Abstract
Neuroendocrine prostate cancer (NEPC) presents a formidable clinical challenge owing to its aggressive progression and resistance to conventional therapies. A key driver of NEPC is the overexpression of MYCN, a well-established oncogene associated with neuroendocrine tumors. However, efforts to directly inhibit the N-Myc protein encoded by this gene have resulted in limited success, thereby hindering therapeutic advancements. To overcome this obstacle, we conducted unbiased genome-wide screening using isogenic prostate cancer cell lines to identify the synthetic vulnerabilities of MYCN. Among the identified candidates, NEUROG2 emerged as a significant candidate. Neurog2 is a proneural transcription factor (PTF) known for its role in developmental processes and trans-differentiation of adult cells. Our findings demonstrate that Neurog2 depletion does not affect non-malignant cells but significantly suppresses the growth of MYCN-overexpressing cells and tumors in orthotopic NEPC models. Furthermore, our observations indicate that Neurog2-driven modulation of PTFs potentially contribute to NEPC development. Thus, targeting Neurog2 holds promise as an effective therapeutic strategy for MYCN-overexpressing NEPC.
Similar content being viewed by others
Introduction
Prostate cancer (PC) is a common disease in the male population, exerting a profound impact on the public health systems worldwide [1]. Despite advances in diagnostic techniques and therapeutic modalities, managing advanced PC subtypes continues to present clinical hurdles. Androgen deprivation therapy (ADT), which reduces testosterone levels to castration levels, is the cornerstone treatment for primary prostate adenocarcinoma in patients with inoperable tumors [2,3,4]. However, the emergence of castration-resistant prostate cancer (CRPC) after ADT underscores the urgent need for additional therapeutic strategies [5]. The transition from adenocarcinoma to CRPC is orchestrated by several mechanisms, including intratumoral androgen synthesis, mutation and overexpression of androgen receptor (AR), and upregulation of AR coactivators, perpetuating AR signaling despite castration levels of testosterone [5, 6]. While new-generation anti-androgen therapies have demonstrated some improvements in the overall survival of CRPC patients, therapeutic resistance and development of neuroendocrine prostate cancer (NEPC) remain major challenges, necessitating continued exploration of novel therapeutic avenues [4, 7–10].
Among the spectrum of PC subtypes, NEPC has emerged as particularly aggressive, characterized by treatment resistance and rapid disease progression. Although NEPC may develop de novo, it is frequently triggered by hormonal (anti-androgen) therapies, resulting in dismal prognosis with limited therapeutic options [4, 7]. Molecular alterations, including amplification and overexpression of the MYCN oncogene, frequently accompany the transition of PC into NEPC, underlining the pivotal role of MYCN and the N-Myc transcription factor it encodes in driving this aggressive PC subtype [9, 11, 12].
The pursuit of therapeutic interventions targeting N-Myc has witnessed significant strides in recent years, despite its initial classification as an ‘undruggable’ molecule. Various strategies, including small molecules targeting the N-Myc-MAX interaction or direct modulation of N-Myc function, have shown promise in preclinical models [13]. Additionally, innovative approaches such as therapeutic mini-proteins and targeted degradation strategies offer novel avenues for inhibiting N-Myc activity. Encouragingly, the development of MRT-2359, a phase I/II clinical trial candidate that indirectly causes N-Myc downregulation, underscores the growing momentum of exploring anti-N-Myc therapies [14]. Unfortunately, despite these efforts, no therapy targeting N-Myc is currently available for clinical application.
Here, instead of targeting N-Myc, we aimed to elucidate the genetic dependencies of MYCN in prostate cancer cells. Specifically, we applied a genetic principle known as Synthetic Dosage Lethality (SDL), where the loss of function of a gene leads to lethality only when another gene, such as MYCN, is overexpressed [15, 16]. This paradigm holds immense potential not only in indirectly targeting overexpressed oncogenes but also in facilitating the development of personalized medicine based on the functional status of an oncogene in individual patients. Expanding on this genetic principle, we conducted extensive genome-wide loss-of-function screens employing both sgRNA/CRISPR/Cas9 and shRNA platforms and isogenic cell line models of NEPC. Our findings revealed, for the first time, a novel genetic interaction between NEUROG2 and MYCN. Subsequent in vitro studies have confirmed that Neurog-2 is not expressed and is dispensable for the survival of non-malignant adult cells. However, in vitro viability studies and in vivo studies in orthotopic tumor models suggest that the loss of function of Neurog-2 suppresses the growth of MYCN-overexpressing cells and tumors, respectively. Overall, our findings have promising implications for the development of targeted therapies customized for N-Myc-driven NEPC.
Materials and methods
Cell lines and cell culture
LASCPC-01 cells (ATCC® CRL-3356) and LNCaP cells (ATCC® CRL-1740) were obtained from ATCC (Manassas, VA, USA). LASCPC-01 cells were cultured in modified HITES medium as previously described [17]. The isogenic cell line pairs 22Rv1 and 22Rv1-N-Myc (22-MYC) were provided by the Rickman Lab (Cornell Medical College). The non-malignant cell lines BPH1, BHPrE1, NHPrE1, and PNT1B were kindly provided by Dr. Michael Cox at the Vancouver Prostate Center. 22Rv1, BPH1, PNT1B, and LNCaP cells were maintained in RPMI-1640 containing 10% FBS and 1% penicillin/ streptomycin. HEK293T cells were cultured in DMEM containing 10% FBS and 1% penicillin/ streptomycin. NHPrE1 and BHPrE1 were cultured in DMEM/F12 media with 5% FBS, 1% insulin-transferrin-selenium-X, 0.4% Bovine Pituitary Extract, 10 ng/L EGF, and 1% penicillin/streptomycin. Cell lines were incubated at 37 °C in 5% CO2.
Orthotopic prostate cancer model
Mice were housed 5 per cage at 23–25 °C in a humidity-controlled colony room, maintained on a 12 h light/dark cycle (08:00 to 20:00 light on), with standard food and water provided ad libitum along with environmental enrichments. All animals were handled in accordance with the approved protocols by the University of Saskatchewan Animal Research Ethics Board (AREB). Mice used in this study were nude (Crl:NU(NCr)-Foxn1nu) for LASCPC-01 tumor experiments and NSG (NOD.Cg-Prkdscidll2rg) for 22-MYC tumor experiments. The mice were randomly allocated to control or treatment groups.
Adult male mice weighing 20–25 g were used in this study. To perform orthotopic injection of cells (LASCPC-01 or 22-MYC) into the prostate, the mice were anesthetized using isoflurane and provided buprenorphine (0.05 mg/kg) for analgesia. A small incision was made in the lower abdomen, and the urinary bladder was exposed. Orthotopic injection was performed by injecting 200,000 cells in 1:1 media and Cultrex basement membrane extract (R&D Systems) into the exposed prostate. The incision was sutured, and the animals were allowed to survive for four weeks. Tumor sizes were recorded using a Vernier caliper (for LASCPC-01 experiments) or digital calipers (for 22-MYC experiments) at experimental termination. Tumor sizes were calculated using the standard formula LxW2/2, where L is the length and W is the width of the tumor.
Lentivirus production
HEK-293T cells were used to produce lentiviral particles expressing short hairpin RNA (shRNA) or sgRNA. This process involved packaging 5400 ng of psPAX2 and 600 ng of pMD2.G plasmids in conjunction with 6000 ng of target gene plasmids. Next, we used 10 cm plates to transfect the cells with a medium consisting of 540 µL Opti MEM (Gibco Life Technologies) and 36 µL of X-treme GENE DNA Transfection Reagent (Roche, Mississauga, ON, Canada). After 18 h, the medium was replaced with DMEM containing 2% bovine serum albumin (MilliporeSigma). Supernatants containing viral particles were collected 24 and 48 h post-transfection after centrifugation at 185 × G for 5 min. The supernatants were stored at −80o C for subsequent use. Stable 22-Rv1-Cas9 and 22-MYC-Cas9 cells were generated by transducing the cells with Cas9-blasticidin lentivirus and 8 µg/mL polybrene (MilliporeSigma). The medium containing blasticidin (4 µg/ml) was changed every 2–3 days (for 10–12 days) until the non-transduced cells were eliminated.
Pooled shRNA and CRISPR/Cas9 screening and data deconvolution
Lentiviral transduction of 22Rv1 and 22-MYC cells with shRNA or sgRNA libraries at 0.3 MOI was done as previously described [18,19,–20]. The cell lines used for sgRNA library transduction had been previously transduced with lentiviral vectors encoding Cas9 to ensure stable Cas9 expression. These Cas9-expressing cell lines were not used for shRNA library transductions, as shRNA screens do not require Cas9 for gene knockdown. Briefly, 24 h post-transduction, the cells were treated with 2 ug/ml of puromycin for 48 h. Puromycin-selected cells were then passaged for 12 d, and the cells were collected at three time points (Day 0, 7, and 13 for shRNA and Day 0, 6, and 12 for CRISPR screens). For shRNA screening, microarray analysis was performed, and deep sequencing was performed as described previously [18,19,–20]. Genomic DNA was extracted at each time point and the shRNA sequences were amplified. The amplified hairpins were digested with Xhol and purified for probe hybridization. FASTQs were aligned to library sequences using MAGeCK software (version 0.5.9.4) for CRISPR/Cas9 screens [21]. Prior to alignment, the FASTQC package (version 0.12.1) was used for quality assurance. These read counts were then cyclic-loss-normalized before computing the fitness score. shRNA- or sgRNA-weighted differential cumulative change (WDCh/g) between 22Rv1 and 22-MYC was calculated at each corresponding time point using the following equation:
where \({x}_{t,r}^{22{Rv}1}\) is the normalized signal intensity of 22Rv1 cells at time point \(t\in \left(0,..T\right)\) in replicates \(r\in (1..N)\). Similarly, \({x}_{t,r}^{22-{MYC}}\) was detected in the 22-MYC cells. \(\varepsilon\) is a constant that improves the weighting of shRNA or sgRNA hits the earlier the gene of interest drops. The WDC gene level WDCgene was calculated as the average of the top two shRNA/sgRNA with the lowest value for that gene using the following equation:
Significant differences between shRNA or sgRNA and their corresponding genes in 22Rv1 and 22-MYC cells were determined by Student’s t-test in combination with the permutation test p-value by estimating the frequency of randomized, shuffled WDC with lower values in comparison to the observed gene level WDC value [20]. Bayesian analysis of gene essentiality was used to evaluate screen performance [22].
Pathway enrichment analysis
Genes that showed significant WDCgene values from both CRISPR and shRNA screens were combined for comprehensive pathway enrichment analysis. This analysis was conducted using Gene Set Enrichment Analysis (GSEA) software version 4.2.2 [23]. The focus was on identifying enriched gene ontology biological processes (version 2023.2). Pathways were considered enriched if they had a false discovery rate (FDR) of less than 0.001 and a edge similarity cutoff of 0.9 or higher. These enriched pathways were imported into the enrichment map visualization plug-in for further examination [24]. The results were subsequently exported to Cytoscape for detailed network visualization [25]. To provide a clearer understanding of the contributions of each screen, the pathways were segmented and visualized as pie charts. These charts illustrate the proportion of genes identified from the CRISPR screen versus those identified from the shRNA screen, offering a visual representation of the data distribution and relative impact of each screening method on the identified pathways.
Differential expression analyses of CRPC and NEPC patient samples
Published data from Beltran et al. [26], which included 34 castration-resistant PC samples with adenocarcinoma features (CRPC-Adeno) and 15 castration-resistant PC samples with neuroendocrine features (CRPC-NE), were used to identify genes relevant to neuroendocrine features that overlapped in our shRNA and CRISPR screens. From the initial pool of 1120 genes identified by both screening methods, 323 genes were found to be significant (p < 0.05) based on the non-parametric Wilcoxon rank-sum test. The expression levels of these 323 genes were normalized across all CRPC-adeno and CRPC-NE samples. Subsequent clustering was performed using Pearson’s correlation to analyze the expression patterns. This clustering approach allowed for the comparison of gene expression profiles between CRPC-Adeno and CRPC-NE samples, highlighting distinct expression signatures associated with neuroendocrine differentiation.
Expression analysis of TCGA datasets
To investigate the differential expression of neuronal plasticity genes in lung cancer subtypes, we utilized data from The Cancer Genome Atlas (TCGA), which includes samples of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). Tumor patients were categorized into two groups of high and low expression based on the median expression levels of four neuroendocrine genes in normal tissues: MYCN, synaptophysin (SYP), chromogranin A (CHGA), and neuron-specific enolase (NSE). We focused on 17 genes related to neuronal plasticity. The expression levels of these genes were compared between the two groups with high and low neuroendocrine gene expression. The statistical significance of the differences in neuronal plasticity gene expression between the two groups was assessed using a one-tailed unpaired t-test performed in GraphPad Prism. This test allowed us to determine whether there were significant differences in the expression levels of neuronal plasticity genes between the high- and low-expression groups.
Neurog 2 overexpression studies
Adenoviral particles carrying the human Neurog 2 plasmid were purchased from Vector Biolabs (SKU-ADV-216616). 5 × 105 22Rv1 and 22-MYC cells were seeded in 6-well plates containing 1 mL culture media. After 24 h, the medium was replaced with 1 mL fresh media, and the cells were infected with adenoviral particles (50 MOI) in the presence of 8 µg/mL polybrene (MilliporeSigma; TR-1003). Twenty-four hours after transduction, the media was removed, the wells were washed with PBS, and fresh media was added. Brightfield images were taken using a Zeiss Axio Observer under 10x magnification at 24 and 48 h after infection.
qRT-PCR
RNA was isolated and purified using a PureLink RNA isolation kit (ThermoFisher) according to the manufacturer’s protocol. RNA was reverse transcribed to cDNA using a cDNA synthesis kit (Applied Biosystems) in a master cycler. qRT-PCR was carried out using the StepOne Real-Time PCR System (Applied Biosystems) and PowerUpTM SYBRTM Green Master Mix (Applied Biosystems). The primers used to amplify cDNAs are listed in Supplementary Table S1. All reactions were performed in triplicate. All target genes were internally normalized to the housekeeping genes (GAPDH, RPLP, and actin).
Colony formation and cell viability assays
For colony formation assay, the control and target sgRNA treated 22Rv1 and 22-MYC cells were seeded (1000 cells/well) in a 6-well plate and incubated at 37 °C and 5% CO2 for 12 d, replacing the media every 3 days. The plates were then washed with PBS three times, fixed with 4% formalin solution, and stained with 0.5% crystal violet. Images of the colonies were captured using a Bio-Rad Imager, and the colonies were quantified using ImageJ software. For the cell viability assay, metabolically active cells were quantified using a resazurin reduction assay after the knockdown of target genes. Briefly, LASCPC-01 cells or non-malignant BPH1, BHPrE1, NHPrE1, and PNT1B cells were transduced with shNeurog2, shIsl1, or shRFP, followed by puromycin selection for 48 h. Puromycin-selected cells (1000 cells) were seeded into 96 well plates. A 1:10 dilution of resazurin was then added to each well of a 96-well plate and incubated for 4 h at 37 °C and 5% CO2. Fluorescence intensity was measured at 590 nm using a Varioskan LUX microplate reader.
Correlation analysis of gene expression
Published data from Beltran et al. [26] was utilized to perform a comparative analysis of gene expression. The expression levels of SYP, CHGA, and NSE were compared to those of Neurog2. Similarly, the expression levels of BRN2, INSM1, and cJUN were also compared with those of Neurog2. Spearman rank correlation was used to calculate the correlation coefficient (r) and determine the statistical significance of these relationships.
Quantification and statistical analysis
GraphPad prism software was used for all tests. Unless otherwise specified, analyses were done using standard ‘t’ tests, one-way ANOVA (with Tukey’s multiple comparisons test) or two-way ANOVA (with Sidak’s multiple comparisons test). The details of the statistical analysis performed, including the specific test employed, the number of samples considered, and the level of significance achieved, are presented in more detail in the corresponding figure legends.
Results
Integration of CRISPR and shRNA screens identifies genetic dependencies of MYCN overexpressing NEPC cells
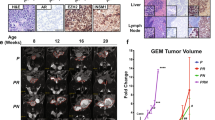
We explored SDL interactions of MYCN to identify novel targets for indirectly inhibiting N-Myc-dependent NEPC tumors. To this end, we performed loss-of-function genetic screens in an isogenic model generated by overexpressing MYCN. We used 22Rv1 and 22Rv1-derived 22-MYC cells (N-Myc-overexpressing 22Rv1, generously provided by the Rickman laboratory), where N-Myc overexpression drives the development of poorly differentiated, invasive prostate cancer phenotype, which exhibits molecular hallmark features of NEPC, including high expression of neuroendocrine markers and low androgen receptor expression [8], to identify MYCN-specific SDL hits. In our experience, screens in isogenic models tend to produce fewer false positives and capture hits specific to the query gene [18,19,–20, 27, 28]. Prior to screening, we confirmed the overexpression of N-Myc in 22-MYC cells by western blotting and qRT-PCR (Fig. 1A, B). We also confirmed the potential of 22-MYC cells to form NEPC type tumors in vivo, as evidenced by the elevated expression of the neuroendocrine differentiation (NED) marker neuron-specific enolase (NSE) in 22-MYC-driven tumors compared to 22Rv1-driven tumors (Fig. 1C).
A N-Myc protein levels in 22Rv1, 22-MYC and LASCPC01 cells. B N-Myc encoding mRNA expression in 22Rv1 and 22-MYC cells. Data presented as mean ± SE, n = 3; Standard ‘t’ test; ***p < 0.001. C Protein levels of N-Myc and the NSE NED marker in 22-MYC and 22Rv1 subcutaneous tumor xenografts harvested at the indicated days. Innately N-Mychigh IMR-32 neuroblastoma cells were used as a positive control for N-Myc and NSE. D, E Volcano plots representing genome-wide screening in 22-Rv1 and 22-MYC cell line pair using pooled shRNA (left) and CRISPR/Cas9 screens (right). The red dots represent SDL hits and the orange dots on the right represent possible growth advantage (GA) hits. Grey dots represent genes whose loss of function did not significantly affect cell viability in the screens. The y-axes show the significance of the hits, and the x-axes represent the fitness score computed for each gene by comparing 22-Rv1 cells with 22-MYC cells. Cut-offs were determined based on data distribution within each screen. F Pathway enrichment analyses of both shRNA and CRISPR screens. GSEA was performed and pathways with FDR ≤ 0.001 and edge similarity cut-off 0.9 were selected for enrichmentMap analysis. Each node is divided into two colors representing the percentage of genes identified in each screen. Enlarged area represents neuron-associated terms. G Dot plot showing the top 58 genes associated with neuronal plasticity (Dark color represents pathway enrichment, light color represents no-enrichment background).
For genome-wide SDL screening, we utilized two independent pooled screening platforms: an shRNA gene knockdown lentiviral library and a CRISPR/Cas9 gene knockout lentiviral library. Each library (shRNA and CRISPR) was designed to individually target approximately 18,000 genes to identify SDL interactions specific to MYCN. The Pearson correlation coefficient showed a strong correlation (r > 0.89) between the replicas considered for each screen (Supplementary Fig. 1). Overall, the shRNA screen identified 581 SDL hits, whereas the CRISPR/Cas9 screen identified 553 hits, whose selective elimination selectively suppressed N-Myc-positive cells (Fig. 1D, E; Supplementary Table S2). Some of the promising hits identified included heat-shock proteins and related co-chaperones, such as HSPA5, HSP90B1, CCT5, HSPB6, and UBXN1, which have critical roles in prostate cancer, including combating ER stress [29, 30]. Interestingly, several of the identified SDL hits, such as CDK6, CERK, TBX2, and MBD3 were also identified in another genome-wide screen performed for MYCN-driven neuroblastoma, which is another cancer of neuroendocrine origin [31], suggesting that some of our hits might also be applicable to other MYCN-driven malignancies. Alternatively, this might also mean that some of our SDL hits may be relevant to the neuroendocrine tumor (NET) phenotype per se. Specifically, among the SDL hits, the shRNA screen identified proneural transcription factors (PTFs), such as NEUROG2 and POU3F2, along with transcriptional targets of N-Myc, including PKMYT1, CKS1B, RBP5, and CHEK2 [32,33,–34]. Similarly, CRISPR/Cas9 screening identified several neuromodulators, including SOX17, FOXG1, PITX3, ONECUT2, and GBX2, which favor NED and potentially contribute to NEPC. Although the overlap between the two screens was anticipated to be modest [35, 36] amalgamating data from both platforms revealed several genes operating within the shared pathways (Fig. 1F). Notably, we identified genes associated with axon guidance (FDR < 0.0001), Wnt signaling (FDR < 0.0001), neurogenesis (FDR < 0.0001), neuron development (FDR < 0.0001), and sensory system development (FDR < 0.0001), all of which are significantly relevant to neuroendocrine tumors, including NEPC [37,38,39,40,–41]. Particularly noteworthy from the enrichment analyses were approximately 58 genes implicated in neuronal plasticity, a trait closely associated with NEPC (Fig. 1G). Thus, the independent identification of NED- and NEPC-relevant hits in both shRNA and CRISPR/Cas9 screens underscores the robustness and advantages of the dual-screening strategy employed in this study.
Coupling differential essentiality with differential expression to prioritize SDL hits
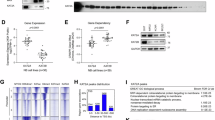
To evaluate the significance of SDL genes specific to NEPC, we also explored genes that are differentially expressed between CRPC and NEPC patient samples using publicly available data [26]. Among the screened hits, 323 genes exhibited significantly increased expression in NEPC samples compared to CRPC samples. Unsupervised clustering of these 323 genes revealed distinct gene expression patterns between NEPC and CRPC (Fig. 2A). Notably, several genes related to neuronal plasticity, including KIT, FOXG1, GBX2, WNT4, SOX1, ONECUT2, NEUROG2 and ISL1, were clustered together. This indicates that the combined evaluation of differentially expressed genes and SDL candidates is a powerful approach to identify potential targets that regulate NEPC biology [42,43,44,45,46,47,48,–49].
A Unsupervised Pearson clustering of gene expression profiles for prostate cancer patient samples from Beltran et al. (Nat. Methods, 2016), highlighting 323 genes with a significant difference (Wilcoxon rank-sum test, p < 0.005) between CRPC and NEPC. The inset image magnifies genes exhibiting the most pronounced differential expression. B Expression analysis of lung cancer patient samples from TCGA, illustrating significant differences (one-tailed unpaired t-test: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001) in the 17 genes associated with neuronal plasticity, after classification based on the expression of the NMYC/SYP/CHGA/NSE signature. Top panel represents lung adenocarcinoma (LUAD) and bottom lung squamous cell carcinoma (LUSC).
To further validate these findings, we examined The Cancer Genome Atlas (TCGA) to assess the expression patterns of some of these genes in lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD), as they can potentially transition to small cell lung cancer that shares more neuroendocrine features [50]. The cellular plasticity of both tumor types has the potential to transform them into neuroendocrine tumours [51, 52]. Therefore, to compare the expression status of SDL hits within these tumor types, we categorized patient data based on the expression of MYCN and NED markers such as SYP, CHGA, and NSE. As noted above, our analysis revealed that several SDL candidates, including ISL1, NEUROG2, EPHA7, EPHA8, ONECUT2, POU3F2, CHRM5, FOXG1 and GBX2 were significantly overexpressed in LUAD and LUSC cases with neuroendocrine features, represented by high expression of MYCN and NED markers (Fig. 2B). These findings not only highlight the potential role of neuronal plasticity genes in NEPC and MYCN-overexpressing lung cancer but also highlight their broader significance in neuroendocrine tumors.
Neurog2 is essential for the survival of N-Myc overexpressing cells
We next chose to validate two SDL hits, NEUROG2 and ISL1 from among the major hits, as they are both involved in neurogenesis [53, 54] and our analyses indicated that they are significantly upregulated in NEPC (Fig. 2A). Although Neurog2 and ISL1 have been studied in other contexts, their therapeutic potentials in NEPC have not previously been reported. To determine their therapeutic potential, we initially assessed the effects of their knockout in the colony formation assay. This revealed a significantly stronger reduction in colony formation in 22-MYC cells than 22Rv1 cells following individual knockouts of NEUROG2 and ISL1 (Fig. 3A). This differential selectivity in N-Myc-overexpressing cells validated the identification of these SDL candidates in our genome-wide screening. To eliminate cell line-specific effects in our observations, we also examined the anti-proliferative effect of the target knockdown in N-Myc+/myrAKT1+ LASCPC-01 cells that demonstrated NEPC features using a resazurin assay and found that individual knockdown of NEUROG2 and ISL1 effectively suppressed cell viability (Fig. 3B). We also confirmed the efficiency of target knockdown in LASPC-01 cells using qRT-PCR (Fig. 3C) [17].
A Colony formation assay and quantitation using 22-Rv1 and 22-MYC cells after NEUROG2 and ISL1 knockouts. Data presented as mean ± SE n = 4; Two-way ANOVA with Sidak’s multiple comparisons test; ***p < 0.001. The same non-targeting control (NTC) data was used to compare with NEUROG2 and ISL1. B Evaluation of cell viability using resazurin assay in LASCPC-01 cells after NEUROG2 and ISL1 knockdown. Percentages of relative viability in knockdown compared to control shRNA transduced cells are shown at the indicated time points. Data presented as mean ± SE; n = 4; Standard ‘t’ test; ****p < 0.0001. The same shRFP was used to compare with shNeurog2 and shISL1. C mRNA levels show efficient knockdown of NEUROG2 and ISL1 in LASCPC-01 cells, compared to non-targeting shRFP controls. Data presented as mean ± SE; n = 9; Standard ‘t’ test; ****p < 0.0001. D Western blot showing protein levels of N-Myc and Neurog2 in non-malignant prostate cells. Vinculin serves as a loading control. E Evaluation of cell viability using resazurin assay in non-malignant cells (BHPrE1, NHPrE1, BPH1 and PNT1B) following NEUROG2 knockdown. Data presented as mean ± SE; n ≥ 3; One-way ANOVA with Tukey’s multiple comparisons test; **p < 0.01, ***p < 0.001.
Our aim was to develop targeted therapies for NEPC that do not impair normal tissue function. In line with this, it is noteworthy that our analysis of the tissue- and cell-specific gene expression dataset from the Genotype-Tissue Expression (GTEx) Portal (Supplementary Fig. 2) revealed a striking difference between NEUROG2 and ISL1 expression. NEUROG2 is poorly expressed in most normal tissues, including the prostate, whereas ISL1 is abundantly expressed, particularly in the prostate. We also examined the innate expression of Neurog2 in non-malignant prostate cell lines BPH1, BHPrE1, NHPrE1, and PNT1B and, consistent with our GTEx analyses, found that its expression is minimal to undetectable in these cells (Fig. 3D). As expected, MYCN was not expressed in non-malignant cells (Fig. 3D). Furthermore, we examined whether NEUROG2 knockdown impaired the viability of these non-malignant prostate cells, including BHPrE1 cells that displayed at least minimally detectable Neurog2 expression, and found that its knockdown did not impair their viability (Fig. 3E). Altogether, these observations indicate that Neurog2 is dispensable to non-malignant cells but is selectively essential for the survival of MYCN overexpressing cancer cells.
Neurog2 plays a key role in neuroendocrine plasticity of prostate cancer cells
NED is a hallmark of NEPC. Therefore, we examined the expression of NED markers in LASCPC-01 cells after NEUROG2 and ISL1 knockdown to evaluate their effect on suppressing NEPC features [55,56,–57]. While ISL1 knockdown had no effect, NEUROG2 knockdown suppressed NED characteristics, as evidenced by the downregulation of all three NED markers tested: SYP, CHGA, and NSE (Fig. 4A). Similar results were observed at the mRNA level (Fig. 4B). These results were further corroborated by identifying positive correlations in gene expression between NED markers and Neurog2 in NEPC but not in CRPC (Fig. 4C). Specifically, while the correlation in gene expression between NEUROG2 and the NED markers appeared poor in CRPC (r = 0.004, p = 0.9 for Neurog2 vs. SYP; r = 0.06, p = 0.71 for Neurog2 vs. CHGA; r = 0.006, p = 0.21 for Neurog2 vs. NSE), the correlation significantly improved in NEPC (r = 0.72, p = 0.003 for Neurog2 vs. SYP; r = 0.33, p = 0.21 for Neurog2 vs. CHGA; r = 0.45, p = 0.09 for Neurog2 vs. NSE), with the exception of CHGA, although its correlation also improved (Fig. 4C). These results suggest that NEUROG2 may play a significant role in NEPC transformation, as evidenced by stronger correlations with NED markers in NEPC compared to CRPC. While these correlations are not definitive evidence of causation, they provide additional support for the functional role of NEUROG2 in promoting neuroendocrine differentiation.
A Reduction of protein levels of NED markers (SYP, CHGA, and NSE) in LASCPC-01 NEPC cells in response to Neurog2 knockdown compared to ISL1 knockdown. Western blot images and corresponding quantifications are shown. Data represented as mean ± SE; n = 3 except for NSE where n = 2; Standard ‘t’ test; *p < 0.05, ***p < 0.001. B Expression of NED markers (SYP, CHGA, and NSE) in LASCPC-01 cells after Neurog2 knockdown compared to ISL1 knockdown. Data presented as mean ± SE; n = 9 except for NSE where n = 8; Two-way ANOVA with Tukey’s multiple comparisons test; ****p < 0.0001. For SYP, the same shRFP data was used to compare in both graphs. C Expression correlation between Neurog2 and NED markers (SYP, CHGA and NSE) in CRPC and NEPC samples from Beltran et al. (Nat. Methods, 2016).
Previous studies indicated that PTFs promote NEPC. For example, Brn2 (also known as Pou3f2) facilitates the transition from PC to NEPC [58]. Similarly, Ascl1, another PTF, has been linked to NEPC development [59]. In addition, INSM1 and NeuroD1 have also been found to contribute to NEPC [60,61,62,–63]. Small cell lung cancer (SCLC), which shows features of neuroendocrine cancers, is characterized by a molecular signature that closely resembles NEPC [51]. Of note, Ascl1, NeuroD1, Pou2f3, and the transcriptional regulator Yap1 have been suggested to be critical players in SCLC, with Ascl1 and NeuroD1 recognized as potential promoters and Pou2f3 and Yap1 recognized as potential suppressors [64]. Strikingly, we found that NEUROG2 knockdown in LASCPC-01 cells downregulated BRN2, ASCL1, INSM1, and NEUROD1, while upregulating TAZ1, a transcriptional co-activator with functions similar to Yap1 [65, 66], indicating that functions of Neurog2 in MYCN expressing cells may be directed towards promoting neuroendocrine plasticity (Fig. 5A).
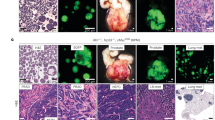
A Knockdown of Neurog2 in LASCPC-01 cells downregulates BRN2 (POU3F2), ASCL1, INSM1, NEUROD1, C-FOS, and C-JUN, while upregulating TAZ1. Data presented as mean ± SE; n ≥ 3; standard ‘t’ test; **p < 0.01; ***p < 0.001, ****p < 0.0001. B Expression correlation between Neurog2 and BRN2, INSM1 and cJUN in CRPC and NEPC samples from Beltran et al. (Nat. Methods, 2016). C SYP, CHGB, NEUROG2 and BRN2 (POU3F2) levels in norepinephrine (NE) supplemented but testosterone (T) withdrawn LNCaP cultures (at 48 h and 72 h) compared to NE + T supplemented cultures at the corresponding time points. Data presented as mean ± SE; n ≥ 3; standard ‘t’ test; *p < 0.05; **p < 0.01. D Ad-Neurog2 transduction and associated Neurog2 overexpression (OE) in 22Rv1 cells promotes cell-cell interaction through longer neurite-like extensions (24h-yellow arrows) and later induces clustered growth characteristics (48h-blue arrows). Ad-Neurog2 transduction and associated Neurog2 OE in 22-MYC cells, while promoting cell-cell contact through neurite-like extensions (24h-yellow arrows), maintains the aggressive growth characteristics, including rapid filling of the growth surface (48h-red arrows) (scale bar, 100 µm). Enlarged insets (green bordered) show the details of the cell phenotypes. E qRT-PCR shows the levels of Neurog2 mRNA in Ad-GFP and Ad-Neurog2 transduced 22Rv1 (top) and 22-MYC (bottom) cells. (Data presented as mean ± SE; n ≥ 3; standard ‘t’ test; *p < 0.05; *** p < 0.001.
The AP-1 transcription factor complex was recently shown to promote CRPC, a subtype of PC that precedes NEPC [67]. Therefore, we next examined the expression of c-JUN and c-FOS, transcription factors that form the AP-1 transcription factor complex, in NEUROG2 knockdown LASCPC-01 cells [68, 69]. Interestingly, we found that both C-JUN and C-FOS were downregulated after NEUROG2 knockdown, further confirming that Neurog2 may be a potential target for tackling NEPC development (Fig. 5A). These results were further corroborated by correlation analyses between the expression of BRN2, INSM1, c-JUN, and Neurog2 in patient samples (Fig. 5B). Notably, although the correlation appeared poor in CRPC (r = 0.244, p = 0.163 for Neurog2 vs. BRN2; r = 0.18, p = 0.28 for Neurog2 vs. INSM1; r = 0.09, p = 0.59 for Neurog2 vs. cJUN), it improved significantly in NEPC (r = 0.534, p = 0.042 for Neurog2 vs. BRN2; r = 0.79, p = 0.0006 for Neurog2 vs. INSM1; r = 0.45, p = 0.08 for Neurog2 vs. cJUN), indicating that Neurog2 may be a critical regulator of NEPC (Fig. 5B).
Suppression of androgen signaling by new-generation anti-androgen therapies is a risk factor for NEPC development [4, 7]. We also recently showed that neurosignaling from the sympathetic neurotransmitter norepinephrine (NE) promotes NED, a hallmark of NEPC [37]. Importantly, prostate tumors in patients receive an adequate supply of NE from sympathetic nerves [37, 70, 71]. During anti-androgen therapies, androgen signaling is severely impaired, whereas intact neurosignaling is maintained. To simulate this scenario, we cultured AR+ LNCaP cells for two weeks in testosterone- and NE-supplemented media and then withdrew testosterone from the cultures. As expected, testosterone depletion increased the levels of the NED markers SYP and CHGB, indicating facilitation of NEPC characteristics (Fig. 5C). Interestingly, this was accompanied by concomitant increases in NEUROG2 and BRN2 levels. These results further substantiate the potential role of Neurog2 as a master regulator of neuroendocrine plasticity.
Next, we examined the morphology of 22Rv1 and 22-MYC cells after transduction with Ad-Neurog2. Interestingly, both 22Rv1 and 22-MYC cells elongated their neurite-like extensions after Neurog2 overexpression, showing enhanced neuroendocrine characteristics (Fig. 5D, E). These cells also appear to form intercellular connections using neurite-like extensions. 22Rv1 cells later grew in large clusters, whereas 22-MYC cells maintained their usual growth characteristics by spreading throughout the growth surface while maintaining intercellular connections with their neurite-like extensions (Fig. 5D, E). Overall, the changes in the growth pattern of Neurog2 overexpressed 22Rv1 and 22-MYC cells indicated that Neurog2 induces diverse cellular fates, including longer neurite-like extensions and increased cell-cell contact, in both the presence and absence of N-Myc. While some of these phenotypes (e.g., neurite-like extensions) are consistent across conditions, others appear distinct (e.g., spreading with retained cellular connections in the presence of N-Myc). These observations emphasize the role of Neurog2 in driving and maintaining neuroendocrine features in PC cells.
Neurog2 knockdown suppresses orthotopic NEPC tumors
To further assess the relevance of Neurog2 in NEPC, we examined the effect of Neurog2 knockdown in in vivo orthotopic NEPC models generated using LASCPC-01 and 22-MYC cells (Fig. 6A, B). These experiments demonstrated significant suppression of NEPC tumor growth in response to NEUROG2 silencing compared to control tumors established by matching shRFP-transduced cells (Fig. 6A, B). These findings confirm that Neurog2 has strong potential as a therapeutic target for treating patients with MYCN overexpressing NEPC tumors.
A (Top) Representative images of tumors established by orthotopic engraftment of LASCPC01 cells transduced by lentiviruses expressing shRFP or shNeurog2; (Bottom left) NEUROG2 knockdown in shNeurog2 transduced LASCPC-01 cells (data presented as mean ± SE; n = 3; standard ‘t’ test; ***p < 0.001); (Bottom right) quantification of tumor sizes (data presented as mean ± SE; n = 6 for shRFP and n = 6 for shNeurog2; one animal did not form measurable tumor in shNeurog2 (not included); standard ‘t’ test; *p < 0.05). B (Top) Representative images of tumors established by orthotopic engraftment of 22-MYC cells transduced by lentiviruses expressing shRFP or shNeurog2; (Bottom left) NEUROG2 knockdown in shNeurog2 transduced 22-MYC cells (data presented as mean ± SE; n = 3; standard ‘t’ test; ***p < 0.001); (Bottom right) quantification of tumor sizes (data presented as mean ± SE; n = 3; standard ‘t’ test; *p < 0.05).
Discussion
Recent advances in high-quality research have paved the way for the better management of PC. Nevertheless, the most aggressive subtype of PC, the NEPC, lacks effective treatment options, which impedes disease management [4, 10, 72]. Although several molecular alterations such as RB1 loss, TP53 loss, and MYCN amplification have been recognized to promote NEPC, the lack of effective ways to directly target these molecules for therapeutic purposes hinders the development of NEPC treatment. To circumvent the complications associated with direct target modulation, we explored SDL interactions of MYCN by performing two independent genome-wide genetic screens using pooled shRNA and CRISPR/Cas9 systems. The SDL approach is a well-established method for identifying optimal therapeutic targets in a completely unbiased manner, as demonstrated by our own and previous work [18, 19, 27, 28, 73, 74]. Using this approach, we shortlisted several potential therapeutic targets for MYCN driven NEPC. As expected, the overlaps between the shRNA and CRISPR/Cas9 screens were poor [15, 35, 75, 76]. While both shRNA and CRISPR screens identify synthetic vulnerabilities in MYCN-overexpressing NEPC cells, the two platforms often yield datasets with limited overlaps, as they capture cell vulnerabilities at different molecular contexts (complete or incomplete gene suppression respectively) [15, 35, 75, 76]. In fact, our recent publication utilizing a graph-based unsupervised learning model demonstrated that neither shRNA nor CRISPR screens are universally superior, indicating that a combination of both is a powerful strategy to capture genetic vulnerabilities [77]. While our specific focus on N-Myc+ NEPC may limit the applicability of the shortlisted potential targets to selected patients, it should help in the development of effective personalized treatments for those who harbor tumors with high levels of Myc.
In this study, we conducted two independent assays in vitro to evaluate cell viability and colony formation using two different N-Myc+ cell lines. Additionally, we assessed the effects of target interventions on NED features of N-Myc+ cells to validate the therapeutic potentials of two SDL hits identified in our screens. This strategy was designed to provide a foundation for future mechanistic studies and in vivo assessments. Ultimately, Neurog2 PTF attracted our attention given that its inhibition selectively inhibited N-Myc+ cell lines and suppressed their NED signatures. Our experiments also confirmed that Neurog2 is a safe target that is not essential for normal cells, as it is not expressed in most normal adult tissues and its loss had no influence on normal cell viability. This agrees well with previously published observations showing that the functions of Neurog2 are mostly confined to developmental stages, particularly contributing to neurogenesis and maturation [53, 78,79,80,81]. According to the SDL concept, inhibition of Neurog2 should preferentially eliminate N-Myc+ cells without affecting cells that lack N-Myc expression. Taken together, our validation experiments demonstrated that this is indeed true for the N-Myc-Neurog2 axis.
The role of Neurog2 in neurogenesis involves inducing transcriptional reprogramming in precursor cells to differentiate them into mature neurons and neuron subtypes [78]. Hence, the abundant expression of Neurog2 in N-Myc+ PC cells displaying NED characteristics may indicate its active role in promoting transcriptional reprogramming in PC cells for terminal transition to NEPC. Several lines of evidence highlight the role of NEUROG2 in adult neuronal differentiation, indicating its potential relevance to NEPC [82, 83].
Neurog2-dependent cell differentiation at developmental stages is driven by epigenetic modifications that promote chromatin accessibility at proneural genetic elements [82, 84, 85]. As NEUROG2 knockdown resulted in the downregulation of BRN2, ASCL1, and NEUROD1 PTFs and upregulation of TAZ1, we expect a potential role for Neurog2 in modulating the heterochromatin landscape in N-Myc+ cells, favoring NEPC transition and maintenance. In addition, previous evidence suggests that epigenetic signatures consistent with NEPC favor chromatin conformations that promote the access of Neurog 2 to its target genes [86]. These findings, coupled with previous observations of Neurog-2’s capacity to transform adult cells into the neuronal phenotype, implicate Neurog-2 reactivation as a potential factor contributing to the transdifferentiation of PC into NEPC [87]. Of the most important to note, Brn2, Ascl1 and NeuroD1 PTFs have been previously reported to promote NEPC, and hence, the collective downregulation of these PTFs downstream of Neurog2 loss observed in our investigation strongly suggests that Neurog2 may be a master regulator of PTFs, and hence, PTF-dependent NEPC. Moreover, Taz1 has been shown to suppress cellular transdifferentiation [88, 89]; hence, the inverse relationship between Neurog2 and Taz1 captured in our experiments further supports a central role for Neurog2 in promoting NED characteristics in N-Myc+ PC cells.
With respect to profiling Neurog2 as a therapeutic target for NEPC, there is precedent for targeting transcription factors to treat NEPC. For example, in addition to the efforts of targeting N-Myc in NEPC, Nouruzi et al., showed that inhibiting the PTF Ascl1 in NEPC results in widespread epigenetic changes modulated by polycomb repressive complex-2 (PRC2) [90]. These changes apparently reversed the neuroendocrine phenotype to a more typical luminal PC phenotype and laid the groundwork around PTFs in developing potential therapies reversing the transformation of CRPC to NEPC [90]. Therefore, targeting Neurog2 in NEPC may offer significant therapeutic promise owing to its close association with other NEPC-related PTFs. Moreover, we discovered that Neurog2 modulates the levels of other transcription factors such as c-Jun and c-Fos, which are linked to tissue growth. Consistent with this finding, Neurog2 knockdown effectively suppressed MYCN-driven NEPC tumors, highlighting the potential of Neurog2 as a therapeutic target for NEPC. Based on our experimental results, we anticipate that Neurog2 inhibitors could either revert NEPC tumors to more clinically manageable prostate cancer subtypes or serve as prophylactic agents to prevent treatment-induced NEPC. We tested Neurog2 inhibition at the preclinical level using genetic depletion approaches due to the lack of effective small-molecule inhibitors. Additional indirect approaches, such as forced induction of MTGR1, a transcriptional repressor, and feedback inhibitor of Neurog2, may be worth exploring [80]. Another potential strategy is to inhibit specific binding of Neurog2 to its heterochromatin landscape. Further mechanistic insights into how Neurog2 orchestrates other PTFs in N-Myc+ NEPC could also reveal additional effective intervention points.
It is also important to note that Neurog2 is critical for neural development in embryonic stages [91]. However, its role and expression in adult tissues, particularly in differentiated non-malignant human cells appear to be limited. Importantly, in our study, Neurog2 depletion did not have any detectable effects on non-malignant cells in vitro, suggesting a preferential essentiality in MYCN-overexpressing cancer cells. While these findings are promising, we acknowledge the potential for neural toxicity and emphasize that further preclinical studies are required to fully evaluate the safety profile of Neurog2 inhibition.
In conclusion, understanding the intricate molecular mechanisms underlying NEPC progression and therapy resistance is crucial for the development of effective targeted therapies. In this study, leveraging the genetic concept of SDL in the context of the major genomic alteration of MYCN overexpression prevalent in NEPC revealed a repertoire of new potential therapeutic targets. Neurog2, one of the targets studied in more detail in this work, shows significant promise as a focal point for developing conceptually novel and effective therapies for treating patients with N-Mychigh NEPC tumors.
Data availability
The data relevant to this study were deposited in the Gene Expression Omnibus (GEO) database. The microarray shRNA screening data can be found under accession number GSE255592, and the CRISPR screening sequencing data are available under accession number GSE255591. The comprehensive dataset was accessible using super series GSE255593. All other data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. All raw unedited western blots are presented in Supplementary Fig. 3. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. 2009;6:76–85.
Agus DB, Cordon-Cardo C, Fox W, Drobnjak M, Koff A, Golde DW, et al. Prostate cancer cell cycle regulators: response to androgen withdrawal and development of androgen independence. J Natl Cancer Inst. 1999;91:1869–76.
Liu S, Alabi BR, Yin Q, Stoyanova T. Molecular mechanisms underlying the development of neuroendocrine prostate cancer. Semin Cancer Biol. 2022;86:57–68.
Mostaghel EA, Zhang A, Hernandez S, Marck BT, Zhang X, Tamae D, et al. Contribution of Adrenal Glands to Intratumor Androgens and Growth of Castration-Resistant Prostate Cancer. Clin Cancer Res. 2019;25:426–39.
Ceraline J, Cruchant MD, Erdmann E, Erbs P, Kurtz JE, Duclos B, et al. Constitutive activation of the androgen receptor by a point mutation in the hinge region: a new mechanism for androgen-independent growth in prostate cancer. Int J Cancer. 2004;108:152–7.
Patel GK, Chugh N, Tripathi M. Neuroendocrine differentiation of prostate cancer-an intriguing example of tumor evolution at play. Cancers. 2019;11:1405.
Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, et al. N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer. Cancer Cell. 2016;30:563–77.
Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–95.
Beltran H, Tagawa ST, Park K, MacDonald T, Milowsky MI, Mosquera JM, et al. Challenges in recognizing treatment-related neuroendocrine prostate cancer. J Clin Oncol. 2012;30:e386–389.
Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229–39.
Bernard D, Pourtier-Manzanedo A, Gil J, Beach DH. Myc confers androgen-independent prostate cancer cell growth. J Clin Invest. 2003;112:1724–31.
Carabet LA, Rennie PS, Cherkasov A. Therapeutic Inhibition of Myc in Cancer. Structural bases and computer-aided drug discovery approaches. Int J Mol Sci. 2018;20:120.
Zhang D, Lin P, Lin J. Molecular glues targeting GSPT1 in cancers: A potent therapy. Bioorg Chem. 2023;143:107000.
Paul JM, Templeton SD, Baharani A, Freywald A, Vizeacoumar FJ. Building high-resolution synthetic lethal networks: a ‘Google map’ of the cancer cell. Trends Mol Med. 2014;20:704–15.
Cunningham CE, MacAuley MJ, Yadav G, Vizeacoumar FS, Freywald A, Vizeacoumar FJ. Targeting the CINful genome: Strategies to overcome tumor heterogeneity. Prog Biophys Mol Biol. 2019;147:77–91.
Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell. 2016;29:536–47.
Chafe SC, Vizeacoumar FS, Venkateswaran G, Nemirovsky O, Awrey S, Brown WS, et al. Genome-wide synthetic lethal screen unveils novel CAIX-NFS1/xCT axis as a targetable vulnerability in hypoxic solid tumors. Sci Adv. 2021;7:eabj0364.
El Zawily A, Vizeacoumar FS, Dahiya R, Banerjee SL, Bhanumathy KK, Elhasasna H, et al. A multipronged unbiased strategy guides the development of an anti-egfr/epha2-bispecific antibody for combination cancer therapy. Clin Cancer Res. 2023;29:2686–701.
Paul JM, Toosi B, Vizeacoumar FS, Bhanumathy KK, Li Y, Gerger C, et al. Targeting synthetic lethality between the SRC kinase and the EPHB6 receptor may benefit cancer treatment. Oncotarget 2016.
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554.
Hart T, Moffat J. BAGEL: a computational framework for identifying essential genes from pooled library screens. BMC Bioinf. 2016;17:164.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14:482–517.
Franz M, Lopes CT, Huck G, Dong Y, Sumer O, Bader GD. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–11.
Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305.
Hanover G, Vizeacoumar FS, Banerjee SL, Nair R, Dahiya R, Osornio-Hernandez AI, et al. Integration of cancer-related genetic landscape of Eph receptors and ephrins with proteomics identifies a crosstalk between EPHB6 and EGFR. Cell Rep. 2023;42:112670.
Vizeacoumar FJ, Arnold R, Vizeacoumar FS, Chandrashekhar M, Buzina A, Young JT, et al. A negative genetic interaction map in isogenic cancer cell lines reveals cancer cell vulnerabilities. Mol Syst Biol. 2013;9:696.
Cornford PA, Dodson AR, Parsons KF, Desmond AD, Woolfenden A, Fordham M, et al. Heat shock protein expression independently predicts clinical outcome in prostate cancer. Cancer Res. 2000;60:7099–105.
Roller C, Maddalo D. The Molecular Chaperone GRP78/BiP in the development of chemoresistance: mechanism and possible treatment. Front Pharm. 2013;4:10.
Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet. 2018;50:1240–6.
Chayka O, D’Acunto CW, Middleton O, Arab M, Sala A. Identification and pharmacological inactivation of the MYCN gene network as a therapeutic strategy for neuroblastic tumor cells. J Biol Chem. 2015;290:2198–212.
Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44:1199–206.
Lasorsa VA, Formicola D, Pignataro P, Cimmino F, Calabrese FM, Mora J, et al. Exome and deep sequencing of clinically aggressive neuroblastoma reveal somatic mutations that affect key pathways involved in cancer progression. Oncotarget. 2016;7:21840–52.
Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34:634–6.
Deans RM, Morgens DW, Okesli A, Pillay S, Horlbeck MA, Kampmann M, et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat Chem Biol. 2016;12:361–6.
Dwivedi S, Bautista M, Shrestha S, Elhasasna H, Chaphekar T, Vizeacoumar FS, et al. Sympathetic signaling facilitates progression of neuroendocrine prostate cancer. Cell Death Discov. 2021;7:364.
Sigorski D, Gulczyński J, Sejda A, Rogowski W, Iżycka-Świeszewska E. Investigation of Neural Microenvironment in Prostate Cancer in Context of Neural Density, Perineural Invasion, and Neuroendocrine Profile of Tumors. Front Oncol. 2021;11:710899.
Bland T, Wang J, Yin L, Pu T, Li J, Gao J, et al. WLS-Wnt signaling promotes neuroendocrine prostate cancer. iScience. 2021;24:101970.
Danza G, Di Serio C, Rosati F, Lonetto G, Sturli N, Kacer D, et al. Notch signaling modulates hypoxia-induced neuroendocrine differentiation of human prostate cancer cells. Mol Cancer Res. 2012;10:230–8.
Huckesfeld S, Schlegel P, Miroschnikow A, Schoofs A, Zinke I, Haubrich AN, et al. Unveiling the sensory and interneuronal pathways of the neuroendocrine connectome in Drosophila. eLife. 2021;10:e65745.
Chen X, Chen Z, Li Z, Zhao C, Zeng Y, Zou T, et al. Grafted c-kit(+)/SSEA1(-) eye-wall progenitor cells delay retinal degeneration in mice by regulating neural plasticity and forming new graft-to-host synapses. Stem Cell Res Ther. 2016;7:191.
Yu B, Liu J, Su M, Wang C, Chen H, Zhao C. Disruption of Foxg1 impairs neural plasticity leading to social and cognitive behavioral defects. Mol Brain. 2019;12:63.
Mallika C, Guo Q, Li JY. Gbx2 is essential for maintaining thalamic neuron identity and repressing habenular characters in the developing thalamus. Dev Biol. 2015;407:26–39.
Zhang Q, Pan Y, Ji J, Xu Y, Zhang Q, Qin L. Roles and action mechanisms of WNT4 in cell differentiation and human diseases: a review. Cell Death Discov. 2021;7:287.
Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci USA. 2011;108:7838–43.
Kabayiza KU, Masgutova G, Harris A, Rucchin V, Jacob B, Clotman F. The onecut transcription factors regulate differentiation and distribution of dorsal interneurons during spinal cord development. Front Mol Neurosci. 2017;10:157.
Liu F, Zhang Y, Chen F, Yuan J, Li S, Han S, et al. Neurog2 directly converts astrocytes into functional neurons in midbrain and spinal cord. Cell Death Dis. 2021;12:225.
Filova I, Pysanenko K, Tavakoli M, Vochyanova S, Dvorakova M, Bohuslavova R, et al. ISL1 is necessary for auditory neuron development and contributes toward tonotopic organization. Proc Natl Acad Sci USA. 2022;119:e2207433119.
Quintanal-Villalonga A, Taniguchi H, Zhan YA, Hasan MM, Chavan SS, Meng F, et al. Multiomic analysis of lung tumors defines pathways activated in neuroendocrine transformation. Cancer Discov. 2021;11:3028–47.
Rubin MA, Bristow RG, Thienger PD, Dive C, Imielinski M. Impact of lineage plasticity to and from a neuroendocrine phenotype on progression and response in prostate and lung cancers. Mol Cell. 2020;80:562–77.
Ahmed T, Vial MR, Ost D, Stewart J, Hasan MA, Grosu HB. Non-small cell lung cancer transdifferentiation into small cell lung cancer: A case series. Lung Cancer. 2018;122:220–3.
Aslanpour S, Han S, Schuurmans C, Kurrasch DM. Neurog2 Acts as a classical proneural gene in the ventromedial hypothalamus and is required for the early phase of neurogenesis. J Neurosci. 2020;40:3549–63.
Zhang Q, Huang R, Ye Y, Guo X, Lu J, Zhu F, et al. Temporal requirements for ISL1 in sympathetic neuron proliferation, differentiation, and diversification. Cell Death Dis. 2018;9:247.
Sreekumar A, Saini S. Role of transcription factors and chromatin modifiers in driving lineage reprogramming in treatment-induced neuroendocrine prostate cancer. Front Cell Dev Biol. 2023;11:1075707.
Bery F, Cancel M, Chantome A, Guibon R, Bruyere F, Rozet F, et al. The Calcium-sensing receptor is a marker and potential driver of neuroendocrine differentiation in prostate cancer. Cancers. 2020;12:860.
Komiya A, Suzuki H, Imamoto T, Kamiya N, Nihei N, Naya Y, et al. Neuroendocrine differentiation in the progression of prostate cancer. Int J Urol. 2009;16:37–44.
Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, et al. The master neural transcription factor brn2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7:54–71.
Chen CC, Tran W, Song K, Sugimoto T, Obusan MB, Wang L, et al. Temporal evolution reveals bifurcated lineages in aggressive neuroendocrine small cell prostate cancer trans-differentiation. Cancer Cell. 2023;41:2066–82.e2069.
Gao Z, Ure K, Ables JL, Lagace DC, Nave KA, Goebbels S, et al. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci. 2009;12:1090–2.
Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, et al. Clinical and Genomic Characterization of Treatment Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J Clin Oncol. 2018;36:2492–506.
Chakraborty G, Gupta K, Kyprianou N. Epigenetic mechanisms underlying subtype heterogeneity and tumor recurrence in prostate cancer. Nat Commun. 2023;14:567.
Asrani K, Torres AF, Woo J, Vidotto T, Tsai HK, Luo J, et al. Reciprocal YAP1 loss and INSM1 expression in neuroendocrine prostate cancer. J Pathol. 2021;255:425–37.
Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19:289–97.
Hicks-Berthet J, Ning B, Federico A, Tilston-Lunel A, Matschulat A, Ai X, et al. Yap/Taz inhibit goblet cell fate to maintain lung epithelial homeostasis. Cell Rep. 2021;36:109347.
Gao Y, Zhang W, Han X, Li F, Wang X, Wang R, et al. YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat Commun. 2014;5:4629.
Tang F, Xu D, Wang S, Wong CK, Martinez-Fundichely A, Lee CJ, et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science. 2022;376:eabe1505.
Raivich G, Bohatschek M, Da Costa C, Iwata O, Galiano M, Hristova M, et al. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67.
Ko KR, Lee J, Nho B, Kim S. c-Fos is necessary for HGF-mediated gene regulation and cell migration in Schwann cells. Biochem Biophys Res Commun. 2018;503:2855–60.
Magnon C, Hall SJ, Lin J, Xue X, Gerber L, Freedland SJ, et al. Autonomic nerve development contributes to prostate cancer progression. Science. 2013;341:1236361.
Zahalka AH, Arnal-Estape A, Maryanovich M, Nakahara F, Cruz CD, Finley LWS, et al. Adrenergic nerves activate an angio-metabolic switch in prostate cancer. Science. 2017;358:321–6.
Kemble J, Kwon ED, Karnes RJ. Addressing the need for more therapeutic options in neuroendocrine prostate cancer. Expert Rev Anticancer Ther. 2023;23:177–85.
Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102.
Auslander N, Cunningham CE, Toosi BM, McEwen EJ, Yizhak K, Vizeacoumar FS, et al. An integrated computational and experimental study uncovers FUT9 as a metabolic driver of colorectal cancer. Mol Syst Biol. 2017;13:956.
Evers B, Jastrzebski K, Heijmans JP, Grernrum W, Beijersbergen RL, Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34:631–3.
Housden BE, Perrimon N. Comparing CRISPR and RNAi-based screening technologies. Nat Biotechnol. 2016;34:621–3.
Ding Y, Denomy C, Freywald A, Pan Y, Vizeacoumar FJ, Vizeacoumar FS, et al. Systematic Comparison of CRISPR and shRNA screens to identify essential genes using a graph-based unsupervised learning model. Cells. 2024;13:1653.
Kovach C, Dixit R, Li S, Mattar P, Wilkinson G, Elsen GE, et al. Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb Cortex. 2013;23:1884–1900.
Sommer L, Ma Q, Anderson DJ. neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–41.
Aaker JD, Patineau AL, Yang HJ, Ewart DT, Gong W, Li T, et al. Feedback regulation of NEUROG2 activity by MTGR1 is required for progression of neurogenesis. Mol Cell Neurosci. 2009;42:267–77.
Hufnagel RB, Le TT, Riesenberg AL, Brown NL. Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev Biol. 2010;340:490–503.
Smith DK, Yang J, Liu ML, Zhang CL. Small Molecules Modulate Chromatin Accessibility to Promote NEUROG2-mediated fibroblast-to-neuron reprogramming. Stem Cell Rep. 2016;7:955–69.
Liu ML, Zang T, Zou Y, Chang JC, Gibson JR, Huber KM, et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nature Commun. 2013;4:2183.
Noack F, Vangelisti S, Raffl G, Carido M, Diwakar J, Chong F, et al. Multimodal profiling of the transcriptional regulatory landscape of the developing mouse cortex identifies Neurog2 as a key epigenome remodeler. Nat Neurosci. 2022;25:154–67.
Aydin B, Kakumanu A, Rossillo M, Moreno-Estelles M, Garipler G, Ringstad N, et al. Proneural factors Ascl1 and Neurog2 contribute to neuronal subtype identities by establishing distinct chromatin landscapes. Nat Neurosci. 2019;22:897–908.
Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science. 2018;362:91–95.
Kaarijarvi R, Kaljunen H, Ketola K. Molecular and functional links between neurodevelopmental processes and treatment-induced neuroendocrine plasticity in prostate cancer progression. Cancers. 2021;13:692.
Ito T, Matsubara D, Tanaka I, Makiya K, Tanei ZI, Kumagai Y, et al. Loss of YAP1 defines neuroendocrine differentiation of lung tumors. Cancer Sci. 2016;107:1527–38.
Horie M, Saito A, Ohshima M, Suzuki HI, Nagase T. YAP and TAZ modulate cell phenotype in a subset of small cell lung cancer. Cancer Sci. 2016;107:1755–66.
Nouruzi S, Ganguli D, Tabrizian N, Kobelev M, Sivak O, Namekawa T, et al. ASCL1 activates neuronal stem cell-like lineage programming through remodeling of the chromatin landscape in prostate cancer. Nat Commun. 2022;13:2282.
Venteo S, Desiderio S, Cabochette P, Deslys A, Carroll P, Pattyn A. Neurog2 deficiency uncovers a critical period of cell fate plasticity and vulnerability among neural-crest-derived somatosensory progenitors. Cell Rep. 2019;29:2953–60.e2952.
Acknowledgements
We sincerely thank the Rickman lab for kindly providing the 22Rv1 and 22Rv-1 N-Myc cell lines. We also thank Dr. Michael Cox for his kind contribution to BPH1, BHPrE1, NHPrE1, and PNT1B cell lines. Financial Support: This work was supported by operating grants from the Canadian Institutes of Health Research (PJT-156401; PJT-156309) to F.J.V and A.F; the Canadian Foundation for Innovation (CFI-33364) and Saskatchewan Cancer Agency operating grants with funds donated to the Cancer Foundation of Saskatchewan to F.J.V. F.S.V. is supported by the College of Medicine, U of S. H.E. was supported by the CoMGRAD award, and the U of S. J.D.W.P. is supported by grant 1160056 from the Cancer Research Society. V.M. is supported by the Canada Graduate Scholarship - Doctoral from Canadian Institutes of Health Research (FBD-187665) as well as the Health Sciences Graduate Scholarship from the College of Medicine at the University of Saskatchewan. This work was also supported by the CoMBRIDGE award (A.K, F.J.V, A.F), and funding from the Saskatchewan Health Research Foundation (Establishment Grant), and Prostate Cancer Fight Foundation and Ride for Dad to AK.
Funding
This work was supported by operating grants from the Canadian Institutes of Health Research (PJT-156401; PJT-156309) to F.J.V and A.F; the Canadian Foundation for Innovation (CFI-33364) and Saskatchewan Cancer Agency operating grants with funds donated to the Cancer Foundation of Saskatchewan to F.J.V. This work was also supported by the CoMBRIDGE award (A.K, F.J.V, A.F), and funding from the Saskatchewan Health Research Foundation (Establishment Grant), and Prostate Cancer Fight Foundation and Ride for Dad to AK.
Author information
Authors and Affiliations
Contributions
AK, AF, FSV and FJV were responsible for the conceptualization of the project. AK, AF, FSV, YW, YW and FJV supervised the research. AK, AF and FJV acquired the funding. PW, JDWP, FSV, NJ, VM, BC, JP, YZ, HD, LN, AG, LHG, MLW, HE, and KKB performed the experiments and performed data analysis. All authors contributed to writing, editing and reviewing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The authors confirm that all methods were performed in accordance with the relevant guidelines and regulations. All animals were handled in accordance with the approved protocols by the University of Saskatchewan Animal Research Ethics Board (AREB) (Permit 20190119).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Walke, P., Price, J.D.W., Vizeacoumar, F.S. et al. A novel role for Neurog2 in MYCN driven neuroendocrine plasticity of prostate cancer. Oncogene 44, 2460–2473 (2025). https://doi.org/10.1038/s41388-025-03413-0
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03413-0