Med. 2017;14:1–21.
Ferretti MT, Iulita MF, Cavedo E, Chiesa PA, Dimech AS, Chadha AS, et al. Sex differences in Alzheimer disease—the gateway to precision medicine. Nat Rev Neurol. 2018;14:457–69.
Arenaza-Urquijo EM, Boyle R, Casaletto K, Anstey KJ, Vila-Castelar C, Colverson A, et al. Sex and gender differences in cognitive resilience to aging and Alzheimer’s disease. Alzheimers Dement. 2024;20:5695–719.
Ferretti MT, Martinkova J, Biskup E, Benke T, Gialdini G, Nedelska Z, et al. Sex and gender differences in Alzheimer’s disease: current challenges and implications for clinical practice: position paper of the Dementia and Cognitive Disorders Panel of the European Academy of Neurology. Eur J Neurol. 2020;27:928–43.
Tifratene K, Robert P, Metelkina A, Pradier C, Dartigues JF. Progression of mild cognitive impairment to dementia due to AD in clinical settings. Neurology. 2015;85:331–8.
Lin KA, Choudhury KR, Rathakrishnan BG, Marks DM, Petrella JR, Doraiswamy PM. Marked gender differences in progression of mild cognitive impairment over 8 years. Alzheimers Dement Transl Res Clin Interv. 2015;1:103–10.
Davis EJ, Broestl L, Williams G, Garay BI, Lobach I, Devidze N, et al. A second X chromosome contributes to resilience in a mouse model of Alzheimer’s disease. Sci Transl Med. 2020;12:eaaz5677.
Zheng X, Wang S, Huang J, Li C, Shang H. Predictors for survival in patients with Alzheimer’s disease: a large comprehensive meta-analysis. Transl Psychiatry. 2024;14:1–9.
Davis EJ, Solsberg CW, White CC, Miñones-Moyano E, Sirota M, Chibnik L, et al. Sex-specific association of the X chromosome with cognitive change and tau pathology in aging and Alzheimer disease. JAMA Neurol. 2021;78:1249–54.
Yan Y, Wang X, Chaput D, Pieper AA, Woo JA, Kang DE, et al. X-linked ubiquitin-specific peptidase 11 increases tauopathy vulnerability in women increases tauopathy vulnerability in women. Cell. 2022;185:3913–30.e19.
Liu JZ, Erlich Y, Pickrell JK. Case-control association mapping by proxy using family history of disease. Nat Genet. 2017;49:325–31.
Jansen IE, van der Lee SJ, Gomez-Fonseca D, de Rojas I, Dalmasso MC, Grenier-Boley B, et al. Genome-wide meta-analysis for Alzheimer’s disease cerebrospinal fluid biomarkers. Acta Neuropathol. 2022;144:821–42.
Luck T, Riedel-Heller SG, Kaduszkiewicz H, Bickel H, Jessen F, Pentzek M, et al. Mild cognitive impairment in general practice: age-specific prevalence and correlate results from the German study on ageing, cognition and dementia in primary care patients (AgeCoDe). Dement Geriatr Cogn Disord. 2007;24:307–16.
Grande G, Vetrano DL, Fratiglioni L, Marseglia A, Vanacore N, Laukka EJ, et al. Disability trajectories and mortality in older adults with different cognitive and physical profiles. Aging Clin Exp Res. 2020;32:1007–16.
Artero S, Ancelin ML, Portet F, Dupuy A, Berr C, Dartigues JF, et al. Risk profiles for mild cognitive impairment and progression to dementia are gender specific. J Neurol Neurosurg Psychiatry. 2008;79:979–84.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023:613;508–18.
Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–9.
Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Loh PR, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48:1443–8.
Walter K, Min JL, Huang J, Crooks L, Memari Y, McCarthy S, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–89.
Clayton DG. Sex chromosomes and genetic association studies. Genome Med. 2009;1:1–7.
Clayton DG. Testing for association on the X chromosome. Biostatistics. 2008;9:593–600.
Ghosh A, Hartge P, Kraft P, Joshi AD, Ziegler RG, Chanock SJ, et al. Leveraging family history in population-based case-control association studies. Genet Epidemiol. 2014;38:114–22.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1.
Magi R, Lindgren CM, Morris AP. Meta-analysis of sex-specific genome-wide association studies. Genet Epidemiol. 2010;34:846–53.
Satizabal CL, Adams HH, Hibar DP, White CC. Genetic architecture of subcortical brain structures in 38,851 individuals. Physiol Behav. 2019;176:139–48.
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6.
Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13.
Philipps V, Amieva H, Andrieu S, Dufouil C, Berr C, Dartigues JF, et al. Normalized mini-mental state examination for assessing cognitive change in population-based brain aging studies. Neuroepidemiology. 2014;43:15–25.
Kleineidam L, Chouraki V, Próchnicki T, van der Lee SJ, Madrid-Márquez L, Wagner-Thelen H, et al. PLCG2 protective variant p.P522R modulates tau pathology and disease progression in patients with mild cognitive impairment. Acta Neuropathol. 2020;139:1025–44.
Wingo AP, Liu Y, Gerasimov ES, Vattathil SM, Liu J, Cutler DJ, et al. Sex differences in brain protein expression and disease. Nat Med. 2023;29:2224–32.
Lukacsovich D, O’Shea D, Huang H, Zhang W, Youn JI, Chen XS, et al. MIAMI-AD (methylation in aging and methylation in AD): an integrative knowledgebase that facilitates explorations of DNA methylation across sex, aging, and Alzheimer’s disease. MedRxiv. 2023.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:1–19.
Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371:64–74.
Group WW. Glucose-6-phosphate dehydrogenase deficiency. Bull World Health Organ. 1989;67:601–11.
Lee HW, Jeonghoon C, Hyewon S, Karam K, Jinhee Y, Moonseok N, et al. Preso, a novel PSD-95-interacting FERM and PDZ domain protein that regulates dendritic spine morphogenesis. J Neurosci. 2008;28:14546–56.
Piard J, Hu JH, Campeau PM, Rzońca S, Van Esch H, Vincent E, et al. FRMPD4 mutations cause X-linked intellectual disability and disrupt dendritic spine morphogenesis. Hum Mol Genet. 2018;27:589–600.
Okbay A, Wu Y, Wang N, Jayashankar H, Bennett M, Nehzati SM, et al. Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals. Nat Genet. 2022;54:437–49.
Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–13.
Smith ER, Cayrou C, Huang R, Lane WS, Côté J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–88.
Hayward GC, Caceres D, Copeland EN, Baranowski BJ, Mohammad A, Whitley KC, et al. Characterization of Alzheimer’s disease-like neuropathology in Duchenne’s muscular dystrophy using the DBA/2J mdx mouse model. FEBS Open Bio. 2022;12:154–62.
Wang K-W, Yuan Y-X, Zhu B, Zhang Y, Wei Y-F, Meng F-S, et al. X chromosome-wide association study of quantitative biomarkers from the Alzheimer’s Disease Neuroimaging Initiative study. Front Aging Neurosci. 2023;15:1–15.
Begum G, Yuan H, Kahle KT, Li L, Wang S, Shi Y, et al. Inhibition of WNK3 kinase signaling reduces brain damage and accelerates neurological recovery after stroke. Stroke. 2015;46:1956–65.
Wu D, Lai N, Deng R, Liang T, Pan P, Yuan G, et al. Activated WNK3 induced by intracerebral hemorrhage deteriorates brain injury maybe via WNK3/SPAK/NKCC1 pathway. Exp Neurol. 2020;332:113386.
Zhu J, Lin X, Chen C, Tan H, Gao Y, Li D, et al. WNK3 promotes neuronal survival after traumatic brain injury in rats. Neuroscience. 2021;477:76–88.
Naqvi S, Sleyp Y, Hoskens H, Indencleef K, Spence JP, Bruffaerts R, et al. Shared heritability of human face and brain shape. Nat Genet. 2021;53:830–9.
Sha Z, Schijven D, Fisher SE, Francks C. Genetic architecture of the white matter connectome of the human brain. Sci Adv. 2023;9:eadd2870.
Belloy ME, Guen YL, Stewart I, Herz J, Sherva R, Zhang R, et al. The role of X chromosome in Alzheimer′s disease genetics. JAMA Neurol. 2024;11:2024.04.22.24306094.
Gorlov IP, Amos CI. Why does the X chromosome lag behind autosomes in GWAS findings? PLoS Genet. 2023;19:1–19.
Seto M, Hohman TJ, Mormino EC, Papp KV, Rentz READM, Johnson KA, et al. Parental history of memory impairment and β-amyloid in cognitively unimpaired older adults. JAMA Neurol. 2024;81:798–804.
Dubal DB, Elser HC. β-Amyloid in cognitively unimpaired individuals-blame mom? JAMA Neurol. 2024;81:795–7.
Acknowledgements
EADB: This study was supported by grants from the Fondation pour la Recherche sur Alzheimer (convention 2022-A-01 and cluster grant), and the JPco-fuND-2 ‘Multinational research projects on Personalized Medicine for Neurodegenerative Diseases’ PREADAPT project (ANR-19-JPW2-0004). We thank the many study participants, researchers and staff for collecting and contributing to the data, the high-performance computing service at the University of Lille and the staff at CEA-CNRGH for their help with sample preparation and genotyping and excellent technical assistance. We thank Antonio Pardinas for his help. We thank the Netherlands Brain Bank. This research was conducted using the UKBB resource (application number 61054). This work was funded by a grant (EADB) from the EU Joint Programme—Neurodegenerative Disease Research. Inserm UMR1167 is also funded by the Inserm, Institut Pasteur de Lille, Lille Métropole Communauté Urbaine and French government’s LABEX DISTALZ program (Development of Innovative Strategies for a Transdisciplinary Approach to ALZheimer’s disease). This work was also supported by the Research Council of Finland grants 338182 and 334802, the Sigrid Jusélius Foundation, and the Strategic Neuroscience Funding of the University of Eastern Finland. ADGC: The National Institutes of Health, National Institute on Aging (NIH-NIA) supported this work through the following grants: ADGC, U01 AG032984, RC2 AG036528; Samples from the National Cell Repository for Alzheimer’s Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study. We thank contributors who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible. Data for this study were prepared, archived, and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (U24-AG041689-01) in cooperation with NACC (U24 AG072122) and additional studies described in the Supplementary Materials. Full consortium acknowledgments and funding are in the Supplementary Materials.
Author information
Authors and Affiliations
Consortia
Contributions
Coordination: BK and CB. Data analyses: JLB, LG, SH, NA, SA, SHC, JB, BG-B, OGR, LK, JY, KPT, LW, AV, RC-M, SvdL, VD, IdR, SP, AD, JD, BK and CB. Core writing group: JLB, J-CL, BK and CB. All other authors contributed samples and data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Written informed consent was obtained from study participants or, for those with substantial cognitive impairment, a caregiver, legal guardian or other proxy. Study protocols for all cohorts were reviewed and approved by the appropriate institutional review boards.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Le Borgne, J., Gomez, L., Heikkinen, S. et al. X‐chromosome-wide association study for Alzheimer’s disease. Mol Psychiatry 30, 2335–2346 (2025). https://doi.org/10.1038/s41380-024-02838-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-024-02838-5