Abstract
Suicidality phenotypes, consisting of suicidal ideation (SI), suicide attempt (SA), and suicide death (SD), are all heritable but present unique challenges in genome-wide association studies (GWAS) due to their individual complexity, overlap with each other and with related self-harm phenotypes, and varying associations with psychiatric disorders. GWAS have uncovered several loci associated with suicidality phenotypes by meta-analyzing data from multiple cohorts. However, combining datasets from many research groups, where each group may use different study designs, phenotyping instruments, and definitions of suicidality phenotypes, presents challenges. Heterogeneity resulting from these differences can limit genetic discovery; harmonizing phenotype definitions to ensure consistency will greatly improve results. Here, we describe a standardized phenotyping protocol that draws on the expertise of a subgroup of clinicians, researchers, and experts from the Psychiatric Genomics Consortium Suicide Working Group to propose consensus definitions for SI, SA, and SD for genetic studies.
Similar content being viewed by others
Introduction
Suicidality phenotypes are multidimensional, representing a range of thoughts and behaviors directed toward intentional self-injurious acts with at least partial intent to die. Suicidality phenotypes are defined within three major categories [1]: (1) suicidal ideation (SI), wherein an individual contemplates ending their own life with or without a specific plan; (2) suicide attempt (SA), wherein an individual takes action to cause harm to themselves with at least some intent to die; (3) suicide death (SD), or death caused by intentional action to take one’s own life. Consistent phenotype definitions remain a challenge in suicide genetics, however, for several reasons. Suicidality phenotypes lack standardized diagnostic criteria, have considerable overlap with other self-harm phenotypes, and are often considered psychiatric symptoms rather than a distinct clinical category. Several lines of evidence support the view that suicidality phenotypes are distinct clinical categories, specifically studies demonstrating substantial unique heritability independent of psychiatric disorders [2, 3], recent recognition of suicidal behavior as a standalone diagnostic code in the DSM-5-TR [4], and the significant clinical and public health relevance of SD and SA as leading causes of preventable death and injury. Moreover, suicidality can be directly reduced or prevented without fully resolving potentially underlying psychiatric conditions. For example, clozapine and ketamine have been associated with a reduction in suicidality that is not solely attributable to their effects on psychotic symptoms [5, 6] or depression and anxiety [7], respectively. These studies suggest that suicidality phenotypes can have partially distinct mechanisms and treatment responses, supporting their consideration as clinically meaningful phenotypes in their own right.
Genome-wide association studies (GWAS) of SI, SA, and SD have uncovered several associated genetic loci. Although the most recent GWAS of SI [8] and SD [9] were conducted in single cohorts, the formation of the Psychiatric Genomics Consortium Suicide Working Group (PGC SUI, formerly the International Suicide Genetics Consortium (ISGC)) facilitated the first organized effort to conduct GWAS meta-analyses of SA [3, 10], including over 43 000 cases from 22 diverse cohorts. GWAS meta-analyses are sensitive to the heterogeneity of the contributing cohorts. Solutions are available for handling some sources of heterogeneity, such as using inverse-variance weighted methods to meta-analyze cohorts with sample size disparities and implementing a standard analytic protocol to ensure that GWAS within cohorts apply consistent data processing pipelines, statistical models, and covariates. However, heterogeneity resulting from inconsistent phenotype definitions remains a challenge in suicide genetics, as for the full range of psychiatric and substance use traits. Such heterogeneity can diminish the ability to detect genetic variations uniquely associated with a given suicidality phenotype [11, 12].
To best analyze suicidality data from varied cohorts, careful consideration of phenotyping is required to maximize comparability of these complex phenotypes across study samples to ensure robust genetic analyses. Developing a protocol for consistent suicidality phenotype definitions across cohorts in genetic studies will, therefore, be of great value. The ideal protocol will facilitate harmonization and ensure that phenotype definitions are accurate, easy to implement, and provide guidance for incorporating the varied phenotyping methods commonly used in psychiatric genetics. Definitions should also be designed to allow effective application to existing datasets and to guide the development and inclusion of new cohorts.
To address these challenges, members of PGC SUI describe here a set of guidelines for suggested best practices in defining suicidality case and control phenotypes. This protocol is implemented within PGC SUI and can be applied more broadly to research on the genetics and biology of suicidality phenotypes. Specifically, we make recommendations to derive standardized phenotypes from a variety of information sources, including clinical interviews, self-report questionnaires, suicide-specific rating scales and electronic health records (EHR). We also provide guidance on how to handle missing phenotype information, co-occurring phenotypes, and time-limited measures. Utilization of these recommendations will substantially benefit collaborative efforts by increasing participation and statistical power, improving comparability and reproducibility, and enhancing the overall quality of meta-analyses across studies.
Aim and objectives
PGC SUI conducts large-scale genomic analyses of suicidality phenotypes by combining data from studies worldwide. Our current priorities are to perform separate GWAS of SI, SA, and SD, dissect their shared and distinct genetic etiologies and quantify the extent to which their genetic liabilities may overlap with, or be independent from, those of co-occurring psychiatric disorders. PGC SUI designed a phenotyping protocol to enable these objectives by ensuring rigor and comparability of phenotype definitions across the cohorts in our GWAS, allowing us to study the genetics of these suicidality phenotypes both separately and together, and control for bias that may arise from the frequent co-occurrence of psychiatric disorders.
Protocol development
The recommendations presented in this protocol reflect a consensus reached by PGC SUI, based on literature review, expert opinion and workgroup discussions. An initial evaluation of phenotypes used in prior GWAS and GWAS meta-analyses of suicidality was conducted by a smaller phenotyping task force, comprising clinical experts in the field along with core PGC SUI analysts. This evaluation served as the foundation for the development of the present protocol.
Specifically, we considered GWAS meta-analyses of suicide attempt conducted by ISGC [3, 10], as well as single-cohort GWAS from population-based studies like MVP [8, 13, 14] and iPSYCH [15] or cohorts specifically ascertained for suicidality, such as the Columbia University cohort [16] and the Genetic Investigation of Suicide and SA (GISS) cohort [17]. The phenotyping task force assessed the strengths and weaknesses of various definitions, ascertainment methods, and measures. We also considered how the data generated from GWAS meta-analyses of suicidality phenotypes may be used in downstream genetic analyses. We incorporated insights from literature on phenotyping in genetic research [12], with the goal of constructing phenotype definitions that are both robust and compatible with follow-up analyses. Leveraging all of this information, the phenotyping task force developed a preliminary phenotyping protocol.
This protocol was then iteratively refined in close collaboration with the broader PGC SUI working group during monthly workgroup meetings, where feedback was actively solicited and incorporated. Once a full draft of the protocol was completed, it was circulated to the working group mailing list and underwent several rounds of review and revision. All members of the working group were encouraged to provide questions, concerns, or suggestions. All members’ input was weighted equally and feedback could be provided anonymously if desired. Discussions were held on monthly working group calls to resolve differences of opinion on specific recommendations. In cases where consensus could not be fully reached, solutions that accounted for multiple perspectives were adopted. The protocol was revised until all concerns from working group members were addressed. All named authors approved the final version of the protocol presented here.
Phenotype definitions
Cases: Table 1 presents the current international standard phenotype definitions for SI, SA, and SD along with non-suicidal self-injury (NSSI) [1, 18]. The present strategy of PGC SUI is to conduct GWAS focusing on each of the suicidality phenotypes, rather than combining all of them into one broad suicidality phenotype. This approach serves to maximize specificity and minimizes heterogeneity, as substantial genetic differences exist between these phenotypes. For example, molecular genetics studies estimate moderate to high genetic correlations among these four phenotypes (rg = 0.53–0.84), but all are significantly below 1 [19]. Similarly, twin studies estimate substantial but incomplete genetic correlations between SA and SD [20] and show differing heritability estimates among these phenotypes [21, 22]. To further maintain specificity in GWAS analyses, we propose that each suicidality case phenotype be directly assessed for the phenotype of interest using validated measures, such as clinical psychiatric interviews or self-report instruments, rather than inferred by proxy. This study design aims to increase the specificity of the GWAS of each suicidality phenotype by minimizing bias that could arise from including cohorts specifically collected for the study of a more severe suicidality phenotype. For example, the definition of SA used here specifically describes non-fatal acts, thus SD cases should not be considered as SA cases unless a previous non-fatal SA is known. Similarly, when using data from a cohort which specifically collected individuals who made a suicide attempt or died by suicide, only individuals with phenotypic information available indicating that they also meet criteria for SI should be included in an SI GWAS. Thus, all SA or SD cohorts should not automatically be meta-analyzed with SI cohorts. Although SI logically precedes SA and SD, this approach avoids constructing an overly broad ‘suicidality phenotype’ that conflates distinct clinical presentations and enriches SI samples with SA/SD cases. However, SA and SD are not exclusion criteria for SI case status. If individuals meet the criteria for SI case status outlined here, they should still be included, even if it is known that they made a suicide attempt or died by suicide. Together, these criteria ensure that each GWAS reflects the expected prevalence of more severe suicidality phenotypes in these populations. Thus, for example, an SI GWAS will proportionally reflect the full spectrum of individuals who experience SI, such that most individuals with SI do not go on to attempt or die by suicide, while some do [23, 24].
Similarly, the presence or absence of a psychiatric disorder does not impact case status, such that the sample used in a GWAS of a specific suicidality phenotype will accurately resemble the general population in relation to the prevalence of psychiatric disorders in suicidality cases. Moreover, many cohorts lack information on all three major suicidality phenotypes or complete psychiatric histories; therefore, requiring case phenotype definitions to exclude individuals with a more severe suicidality phenotype or a psychiatric disorder would likely result in a sample size that is too small to conduct a GWAS with reasonable statistical power. Finally, conducting separate and specific GWAS of each suicidality phenotype still allows subsequent genetic analysis across all suicidality phenotypes together via meta-analysis, common factor GWAS, or similar approaches.
Controls: Controls are individuals without the case suicidality phenotype. Table 2 describes the criteria for controls in suicidality GWAS. All controls should be screened for the case phenotype and any more severe suicidality phenotypes using available information, with affected individuals being removed from the analyses. For example, controls in a GWAS of SA should be screened for SA and SD, but not SI. Individuals ascertained for having psychiatric disorders should be included within the control group, however, they should be screened for the absence of the case suicidality phenotype. Otherwise, the higher prevalence of these phenotypes amongst individuals with psychiatric disorders [25] could lead to a higher possibility of misclassification of controls. For example, individuals ascertained for psychiatric diagnoses should also be screened for SI before inclusion as controls in a GWAS of SI and should be screened for SA before inclusion as controls in a GWAS of SA and GWAS of SD. Evidence of SA is used as an additional exclusion criterion for controls in SD GWAS, since most potential controls are living. When suicidality screening is missing only for psychiatrically healthy individuals, however, the likelihood of misclassification is low, and retaining these individuals helps preserve sample size and reduce potential bias in cohorts that do not assess suicidality in all participants. When data on deceased controls and cause of death are available, individuals classified as having died by undetermined intent (UDI) should be excluded from the control group, as several studies have suggested that a proportion of UDI deaths are SDs [26,27,28]. When screening controls, any individual with a more severe phenotype should be excluded (e.g., any individual with evidence of SA or SD should be excluded as controls in a GWAS of SI regardless of whether there is evidence of SI). Individuals ascertained for having psychiatric disorders who are missing information on the case suicidality phenotype (e.g., they were not asked or declined to answer during their interview) should be excluded.
Typical phenotyping sources
SI and SA data may be available from structured psychiatric interviews, and other forms of clinical instruments, scales, and questionnaires. While several suicide-specific instruments exist (e.g., the Columbia Suicide Severity Rating Scale (C-SSRS) [29]), general psychiatric instruments (e.g., the Composite International Diagnostic Interview [30], the Schedules for Clinical Assessment in Neuropsychiatry [31]) also often include items that assess suicidality phenotypes (Supplementary Tables 1–2). SI and SA phenotypes may also be derived from EHRs in the form of International Classification of Diseases (ICD) codes or clinical notes. SD can be identified from coroners’ or medical examiners’ reports and death registries (such as the U.S. National Death Index or state-based registries). Here, we suggest guidelines for how these different information sources can be used to define suicidality phenotype cases and controls according to our phenotyping definitions.
SI/SA instrument guidelines
PGC SUI has developed basic guidelines for determining whether a particular item from an instrument should be used to define cases and controls for SI and SA. Most importantly, questions/items that are acceptable for use in defining SI should include the specification of thoughts of suicide or death, including terminology such as “suicidal thoughts”, “better off dead”, or “thoughts that life was not worth living”. Questions or items that are used for identifying prior SA should include specific language regarding the attempt and, ideally, assess for a history of prior attempts at any time in the individual’s life. Language for suicide attempt assessment should include phrases such as “suicide attempt”, “tried to kill yourself”, “intent to die”, or “result in death” and avoid language that conflates SA and NSSI, such as “harm yourself” or “injure yourself.” For both SI and SA questions/items, only one phenotype should be included within a question with a binary response. For example, a question that asks whether an individual has “considered or done anything to hurt yourself” with a binary “yes-no” response is unable to differentiate cases of SI, SA, and NSSI.
Time frame
Additionally, caution should be exercised with questions that assess a specified time period (e.g., the past week or past year). Although it is reasonable to include an individual as an SA case who answered “yes” to an attempt in the past year, another individual that answered negatively to such a question cannot be easily ruled out as a case, in contrast to individuals assessed with lifetime measures. When a response to a single time-limited question may be inconclusive, other factors such as psychiatric diagnostic status should be considered to determine the likelihood of a false negative. This is not to say that time-limited questions have no utility; several studies suggest that when time-limited questions are assessed repeatedly at multiple time points (as is often done in longitudinal studies), the cumulative response captures mental health conditions more accurately than a single lifetime measurement which is susceptible to recall bias [32, 33]. Therefore, the use case for time-limited questions may depend on the specific study design and the other information available. Some instruments, for example the C-SSRS, have both a time-limited and lifetime history version, and in such cases, the lifetime version should be employed at the first assessment in new cohorts ascertained for GWAS. If both time-limited and lifetime measures are available in an existing dataset, it is recommended that the most recent measure of lifetime history of suicidality be used first to determine cases and controls, and any potential missed cases can be identified using time-limited measures or earlier assessments of lifetime measures.
Inconsistencies
Individuals positive for one item but negative for another should be included as a case. Many valid inconsistencies often arise when assessments are conducted at different times or measure different periods of time. For example, a negative response at baseline but a positive response at follow-up likely indicates suicidality during the follow-up period and should result in the individual being classified as a case. Similarly, endorsement of suicidality on a lifetime assessment, but not on a time-limited (e.g., “past year”) assessment, should warrant case status.
Importantly, inconsistencies across instruments assessing the same time period do not necessarily indicate measurement error. Evidence suggests that a single positive report, regardless of modality, likely still reflects true suicidality. For instance, one study comparing ecological momentary assessments (EMA) of SI collected every day over one week with retrospective ratings from a clinician-administered interview at week’s end, showed that individuals who reported SI only during EMA, but not in the interview, were no less likely to have a valid history of suicidality than those who reported consistently [34].
A related and common issue involves inconsistencies in responses to the same instrument across time points that cannot be explained by the aforementioned factors. Studies have shown that 23–43% of individuals who report a lifetime history of suicidality at baseline fail to report the same history at follow-up [35,36,37]. Furthermore, longitudinal studies show stable rates of lifetime suicidality across waves, despite the expectation that such rates should increase within a closed cohort over time [38]. This phenomenon is not unique to suicidality; for example, cross-sectional studies consistently report declining rates of lifetime depression and anxiety across age groups [39]. These inconsistencies likely reflect recall bias, mood congruent memory, and psychological distancing from past SI/SA, rather than initial false endorsements.
Altogether, these findings support classifying individuals as cases based on any credible endorsement of the suicidality phenotype, regardless of consistency across measures or time points. Variability in reporting across modalities or assessment windows does not undermine the validity of a positive response. However, when possible, inconsistencies should be periodically reevaluated, and the reliability and validity of assessment tools should be empirically reassessed.
Dichotomization
Several instruments, such as those which employ scales, do not use binary “yes-no” questions. Continuous traits can yield more powerful GWAS because they contain more information. However, for the purposes of contributing to a consortium or participating in a meta-analysis, it is usually best to code suicidality phenotypes as binary variables, for consistency with most other studies. Items on scales should therefore be dichotomized based on any reported evidence of a phenotype (case) and evidence of the absence of a phenotype (control). For example, the Beck Depression Inventory [40] assesses SI on a scale: 0 = “I don’t have any thoughts of killing myself”, 1 = “I have thoughts of killing myself, but I would not carry them out”, 2 = “I would like to kill myself”, 3 = “I would kill myself if I had the chance”. In this item, responses 1–3 indicate varying degrees of SI, and all these scores would be defined as SI cases if the responses were dichotomized. The complete absence of SI, indicated by those who responded 0, would be used to define controls.
Minimal phenotypes
Lastly, in some scenarios, research groups may only have access to rapid screening measures that are designed to quickly identify the need for further evaluation and inform disposition decisions, resulting in a “minimal” or less specific phenotype. For example, item 9 in the popular Patient Health Questionnaire (PHQ)-9 (“Over the last 2 weeks, how often have you been bothered by thoughts that you would be better off dead, or of hurting yourself?”) is a broad assessment of thoughts of self-harm that does not separate SI from thoughts of NSSI, making it less specific than more detailed evaluations [41, 42]. The PHQ-9 is widely used in healthcare and research settings, meaning that this item is often available for large cohorts such as biobanks. The use of such “minimal” phenotypes and particularly their inclusion within larger consortia efforts, should balance the trade-offs between sample size, statistical power, and potential loss of specificity. Additionally, the impact of including less-specific phenotypes can and should be assessed in many ways. The optimal benchmarking method will depend on the characteristics of the specific GWAS. One common approach is to compare SNP-heritability estimates, as previous studies have shown that GWAS using minimal phenotypes tend to yield lower SNP-heritability estimates than those using more strictly defined phenotypes [11, 43]. However, this method is best suited for large, well-powered GWAS that yield significant SNP-heritability estimates. In the context of suicidality GWAS, benchmarking may involve comparing SNP-heritability and pairwise genetic correlations with previously validated GWAS, such as the ISGC1 GWAS of SA [3], as well as with other contributing cohorts. In cases where GWAS are underpowered due to small sample size, the variance explained (R2) by polygenic risk scores for the suicidality phenotype in question, trained on a validated GWAS, can instead be compared across cohorts and phenotype definitions. Additionally, leave-one-out and subgroup meta-analyses, in which GWAS of minimal phenotypes are excluded, can help evaluate changes in heterogeneity statistics, offering insight based on the influence of minimal phenotypes on meta-analytic results.
Although we promote initially analyzing minimal and strict phenotype definitions separately, we encourage collecting both so that future meta-analysis efforts can evaluate how to best use different kinds of data to curate appropriate phenotype definitions for their specific purposes and goals. Given the small number of large-scale suicidality GWAS to date, our current understanding of how these “minimal” phenotypes differ genetically from more stringent definitions remains incomplete. For this reason, we do not recommend the use of rigid benchmarks at this stage. Instead, we emphasize the importance of characterizing this potential heterogeneity as a key focus of future genetic studies of suicidality. Critically, researchers should clearly and thoroughly report any benchmarking and sensitivity analyses they conduct, not only to support interpretation and discussion of their results, but also to enable other researchers to independently evaluate the reliability and robustness of the findings.
SI/SA instrument recommendations
We applied the above guidelines to a collection of instruments used in psychiatric genetic studies and/or suicide research identified by group consensus and literature review of large meta-analyses performed by various working groups of the PGC [3, 44,45,46]. Specific guidelines for all evaluated questionnaires, including exact questions/items and acceptable responses are provided in Supplementary Tables 1 and 2 for SA and SI, respectively. Frequently used instruments that define a minimal phenotype are also included. The instruments assessed do not represent a comprehensive evaluation of every psychiatric instrument available but rather are presented as examples of commonly used instruments that meet the best practice guidelines set in this protocol and are likely to be useful in studies that aim to construct clear and accurate definitions of SI and SA using pre-existing phenotype collections.
For new studies that aim to collect data on suicidality phenotypes, it is strongly recommended to use an instrument that provides a detailed assessment of suicidality phenotypes, has wide distribution and accessibility, offers flexible language and licensing options (preferably validated in the languages used), and adheres to broadly accepted phenotype definitions. In addition, it is recommended to use versions of instruments that assess lifetime history, whenever possible. The C-SSRS [29] meets these criteria, and it is recommended that this or a similarly constructed instrument be used in new datasets.
EHR data guidelines
Currently, most genetic studies of suicidality phenotypes rely on clinical instruments for phenotyping; however, with the advent of large-scale, EHR-linked biobanks, it is anticipated that ICD codes and other types of EHR data (e.g., clinical notes) will become increasingly important in defining suicidality phenotypes. Several studies have compared ICD code-based definitions with natural language processing (NLP) algorithms developed to identify suicidality phenotypes from clinical notes in EHR data. These show that standard SI/SA ICD codes alone perform poorly, and phenotyping is improved by using information from both ICD codes and clinical notes [47,48,49,50,51]. Efforts to develop novel ICD-9 and ICD-10 diagnostic code lists for SI and SA based on literature review and expert consensus [52, 53] still show that ICD codes underperform relative to instrument and clinical data. While ICD codes are the least accurate among phenotyping sources, abandoning their use, or EHR data altogether, would overlook the substantial value of EHR-based studies, which offer scalable, cost-efficient access to diverse real-world clinical populations. Thus, it is recommended that ICD code data be coupled with data from instruments or clinical notes when possible to enhance phenotyping, as in previous genetic studies of suicidality using EHR data [14, 49, 54]. For studies with ICD code data available, we recommend the use of the ICD code lists provided by Monson et al. [52] to define SI and SA. While NLP algorithms have shown promise in defining suicidality phenotypes in certain healthcare systems [47, 55], a standard consensus on their application has not yet been established. Thus, their use in cohorts contributing to meta-analyses should be considered on a case-by-case basis to ensure that they have been properly validated and adhere to the guidelines set above, and in particular, that the NLP algorithm can differentiate suicidality phenotypes from one another and from NSSI.
Death records
Suicide is conservatively attributed as a cause of death worldwide [56] so the possibility of false positive classification from death records is considered minimal. However, in some cases national death registries may serve as a more accurate source of cause of death than local or state registries, as was shown in a previous evaluation of the accuracy of firearm death determination [57]. Although some studies suggest that a substantial proportion of deaths of undetermined intent (UDIs) are SDs, and genetic epidemiology studies indicate minimal genetic differences between SD and UDI [58], these findings have yet to be confirmed in molecular genetic studies. Given our current preference for conservative phenotype definitions which allow us to better examine genetic similarities and differences among specific suicidality phenotypes, we do not consider UDIs suitable for inclusion in SD GWAS.
Co-occurring psychiatric disorders
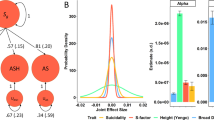
Psychiatric disorders are major risk factors for suicidality phenotypes and are often comorbid with them. The prevalence of psychiatric disorders is estimated to range between 43–52% among SI cases, 55–66% among SA cases [25], and 60–98% among SD cases [59, 60]. The high comorbidity of psychiatric disorders with suicidality phenotypes can bias GWAS towards detecting associations with psychiatric disorders if not appropriately controlled. Because the lifetime prevalence of psychiatric disorders in the general population is ~30% [61], retaining individuals with psychiatric diagnoses in the control group may increase statistical power to detect associations specific to the suicidality phenotype (Fig. 1A). Conversely, removing all individuals with psychiatric disorders from the control group, while retaining individuals with psychiatric disorders in the case group (Fig. 1C), would increase the likelihood of identifying associations with psychiatric phenotypes generally rather than the suicidality phenotype [12], distort estimates of variance explained [62], and bias genetic correlations [63]. PGC SUI recommends screening for the absence of the case suicidality phenotype and any more severe suicidality phenotype, and retaining individuals with psychiatric diagnoses in the control group (Fig. 1B). The use of such controls, as opposed to completely unscreened controls or only psychiatrically healthy controls, maximizes statistical power without introducing substantial bias to the GWAS [12]. While psychiatrically healthy controls are often used in psychiatric genetic studies, we advise against removing controls with psychiatric disorders in suicidality studies. Such exclusion risks producing GWAS results that would likely measure differences not just between individuals with and without suicidality, but also differences between those with and without psychiatric disorders more broadly [63]. In turn, this can inflate genetic correlations or generate spurious associations between a target phenotype and any secondary phenotype screened out of the control group, as has been seen in studies on other psychiatric phenotypes [63, 64].
A–C The left panels represent SA cases, and the right panels represent the control group. Amongst SA cases, the prevalence of psychiatric disorders is 0.9. A The population control group displays psychiatric disorders at a prevalence of 0.3 and SA at a prevalence of 0.02. B The SA-screened control group displays psychiatric disorders at a prevalence of 0.3. C The non-psychiatric control group assumes a prevalence of 0 for both psychiatric disorders and SA.
Limitations
Some potential limitations of our phenotyping protocol should be noted. As described in the Phenotype Definitions section, our case phenotype definitions do not exclude individuals with a more severe suicidality phenotype, a strategy used in some previous GWAS [8]. Although this stricter exclusion approach may enhance specificity, it is constrained by the availability of detailed phenotypic data for all suicidality phenotypes. Most datasets lack comprehensive information on all suicidality phenotypes, and imposing strict exclusions would substantially reduce sample sizes to levels that make GWAS infeasible. Our protocol was designed to balance the ideal phenotype definitions with those that are practical given existing data, and to provide the flexibility to investigate the distinct and shared genetic etiology of suicidality phenotypes. Additionally, although PGC SUI is an international working group, the nuances in terminology and phenotype definitions focused on in this protocol may differ or not be relevant in specific languages, cultures, and contexts. When this is the case, we suggest that the broad ideas of this protocol be considered while relying primarily on the expert opinion of clinicians and scientists familiar with the specific context. Finally, our perspectives are based on current knowledge and best practices in the field. As our understanding of the components of suicidality and their phenotypic definitions continues to evolve, these recommendations may need to be refined. Therefore, we encourage consortia to prioritize the collection of comprehensive phenotypic data at the individual level whenever possible, to allow for centralized phenotype reconstruction as needed.
Conclusion
Here we provide the perspectives of PGC SUI on defining SI, SA, and SD phenotypes for genetic studies and comprehensive phenotyping protocols. Recognizing the many complexities in these phenotypes and the sources from which they are derived, we present proposed standard definitions and guidelines to address these challenges and ensure consistency. By harmonizing phenotypes across cohorts, this protocol aims to reduce heterogeneity, increase power in meta-analyses, and improve the comparability and reproducibility of genetic studies. Use of this protocol by PGC SUI and the greater suicide research community is expected to increase collaborative research efforts and advance understanding of the genetic underpinnings of suicidality.
References
Crosby A, Ortega LV, Melanson C. Self-directed violence surveillance; uniform definitions and recommended data elements. National Center for Injury Prevention and Control (U.S.). Division of Violence Prevention.; 2011.
Fu Q, Heath AC, Bucholz KK, Nelson EC, Glowinski AL, Goldberg J, et al. A twin study of genetic and environmental influences on suicidality in men. Psychol Med. 2002;32:11–24.
Mullins N, Kang J, Campos AI, Coleman JRI, Edwards AC, Galfalvy H, et al. Dissecting the shared genetic architecture of suicide attempt, psychiatric disorders, and known risk factors. Biol Psychiatry. 2022;91:313–27.
American Psychiatric Association, issuing body. Diagnostic and statistical manual of mental disorders: DSM-5-TR. American Psychiatric Association Publishing; 2022. https://doi.org/10.1001/archpsyc.60.1.82.
Meltzer HY, Alphs L, Green AI, Altamura AC, Anand R, Bertoldi A, et al. Clozapine treatment for suicidality in schizophrenia: International Suicide Prevention Trial (InterSePT). Arch Gen Psychiatry. 2003;60:82–91.
Masdrakis VG, Baldwin DS. Prevention of suicide by clozapine in mental disorders: systematic review. Eur Neuropsychopharmacol. 2023;69:4–23.
Ballard ED, Ionescu DF, Vande Voort JL, Niciu MJ, Richards EM, Luckenbaugh DA, et al. Improvement in suicidal ideation after ketamine infusion: relationship to reductions in depression and anxiety. J Psychiatr Res. 2014;58:161–6.
Ashley-Koch AE, Kimbrel NA, Qin XJ, Lindquist JH, Garrett ME, Dennis MF, et al. Genome-wide association study identifies four pan-ancestry loci for suicidal ideation in the Million Veteran Program. PLoS Genet. 2023;19:e1010623.
Docherty AR, Shabalin AA, DiBlasi E, Monson E, Mullins N, Adkins DE, et al. Genome-wide association study of suicide death and polygenic prediction of clinical antecedents. Am J Psychiatry. 2020;177:917–27.
Docherty AR, Mullins N, Ashley-Koch AE, Qin X, Coleman JRI, Shabalin A, et al. GWAS meta-analysis of suicide attempt: identification of 12 genome-wide significant loci and implication of genetic risks for specific health factors. Am J Psychiatry. 2023;180:723–38.
Cai N, Revez JA, Adams MJ, Andlauer TFM, Breen G, Byrne EM, et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat Genet. 2020;52:437–47.
Cai N, Verhulst B, Andreassen OA, Buitelaar J, Edenberg HJ, Hettema JM, et al. Assessment and ascertainment in psychiatric molecular genetics: challenges and opportunities for cross-disorder research. Mol Psychiatry. 2025;30:1627–38.
Kimbrel NA, Ashley-Koch AE, Qin XJ, Lindquist JH, Garrett ME, Dennis MF, et al. Identification of novel, replicable genetic risk loci for suicidal thoughts and behaviors among US military veterans. JAMA Psychiatry. 2023;80:135–45.
Kimbrel NA, Ashley-Koch AE, Qin XJ, Lindquist JH, Garrett ME, Dennis MF, et al. A genome-wide association study of suicide attempts in the million veterans program identifies evidence of pan-ancestry and ancestry-specific risk loci. Mol Psychiatry. 2022;27:2264–72.
Erlangsen A, Appadurai V, Wang Y, Turecki G, Mors O, Werge T, et al. Genetics of suicide attempts in individuals with and without mental disorders: a population-based genome-wide association study. Mol Psychiatry. 2020;25:2410–21.
Galfalvy H, Haghighi F, Hodgkinson C, Goldman D, Oquendo MA, Burke A, et al. A genome-wide association study of suicidal behavior. Am J Med Genet B Neuropsychiatr Genet. 2015;168:557–63.
Sokolowski M, Wasserman J, Wasserman D. Polygenic associations of neurodevelopmental genes in suicide attempt. Mol Psychiatry. 2016;21:1381–90.
Cipriano A, Cella S, Cotrufo P. Nonsuicidal self-injury: a systematic review. Front Psychol. 2017;8:1946.
Colbert SMC, Mullins N, Chan G, Meyers JL, Schulman J, Kuperman S, et al. Polygenic contributions to suicidal thoughts and behaviors in a sample ascertained for alcohol use disorders. Complex Psychiatry. 2023;9:11–23.
Edwards AC, Ohlsson H, Mościcki E, Crump C, Sundquist J, Lichtenstein P, et al. On the genetic and environmental relationship between suicide attempt and death by suicide. Am J Psychiatry. 2021;178:1060–9.
Voracek M, Loibl LM. Genetics of suicide: a systematic review of twin studies. Wien Klin Wochenschr. 2007;119:463–75.
Brent DA, Mann JJ. Family genetic studies, suicide, and suicidal behavior. Am J Med Genet C Semin Med Genet. 2005;133C:13–24.
Beck AT, Steer RA, Kovacs M, Garrison B. Hopelessness and eventual suicide: a 10-year Prospective study of patients hospitalized with suicidal ideation. Am J Psychiatry. 1985;142:559–63.
Nock MK, Borges G, Bromet EJ, Alonso J, Angermeyer M, Beautrais A, et al. Cross-national prevalence and risk factors for suicidal ideation, plans and attempts. Br J Psychiatry. 2008;192:98–105.
Nock MK, Hwang I, Sampson N, Kessler RC, Angermeyer M, Beautrais A, et al. Cross-national analysis of the associations among mental disorders and suicidal behavior: findings from the WHO World Mental Health Surveys. PLoS Med. 2009;6:e1000123.
Fernandez JM, Jayawardhana J. Are suicides underreported? The impact of coroners versus medical examiners on suicide reporting. Health Serv Res. 2025;60:e14381.
Snowdon J, Choi NG. Undercounting of suicides: where suicide data lie hidden. Glob Public Health. 2020;15:1894–901.
Pritchard C, Iqbal W, Dray R. Undetermined and accidental mortality rates as possible sources of underreported suicides: population-based study comparing Islamic countries and traditionally religious Western countries. BJPsych Open. 2020;6:e56.
Posner K, Brown GK, Stanley B, Brent DA, Yershova KV, Oquendo MA, et al. The Columbia-Suicide Severity Rating Scale: initial validity and internal consistency findings from three multisite studies with adolescents and adults. Am J Psychiatry. 2011;168:1266–77.
Robins LN, Wing J, Wittchen HU, Helzer JE, Babor TF, Burke J, et al. The Composite International Diagnostic Interview. An epidemiologic Instrument suitable for use in conjunction with different diagnostic systems and in different cultures. Arch Gen Psychiatry. 1988;45:1069–77.
Wing JK, Babor T, Brugha T, Burke J, Cooper JE, Giel R, et al. SCAN. Schedules for Clinical Assessment in Neuropsychiatry. Arch Gen Psychiatry. 1990;47:589–93.
Haeny AM, Littlefield AK, Sher KJ. Repeated diagnoses of lifetime alcohol use disorders in a prospective study: insights into the extent and nature of the reliability and validity problem. Alcohol Clin Exp Res. 2014;38:489–500.
Moffitt TE, Caspi A, Taylor A, Kokaua J, Milne BJ, Polanczyk G, et al. How common are common mental disorders? Evidence that lifetime prevalence rates are doubled by prospective versus retrospective ascertainment. Psychol Med. 2010;40:899–909.
Gratch I, Choo T-H, Galfalvy H, Keilp JG, Itzhaky L, Mann JJ, et al. Detecting suicidal thoughts: the power of ecological momentary assessment. Depress Anxiety. 2021;38:8–16.
Klimes-Dougan B, Safer MA, Ronsaville D, Tinsley R, Harris SJ. The value of forgetting suicidal thoughts and behavior. Suicide Life Threat Behav. 2007;37:431–8.
Goldney RD, Winefield AH, Winefield HR, Saebel J. The benefit of forgetting suicidal ideation. Suicide Life Threat Behav. 2009;39:33–7.
Eikelenboom M, Smit JH, Beekman ATF, Kerkhof AJFM, Penninx BWJH. Reporting suicide attempts: consistency and its determinants in a large mental health study: reporting suicide attempts. Int J Methods Psychiatr Res. 2014;23:257–66.
Hart SR, Musci RJ, Ialongo N, Ballard ED, Wilcox HC. Demographic and clinical characteristics of consistent and inconsistent longitudinal reporters of lifetime suicide attempts in adolescence through young adulthood: research article: consistent reporting of A suicide attempt. Depress Anxiety. 2013;30:997–1004.
Streiner DL, Patten SB, Anthony JC, Cairney J. Has âlifetime prevalenceâ reached the end of its life? An examination of the concept: lifetime prevalence. Int J Methods Psychiatr Res. 2009;18:221–8.
Beck A, Steer R, Carbin MG. Psychometric properties of the Beck Depression Inventory: twenty-five years of evaluation. Clinical Psychology Review. 1988;8:77–100.
Na PJ, Yaramala SR, Kim JA, Kim H, Goes FS, Zandi PP, et al. The PHQ-9 Item 9 based screening for suicide risk: a validation study of the Patient Health Questionnaire (PHQ)-9 Item 9 with the Columbia Suicide Severity Rating Scale (C-SSRS). J Affect Disord. 2018;232:34–40.
Chung TH, Hanley K, Le Y-C, Merchant A, Nascimento F, De Figueiredo JM, et al. A validation study of PHQ-9 suicide item with the Columbia Suicide Severity Rating Scale in outpatients with mood disorders at National Network of Depression Centers. J Affect Disord. 2023;320:590–4.
O’Connell KS, Koromina M, van der Veen T, Boltz T, David FS, Yang JMK, et al. Genomics yields biological and phenotypic insights into bipolar disorder. Nature. 2025;639:968–75.
Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29.
Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–8.
Bejan CA, Ripperger M, Wilimitis D, Ahmed R, Kang J, Robinson K, et al. Improving ascertainment of suicidal ideation and suicide attempt with natural language processing. Sci Rep. 2022;12:15146.
Callahan ST, Fuchs DC, Shelton RC, Balmer LS, Dudley JA, Gideon PS, et al. Identifying suicidal behavior among adolescents using administrative claims data: IDENTIFYING SUICIDAL BEHAVIOR USING CLAIMS DATA. Pharmacoepidemiol Drug Saf. 2013;22:769–75.
Colbert SMC, Lepow L, Fennessy B, Iwata N, Ikeda M, Saito T, et al. Distinguishing clinical and genetic risk factors for suicidal ideation and behavior in a diverse hospital population. Transl Psychiatry. 2025;15:63.
Fernandes AC, Dutta R, Velupillai S, Sanyal J, Stewart R, Chandran D. Identifying suicide ideation and suicidal attempts in a psychiatric clinical research database using Natural Language Processing. Sci Rep. 2018;8:7426.
Walkup JT, Townsend L, Crystal S, Olfson M. A systematic review of validated methods for identifying suicide or suicidal ideation using administrative or claims data: METHODS FOR IDENTIFYING SUICIDE USING CLAIMS DATA. Pharmacoepidemiol Drug Saf. 2012;21(Suppl 1):174–82.
Monson ET, Colbert SMC, Barr PB, Bejan CA, Andreassen OA, Ayinde OO, et al. Defining and Assessing International Classification of Disease Suicidality Phenotypes for Genetic Studies. medRxiv:2024.07.27.24311110. 2025. https://doi.org/10.1101/2024.07.27.24311110.
Barak-Corren Y, Castro VM, Javitt S, Hoffnagle AG, Dai Y, Perlis RH, et al. Predicting suicidal behavior from longitudinal electronic health records. Am J Psychiatry. 2017;174:154–62.
Coon H, Shabalin AA, Monson ET, DiBlasi E, Han S, Baird LM, et al. Different genetic liabilities to neuropsychiatric conditions in suicides with no prior suicidality. medRxiv:2025.05.02.25326877. 2025.
Cusick M, Velupillai S, Downs J, Campion TR Jr, Sholle ET, Dutta R, et al. Portability of natural language processing methods to detect suicidality from clinical text in US and UK electronic health records. J Affect Disord Rep. 2022;10:100430.
Tøllefsen IM, Hem E, Ekeberg Ø. The reliability of suicide statistics: a systematic review. BMC Psychiatry. 2012;12:9.
Barber C, Hemenway D. Too many or too few unintentional firearm deaths in official U.S. mortality data? Accid Anal Prev. 2011;43:724–31.
Edwards AC, Ohlsson H, Mościcki EK, Sundquist J, Crump C, Kendler KS, et al. Genetic differences between suicide deaths and deaths of undetermined intent. Suicide Life Threat Behav. 2023;53:100–9.
Bachmann S. Epidemiology of suicide and the psychiatric perspective. Int J Environ Res Public Health. 2018;15:1425.
Cavanagh JTO, Carson AJ, Sharpe M, Lawrie SM. Psychological autopsy studies of suicide: a systematic review. Psychol Med. 2003;33:395–405.
Steel Z, Marnane C, Iranpour C, Chey T, Jackson JW, Patel V, et al. The global prevalence of common mental disorders: a systematic review and meta-analysis 1980–2013. Int J Epidemiol. 2014;43:476–93.
Preacher KJ, Rucker DD, MacCallum RC, Nicewander WA. Use of the extreme groups approach: a critical reexamination and new recommendations. Psychol Methods. 2005;10:178–92.
Kendler KS, Chatzinakos C, Bacanu S-A. The impact on estimations of genetic correlations by the use of super-normal, unscreened, and family-history screened controls in genome wide case-control studies. Genet Epidemiol. 2020;44:283–9.
Schwartz S, Susser E. The use of well controls: an unhealthy practice in psychiatric research. Psychol Med. 2011;41:1127–31.
Acknowledgements
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. 1842169 (PI Colbert) and the National Institute of Mental Health R01MH132733 (PI Mullins). We thank and acknowledge all members of the Psychiatric Genomics Consortium Suicide Working group.
Author information
Authors and Affiliations
Consortia
Contributions
SMCC, ETM, DR, ARD, JJM, and NM designed the protocol and phenotype definitions. SMCC drafted the manuscript with assistance from ETM. SMCC conducted the review of phenotyping sources and instruments. JJM and NM supervised the work and provided overall direction. All authors (SMCC, ETM, OAA, OOA, PBB, CAB, ZC, HC, ED, HJE, JG, AH, AI, ECJ, EAK, HRK, MK, KL, WM, JIN, AS, JWS, MBS, CCZ, AE, MG, LM, RS, CT, TBB, NAK, DR, ARD, JJM, NM) revised the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
Ole Andreassen: Consultant to Cortechs.ai and Precision-Health.ai, and received speaker’s honorarium from Lundbeck, Sunovion, Janssen and Otsuka. Murray Stein: MBS has in the past 3 years received consulting income from Aptinyx, atai Life Sciences, BigHealth, Biogen, Bionomics, Boehringer Ingelheim, Delix Therapeutics, EmpowerPharm, Engrail Therapeutics, Janssen, Jazz Pharmaceuticals, Karuna Therapeutics, Lykos Therapeutics, NeuroTrauma Sciences, Otsuka US, PureTech Health, Sage Therapeutics, Seaport Therapeutics, and Roche/Genentech. Dr. Stein has stock options in Oxeia Biopharmaceuticals and EpiVario. He has been paid for his editorial work on Depression and Anxiety (Editor-in-Chief), Biological Psychiatry (Deputy Editor), and UpToDate (Co-Editor-in-Chief for Psychiatry). He is on the scientific advisory board of the Brain and Behavior Research Foundation and the Anxiety and Depression Association of America. John Mann: Dr. Mann receives royalties for commercial use of the C-SSRS from the Research Foundation of Mental Hygiene and from Columbia University for the Columbia Pathways App. Jordan Smoller: Dr. Smoller is a member of the Scientific Advisory Board of Sensorium Therapeutics (with options), and has received grant support from Biogen, Inc. He is PI of a collaborative study of the genetics of depression and bipolar disorder sponsored by 23andMe for which 23andMe provides analysis time as in-kind support but no payments. Henry Kranzler: Dr. Kranzler is a member of advisory boards for Altimmune and Clearmind Medicine; a consultant to Sobrera Pharmaceuticals; the recipient of research funding and medication supplies for an investigator-initiated study from Alkermes; a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by Alkermes, Dicerna, Ethypharm, Imbrium, Indivior, Kinnov, Lilly, Otsuka, and Pear; and an inventor on U.S. provisional patent “Multi-ancestry Genome-wide Association Meta-analysis of Buprenorphine Treatment Response”. All other listed authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Colbert, S.M.C., Monson, E.T., Andreassen, O.A. et al. Defining suicidality phenotypes for genetic studies: perspectives of the Psychiatric Genomics Consortium Suicide Working Group. Mol Psychiatry (2025). https://doi.org/10.1038/s41380-025-03271-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-025-03271-y