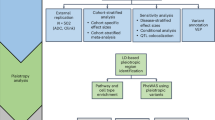
Abstract
Genomic studies of molecular traits have provided mechanistic insights into complex disease, though these lag behind for brain-related traits due to the inaccessibility of brain tissue. We leveraged cerebrospinal fluid (CSF) to study neurobiological mechanisms in vivo, measuring 5543 CSF metabolites, the largest panel in CSF to date, in 977 individuals of European ancestry. Individuals originated from two separate cohorts including cognitively healthy subjects (n = 490) and a well-characterized memory clinic sample, the Amsterdam Dementia Cohort (ADC, n = 487). We performed metabolite quantitative trait loci (mQTL) mapping on CSF metabolomics and found 126 significant mQTLs, representing 65 unique CSF metabolites across 51 independent loci. To better understand the role of CSF mQTLs in brain-related disorders we integrated our CSF mQTL results with pre-existing summary statistics on brain traits, identifying 34 genetic associations between CSF metabolites and brain traits. Over 90% of significant mQTLs demonstrated colocalized associations with brain-specific gene expression, unveiling potential neurobiological pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
GWAS summary statistics on all CSF metabolite levels that were generated as part of this study have been deposited to the European Bioinformatics Institute GWAS Catalog (https://www.ebi.ac.uk/gwas/) under accession no. GCST90286909-GCST90292451. GENCODE v.41 basic gene annotation GTF file from https://www.gencodegenes.org/human/stats_41.html UKBB LD reference panel available from Amazon S3 bucket s3://broad-alkesgroup-ukbb-ld/UKBB_LD/. UKBB functional annotations available from Amazon S3 bucket https://broad-alkesgroup-ukbb-ld.s3.amazonaws.com/UKBB_LD/baselineLF_v2.2.UKB.polyfun.tar.gz. PsychENCODE TWAS weights are available from http://resource.psychencode.org/. PsychENCODE isoTWAS weights are available from https://zenodo.org/record/6795947#.Y8mi2-zMLBI. Summary statistics on Alzheimer’s disease [41], dementia with Lewy bodies [42] and amyotrophic lateral sclerosis [44] are publicly available at the European Bioinformatics Institute GWAS Catalog under accession no. GCST90027158, GCST90001390 and GCST90027163, respectively. Summary statistics on bipolar disorder [45], schizophrenia [46], major depressive disorder [47], attention deficit hyperactivity disorder (ADHD) [48] and alcohol abuse disorder [50] are publicly available on the psychiatric genomics consortium (PGC) website (https://www.med.unc.edu/pgc/results-and-downloads). The MEGASTROKE consortium, launched by the International Stroke Genetics Consortium, has published the stroke [43] summary statistics at https://www.megastroke.org. Full insomnia [49] summary statistics for UKB and the top 10,000 SNPs for 23andMe are available at https://ctg.cncr.nl/software/summary_statistics/.
Change history
16 October 2025
The accession number GCST90286909-GCST90292451 was incorrectly given as GCP000726 and has been corrected. The original article has been corrected.
References
Kraus WE, Muoio DM, Stevens R, Craig D, Bain JR, Grass E, et al. Metabolomic Quantitative Trait Loci (mQTL) mapping implicates the ubiquitin proteasome system in cardiovascular disease pathogenesis. PLoS Genet. 2015;11:e1005553.
Shin S-Y, Fauman EB, Petersen A-K, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–50.
Prakash S. Human metabolic individuality in biomedical and pharmaceutical research. Circ Cardiovasc Genet. 2011;4:714–5.
Long T, Hicks M, Yu H-C, Biggs WH, Kirkness EF, Menni C, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49:568–78.
Nag A, Kurushima Y, Bowyer RCE, Wells PM, Weiss S, Pietzner M, et al. Genome-wide scan identifies novel genetic loci regulating salivary metabolite levels. Hum Mol Genet. 2020;29:864–75.
Schlosser P, Li Y, Sekula P, Raffler J, Grundner-Culemann F, Pietzner M, et al. Genetic studies of urinary metabolites illuminate mechanisms of detoxification and excretion in humans. Nat Genet. 2020;52:167–76.
Yang C, Farias FHG, Ibanez L, Suhy A, Sadler B, Fernandez MV, et al. Genomic atlas of the proteome from brain, CSF and plasma prioritizes proteins implicated in neurological disorders. Nat Neurosci. 2021;24:1302–12.
Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet. 2018;50:538–48.
Nedergaard M. Garbage Truck of the Brain. Science. 2013;340:1529–30.
Tijms BM, Visser PJ. Pathophysiological subtypes of Alzheimer’s disease based on cerebrospinal fluid proteomics. Alzheimer’s & Dementia. 2020;12:3776–92.
Yan J, Kuzhiumparambil U, Bandodkar S, Dale RC, Fu S. Cerebrospinal fluid metabolomics: detection of neuroinflammation in human central nervous system disease. Clin Transl Immunology. 2021;10:e1318.
Dong R, Darst BF, Deming Y, Ma Y, Lu Q, Zetterberg H, et al. CSF metabolites associate with CSF tau and improve prediction of Alzheimer’s disease status. Alzheimers Dement. 2021;13:e12167.
Robinette SL, Holmes E, Nicholson JK, Dumas ME. Genetic determinants of metabolism in health and disease: from biochemical genetics to genome-wide associations. Genome Med. 2012;4:30.
Chen Y, Lu T, Pettersson-Kymmer U, Stewart ID, Butler-Laporte G, Nakanishi T, et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat Genet. 2023;55:44–53.
Surendran P, Stewart ID, Au Yeung VPW, Pietzner M, Raffler J, Wörheide MA, et al. Rare and common genetic determinants of metabolic individuality and their effects on human health. Nat Med. 2022;28:2321–32.
Tahir UA, Katz DH, Avila-Pachecho J, Bick AG, Pampana A, Robbins JM, et al. Whole genome association study of the plasma metabolome identifies metabolites linked to cardiometabolic disease in black individuals. Nat Commun. 2022;13:4923.
Suhre K, Shin S-Y, Petersen A-K, Mohney RP, Meredith D, Wägele B, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60.
Yin X, Chan LS, Bose D, Jackson AU, VandeHaar P, Locke AE, et al. Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat Commun. 2022;13:1644.
Suhre K, Wallaschofski H, Raffler J, Friedrich N, Haring R, Michael K, et al. A genome-wide association study of metabolic traits in human urine. Nat Genet. 2011;43:565–9.
Wang C, Western D, Yang C, Ali M, Wang L, Gorijala P, et al. Unique genetic architecture of CSF and brain metabolites pinpoints the novel targets for the traits of human wellness. Nat Genet. 2024;56:2685–95. https://doi.org/10.1038/s41588-024-01973-7.
Panyard DJ, Kim KM, Darst BF, Deming YK, Zhong X, Wu Y, et al. Cerebrospinal fluid metabolomics identifies 19 brain-related phenotype associations. Commun Biol. 2021;4:63.
Luykx JJ, Bakker SC, Lentjes E, Neeleman M, Strengman E, Mentink L, et al. Genome-wide association study of monoamine metabolite levels in human cerebrospinal fluid. Mol Psychiatry. 2014;19:228–34.
van der Flier WM, Scheltens P. Amsterdam dementia cohort: performing research to optimize care. J Alzheimers Dis. 2018;62:1091–111.
Legdeur N, Badissi M, Carter SF, de Crom S, van de Kreeke A, Vreeswijk R, et al. Resilience to cognitive impairment in the oldest-old: design of the EMIF-AD 90+ study. BMC Geriatr. 2018;18:289.
Boomsma DI, de Geus EJC, Vink JM, Stubbe JH, Distel MA, Hottenga J-J, et al. Netherlands twin register: from twins to twin families. Twin Res Hum Genet. 2006;9:849–57.
Konijnenberg E, Carter SF, Ten Kate M, den Braber A, Tomassen J, Amadi C, et al. The EMIF-AD PreclinAD study: study design and baseline cohort overview. Alzheimers Res Ther. 2018;10:75.
Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270–9.
McKeith IG, Boeve BF, Dickson DW, Halliday G, Taylor JP, Weintraub D, et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology. 2017;89:88–100.
McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–9.
Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456.
Tijms BM, Willemse EAJ, Zwan MD, Mulder SD, Visser PJ, van Berckel BNM, et al. Unbiased approach to counteract upward drift in cerebrospinal Fluid amyloid-β 1-42 analysis results. Clin Chem. 2018;64:576–85.
Duits FH, Teunissen CE, Bouwman FH, Visser P-J, Mattsson N, Zetterberg H, et al. The cerebrospinal fluid ‘Alzheimer profile’: easily said, but what does it mean? Alzheimers Dement. 2014;10:713–723.e2.
Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–46.
Fan S, Kind T, Cajka T, Hazen SL, Tang WHW, Kaddurah-Daouk R, et al. Systematic error removal using random forest for normalizing large-scale untargeted lipidomics data. Anal Chem. 2019;91:3590–6.
Lam M, Awasthi S, Watson HJ, Goldstein J, Panagiotaropoulou G, Trubetskoy V, et al. RICOPILI: rapid imputation for COnsortias PIpeLIne. Bioinformatics. 2020;36:930–3.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Watanabe K, Taskesen E, van Bochoven A. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1–11.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–52.
Bellenguez C, Küçükali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet. 2022;54:412–36.
Chia R, Sabir MS, Bandres-Ciga S, Saez-Atienzar S, Reynolds RH, Gustavsson E, et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet. 2021;53:294–303.
Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–37.
van Rheenen W, van der Spek RAA, Bakker MK, van Vugt JJFA, Hop PJ, Zwamborn RAJ, et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat Genet. 2021;53:1636–48.
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29.
Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–8.
Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75.
Watanabe K, Jansen PR, Savage JE, Nandakumar P, Wang X, Hinds DA, et al. Genome-wide meta-analysis of insomnia prioritizes genes associated with metabolic and psychiatric pathways. Nat Genet. 2022;54:1125–32.
Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, 23andMe Research Team, the Substance Use Disorder Working Group of the Psychiatric Genomics Consortium, Adams MJ, et al. Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population-based cohorts. Am J Psychiatry. 2019;176:107–18.
Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383.
Benner C, Spencer CCA, Havulinna AS, Salomaa V, Ripatti S, Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–501.
Wang G, Sarkar A, Carbonetto P, Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J R Stat Soc Series B Stat Methodol. 2020;82:1273–1300.
Weissbrod O, Hormozdiari F, Benner C, Cui R, Ulirsch J, Gazal S, et al. Functionally informed fine-mapping and polygenic localization of complex trait heritability. Nat Genet. 2020;52:1355–63.
Koromina M, Ravi A, Panagiotaropoulou G, Schilder BM, Humphrey J, Braun A, et al. Fine-mapping genomic loci refines bipolar disorder risk genes. medRxiv [Preprint] https://doi.org/10.1101/2024.02.12.24302716.
Walker RL, Ramaswami G, Hartl C, Mancuso N, Gandal MJ, de la Torre-Ubieta L, et al. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell. 2020;181:745.
Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:eaat8127.
Bhattacharya A, Vo DD, Jops C, Kim M, Wen C, Hervoso JL, et al. Isoform-level transcriptome-wide association uncovers genetic risk mechanisms for neuropsychiatric disorders in the human brain. Nat Genet. 2023;55:2117–28.
Watanabe K, Stringer S, Frei O, Umićević Mirkov M, de Leeuw C, Polderman TJC, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51:1339–48.
Flynn ED, Lappalainen T. Functional characterization of genetic variant effects on expression. Annu Rev Biomed Data Sci. 2022;5:119–39.
Jo B-S, Choi SS. Introns: the functional benefits of introns in genomes. Genomics Inform. 2015;13:112.
Niu H-M, Yang P, Chen H-H, Hao R-H, Dong S-S, Yao S, et al. Comprehensive functional annotation of susceptibility SNPs prioritized 10 genes for schizophrenia. Transl Psychiatry. 2019;9:1–12.
Baslow MH, Guilfoyle DN. N-acetyl-l-histidine, a prominent biomolecule in brain and eye of poikilothermic vertebrates. Biomolecules. 2015;5:635.
Koshiba S, Motoike IN, Saigusa D, Inoue J, Aoki Y, Tadaka S, et al. Identification of critical genetic variants associated with metabolic phenotypes of the Japanese population. Commun Biol. 2020;3:662.
Sall S, Thompson W, Santos A, Dwyer DS. Analysis of major depression risk genes reveals evolutionary conservation, shared phenotypes, and extensive genetic interactions. Front Psychiatry. 2021;12:698029.
Li QX, Liu JC, He MH, Zhou JQ. Kae1 of Saccharomyces cerevisiae KEOPS complex possesses ADP/GDP nucleotidase activity. Biochem J. 2022;479:2433–47.
Edvardson S, Prunetti L, Arraf A, Haas D, Bacusmo JM, Hu JF, et al. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur J Hum Genet. 2017;25:545.
Keller BO, Wu BTF, Li SSJ, Monga V, Innis SM. Hypaphorine is present in human milk in association with consumption of legumes. J Agric Food Chem. 2013;61:7654–60.
Yee SW, Buitrago D, Stecula A, Ngo HX, Chien H-C, Zou L, et al. Deorphaning a Solute Carrier 22 family member, SLC22A15, through functional genomic studies. FASEB J. 2020;34:15734.
Schwarz E, Prabakaran S, Whitfield P, Major H, Leweke FM, Koethe D, et al. High Throughput Lipidomic Profiling of Schizophrenia and Bipolar Disorder Brain Tissue Reveals Alterations of Free Fatty Acids, Phosphatidylcholines, and Ceramides. 2008. 9 September 2008. https://doi.org/10.1021/pr800188y.
MacDonald K, Krishnan A, Cervenka E, Hu G, Guadagno E, Trakadis Y. Biomarkers for major depressive and bipolar disorders using metabolomics: a systematic review. Am J Med Genet B Neuropsychiatr Genet. 2019;180:122–37.
Barupal DK, Fiehn O. Generating the blood exposome database using a comprehensive text mining and database fusion approach. Environ Health Perspect. 2019;127:97008.
Auwerx C, Sadler MC, Woh T, Reymond A, Kutalik Z, Porcu E. Exploiting the mediating role of the metabolome to unravel transcript-to-phenotype associations. Elife. 2023;12:e81097.
Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. 2018;23:639–47.
Yamamoto H, Lee-Okada H-C, Ikeda M, Nakamura T, Saito T, Takata A, et al. GWAS-identified bipolar disorder risk allele in the FADS1/2 gene region links mood episodes and unsaturated fatty acid metabolism in mutant mice. Mol Psychiatry. 2023;28:2848–56. https://doi.org/10.1038/s41380-023-01988-2.
Yin F. Lipid metabolism and Alzheimer’s disease: clinical evidence, mechanistic link and therapeutic promise. FEBS J. 2023;290:1420–53.
Hammouda S, Ghzaiel I, Khamlaoui W, Hammami S, Mhenni SY, Samet S, et al. Genetic variants in FADS1 and ELOVL2 increase level of arachidonic acid and the risk of Alzheimer’s disease in the Tunisian population. Prostaglandins Leukot Essent Fatty Acids. 2020;160:102159.
Guo K, Wang P, Zhang L, Zhou Y, Dai X, Yan Y, et al. Transcription factor POU4F2 promotes colorectal cancer cell migration and invasion through hedgehog‐mediated epithelial‐mesenchymal transition. Cancer Sci. 2021;112:4176–86.
Aguet F, Anand S, Ardlie KG, Gabriel S, Getz GA, Graubert A, et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30. https://doi.org/10.1126/science.aaz1776.
Feofanova EV, Brown MR, Alkis T, Manuel AM, Li X, Tahir UA, et al. Whole-genome sequencing analysis of human metabolome in multi-ethnic populations. Nat Commun. 2023;14:1–12.
Acknowledgements
We are greatly appreciative to those individuals who donated the CSF samples on which this study was based. We appreciate data acquisition and reporting by the UC Davis West Coast Metabolomics Center. We thank Dr. Evan Hurlow for his insightful discussions on the organic chemistry involved in this study. We would also like to thank Nicole Zelster for her contributions in the early stages of the project. We also thank Dr. Jurjen Luykx for his help with the recruitment of the cognitively healthy controls. This work used computational and storage services associated with the Hoffman2 Cluster which is operated by the UCLA Office of Advanced Research Computing’s Research Technology Group. Research of Alzheimer Center Amsterdam is part of the neurodegeneration research program of Amsterdam Neuroscience. Alzheimer Center Amsterdam is supported by Stichting Alzheimer Nederland and Stichting VUmc Steun Alzheimercentrum Amsterdam. The clinical database structure was developed with funding from Stichting Dioraphte. This work has received support from the EU/EFPIA Innovative Medicines Initiative Joint Undertaking (EMIF grant number 115372) and Stichting Dioraphte. Genotyping of the Dutch case-control samples was performed in the context of EADB (European Alzheimer DNA biobank) funded by the JPco-fuND FP-829-029 (ZonMW project number 733051061). This project was funded by the NIH National Institute on Aging (NIA) grant RF1AG058484 and the National Institute of Mental Health (NIMH) grant R01MH115676 to RAO. LMR was funded by the Memorabel fellowship (ZonMW projectnumber: 10510022110012). TB was supported by the NIH (grant no. 5T32HG002536). SJL is recipient of funding from ZonMW (#733050512), Health~Holland, Topsector Life Sciences & Health (PPP-allowance; #LSHM20106). Stichting Dioraphte, the Edwin Bouw Fonds and Stichting VUmc fonds. WMF, SJL, CT are recipients of ABOARD, which is a public-private partnership receiving funding from ZonMW (#73305095007) and Health~Holland, Topsector Life Sciences & Health (PPP-allowance; #LSHM20106). More than 30 partners participate in ABOARD (www.aboard-project.nl). ABOARD also receives funding from de Hersenstichting, Edwin Bouw Fonds and Gieskes-Strijbisfonds. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html.
Author information
Authors and Affiliations
Contributions
LMR and TB led and performed the main analyses and wrote the main draft of the manuscript. MF, NR, MK aided in data QC and analysis. MB prepared and maintained cognitively healthy CSF and genotype samples for processing. YALP, AB, WMF, PJV, SJL, BMT, and CET provided the Amsterdam Dementia Cohort clinical data and sample collection, and contributed to discussion of results. LOL and RAO contributed to the conception, supervised analyses, and the discussion of the results. All authors have reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Reus, L.M., Boltz, T., Francia, M. et al. Quantitative trait loci mapping of circulating metabolites in cerebrospinal fluid to uncover biological mechanisms involved in brain-related phenotypes. Mol Psychiatry 30, 3478–3490 (2025). https://doi.org/10.1038/s41380-025-02934-0
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-02934-0