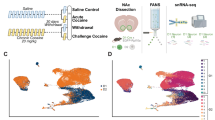
Abstract
Substance use disorders (SUDs) induce widespread molecular dysregulation in nucleus accumbens (NAc), a brain region pivotal for coordinating motivation and reward, which is linked to neural and behavioral disturbances promoting addiction. Despite the overlapping symptomatology of SUDs, different drug classes exert partly unique influences on neural circuits, cell types, physiology, and gene expression. To better understand common and divergent molecular mechanisms governing SUD pathology, we characterized the cell-type-specific restructuring of the NAc transcriptional landscape after psychostimulant or opioid exposure. We combined fluorescence-activated nuclei sorting and deep RNA sequencing to profile NAc D1 and D2 medium spiny neurons (MSNs) across cocaine and morphine exposure paradigms, including initial exposure, prolonged withdrawal after repeated exposure, and re-exposure post-withdrawal. Our analyses reveal that D1 MSNs display many convergent transcriptional responses between the two drug classes, whereas D2 MSNs manifest highly divergent responses, with morphine causing more adaptations in this cell type. Utilizing multiscale embedded gene co-expression network analysis (MEGENA), we discerned transcriptional regulatory networks subserving biological functions altered by cocaine vs. morphine. We observed largely integrative engagement of overlapping gene networks across drug classes in D1 MSNs, but opposite regulation of key D2 networks, highlighting potential therapeutic gene network targets within MSNs. Analysis of gene regulatory systems at the level of enhancers revealed that morphine engages a unique enhancer landscape in D2 MSNs compared to cocaine. Our findings, and future work leveraging this dataset, will open avenues for the development of targeted therapeutic interventions, addressing the urgent need for more effective treatments for SUDs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper or its Supplementary Materials. All RNAseq data reported in this study are publicly available in the Gene Expression Omnibus (GSE272823 and GSE276374). Other supporting scripts/code used in this study are available from the corresponding author upon request.
References
Robinson TE, Berridge KC. The neural basis of drug craving: an incentive-sensitization theory of addiction. Brain Res Brain Res Rev. 1993;18:247–91.
Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–98.
Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–38.
O’Brien CP. Research advances in the understanding and treatment of addiction. Am J Addict. 2003;12:S36–S47.
Nestler EJ, Lüscher C. The molecular basis of drug addiction: linking epigenetic to synaptic and circuit mechanisms. Neuron. 2019;102:48–59.
Lobo MK, Nestler EJ. The striatal balancing act in drug addiction: distinct roles of direct and indirect pathway medium spiny neurons. Front Neuroanat. 2011;5:41.
Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–37.
McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–15.
Ketchesin KD, Becker-Krail DD, Xue X, Wilson RS, Lam TT, Williams KR, et al. Differential effects of cocaine and morphine on the diurnal regulation of the mouse nucleus accumbens proteome. J Proteome Res. 2023;22:2377–90.
Lobo MK, Zaman S, Damez-Werno DM, Koo JW, Bagot RC, DiNieri JA, et al. ΔFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. J Neurosci. 2013;33:18381–95.
Teague CD, Picone JA, Wright WJ, Browne CJ, Silva GM, Futamura R, et al. CREB Binding at the Zfp189 promoter within medium spiny neuron subtypes differentially regulates behavioral and physiological adaptations over the course of cocaine use. Biol Psychiatry. 2023;93:502–11.
Walker DM, Cates HM, Loh Y-HE, Purushothaman I, Ramakrishnan A, Cahill KM, et al. Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry. Biol Psychiatry. 2018;84:867–80.
Mews P, Cunningham AM, Scarpa J, Ramakrishnan A, Hicks EM, Bolnick S, et al. Convergent abnormalities in striatal gene networks in human cocaine use disorder and mouse cocaine administration models. Sci Adv. 2023;9:eadd8946.
Browne CJ, Futamura R, Minier-Toribio A, Hicks EM, Ramakrishnan A, Martínez-Rivera FJ, et al. Transcriptional signatures of heroin intake and relapse throughout the brain reward circuitry in male mice. Sci Adv. 2023;9:eadg8558.
Seney ML, Kim S-M, Glausier JR, Hildebrand MA, Xue X, Zong W, et al. Transcriptional alterations in dorsolateral prefrontal cortex and nucleus accumbens implicate neuroinflammation and synaptic remodeling in opioid use disorder. Biol Psychiatry. 2021;90:550–62.
Chen R, Blosser TR, Djekidel MN, Hao J, Bhattacherjee A, Chen W, et al. Decoding molecular and cellular heterogeneity of mouse nucleus accumbens. Nat Neurosci. 2021;24:1757–71.
Calipari ES, Bagot RC, Purushothaman I, Davidson TJ, Yorgason JT, Peña CJ, et al. In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proc Natl Acad Sci USA. 2016;113:2726–31.
Lobo MK, Covington HE, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–90.
Kupchik YM, Brown RM, Heinsbroek JA, Lobo MK, Schwartz DJ, Kalivas PW. Coding the direct/indirect pathways by D1 and D2 receptors is not valid for accumbens projections. Nat Neurosci. 2015;18:1230–2.
Heimer L, Zahm DS, Churchill L, Kalivas PW, Wohltmann C. Specificity in the projection patterns of accumbal core and shell in the rat. Neuroscience. 1991;41:89–125.
Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–48.
Fox ME, Wulff AB, Franco D, Choi EY, Calarco CA, Engeln M, et al. Adaptations in nucleus accumbens neuron subtypes mediate negative affective behaviors in fentanyl abstinence. Biol Psychiatry. 2023;93:489–501.
Damez-Werno DM, Sun H, Scobie KN, Shao N, Rabkin J, Dias C, et al. Histone arginine methylation in cocaine action in the nucleus accumbens. Proc Natl Acad Sci USA. 2016;113:9623–8.
Godino A, Salery M, Durand-de Cuttoli R, Estill MS, Holt LM, Futamura R, et al. Transcriptional control of nucleus accumbens neuronal excitability by retinoid X receptor alpha tunes sensitivity to drug rewards. Neuron. 2023;111:1453–67.e7.
Cole SL, Chandra R, Harris M, Patel I, Wang T, Kim H, et al. Cocaine-induced neuron subtype mitochondrial dynamics through Egr3 transcriptional regulation. Mol Brain. 2021;14:101.
Chandra R, Engeln M, Schiefer C, Patton MH, Martin JA, Werner CT, et al. Drp1 mitochondrial fission in D1 neurons mediates behavioral and cellular plasticity during early cocaine abstinence. Neuron. 2017;96:1327–41.e6.
Reiner BC, Zhang Y, Stein LM, Perea ED, Arauco-Shapiro G, Ben Nathan J, et al. Single nucleus transcriptomic analysis of rat nucleus accumbens reveals cell type-specific patterns of gene expression associated with volitional morphine intake. Transl Psychiatry. 2022;12:374.
Savell KE, Tuscher JJ, Zipperly ME, Duke CG, Phillips RA, Bauman AJ, et al. A dopamine-induced gene expression signature regulates neuronal function and cocaine response. Sci Adv. 2020;6:eaba4221.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Cahill KM, Huo Z, Tseng GC, Logan RW, Seney ML. Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci Rep. 2018;8:9588.
Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, et al. Gene set knowledge discovery with enrichr. Curr Protoc. 2021;1:e90.
Song W-M, Zhang B. Multiscale embedded gene co-expression network analysis. PLoS Comput Biol. 2015;11:e1004574.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Ramakrishnan A, Wangensteen G, Kim S, Nestler EJ, Shen L. DeepRegFinder: deep learning-based regulatory elements finder. Bioinforma Adv. 2024;4:vbae007.
Pickens CL, Airavaara M, Theberge F, Fanous S, Hope BT, Shaham Y. Neurobiology of the incubation of drug craving. Trends Neurosci. 2011;34:411–20.
Phillips RA, Tuscher JJ, Fitzgerald ND, Wan E, Zipperly ME, Duke CG, et al. Distinct subpopulations of D1 medium spiny neurons exhibit unique transcriptional responsiveness to cocaine. Mol Cell Neurosci. 2023;125:103849.
Salery M, Godino A, Nestler EJ. Drug-activated cells: from immediate early genes to neuronal ensembles in addiction. Adv Pharmacol. 2021;90:173–216.
Kronman H, Richter F, Labonté B, Chandra R, Zhao S, Hoffman G, et al. Biology and bias in cell type-specific RNAseq of nucleus accumbens medium spiny neurons. Sci Rep. 2019;9:8350.
Ludewig S, Korte M. Novel insights into the physiological function of the APP (Gene) family and its proteolytic fragments in synaptic plasticity. Front Mol Neurosci. 2016;9:161.
Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. 2006;31:2304–12.
Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J Neurochem. 2004;88:1211–9.
Cho IH, Panzera LC, Chin M, Hoppa MB. Sodium channel β2 subunits prevent action potential propagation failures at axonal branch points. J Neurosci. 2017;37:9519–33.
Teague CD, Nestler EJ. Key transcription factors mediating cocaine-induced plasticity in the nucleus accumbens. Mol Psychiatry. 2022;27:687–709.
Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–67.
Ray MH, Williams BR, Kuppe MK, Bryant CD, Logan RW. A glitch in the matrix: the role of extracellular matrix remodeling in opioid use disorder. Front Integr Neurosci. 2022;16:899637.
Zea-Redondo L, Pombo A. Mechanisms of enhancer function in neuronal systems in health and disease. Curr Opin Syst Biol. 2023;100443:32–3.
Harrison LJ, Bose D. Enhancer RNAs step forward: new insights into enhancer function. Development. 2022;149:dev200398.
Tan B, Browne CJ, Nöbauer T, Vaziri A, Friedman JM, Nestler EJ. Drugs of abuse hijack a mesolimbic pathway that processes homeostatic need. Science. 2024;384:eadk6742.
Strickland JC, Gipson CD, Dunn KE. Dopamine supersensitivity: a novel hypothesis of opioid-induced neurobiological mechanisms underlying opioid-stimulant co-use and opioid relapse. Front Psychiatry. 2022;13:835816.
Acknowledgements
We thank Stephen Pirpinias, Katherine Beach, Catherine McManus, Kyra Schmidt, and Ezekiell Mouzon for transgenic mouse breeding and genotyping. We also thank Dr. Edgardo Aritzia from the Dean’s Flow Cytometry CoRE at the Icahn School of Medicine at Mount Sinai for assistance with nuclei sorting. Schematics in Figs. 1 and 4 were created with BioRender.com. This work was supported by grants from the National Institute on Drug Abuse (P01DA047233, R01DA007359, and R01DA014133 to E.J.N), National Science and Research Council of Canada (NSERC PDF to C.J.B.), National Institute on Alcohol Abuse and Alcoholism (K99AA027839 to P.M.), and Brain and Behavior Research Foundation (BBRF Young Investigator Grant to P.M.).
Author information
Authors and Affiliations
Contributions
Conceptualization: CJB, PM, and EJN Methodology: ME, XZ, and BZ Investigation: CJB, PM, ME, LMH, XZ, RF Visualization: CJB, PM, ME, XZ, Formal analysis: CJB, PM, ME, XZ Writing: CJB, PM, EJN Writing–review and editing: All authors read and commented on the manuscript. Supervision: LS, BZ, EJN.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All experiments were conducted in accordance with the regulations and guidelines of the Institutional Animal Care and Use Committee (IACUC) at the Icahn School of Medicine at Mount Sinai (protocol number 08-0465).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Browne, C.J., Mews, P., Estill, M. et al. Cocaine and morphine induce shared and divergent transcriptional regulation in nucleus accumbens D1 and D2 medium spiny neurons. Mol Psychiatry 30, 4247–4257 (2025). https://doi.org/10.1038/s41380-025-03004-1
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-03004-1
This article is cited by
-
Cocaine-induced gene regulation in D1 and D2 neuronal ensembles of the nucleus accumbens
Communications Biology (2025)