Correction to: The Pharmacogenomics Journal https://doi.org/10.1038/tpj.2015.99, published online 16 February 2016
The authors identified errors in the published article. Corrections are in bold. These corrections do not affect the results, discussion, or conclusions. The authors regret the errors and sincerely apologize to the Journal and readers of the Journal for any inconvenience.
1. There were errors in Tables 1 and 2. Corrections to the tables have been made, as well as their corresponding entries in the INTRODUCTION section. The “Reference allele” and “Mutant allele” columns in Tables 1 and 2 previously contained the information based on the PharmGKB database and PubMed database. But the authors found that the allele information from different databases can cause discrepancies. Therefore, a correction has been made based on the dbSNP database which is the largest public repository for genetic variation to clarify any confusion or misunderstanding. The corrected INTRODUCTION section, Table 1 and 2 appear below.
Introduction
Although rs776746 T > C (also known as CYP3A5*3), which is a nonfunctioning allele of the CYP3A5 gene, is associated with decreased tacrolimus metabolism4, the role of other genes, including the ABCB1, CYP2C19, POR, UGT1A8, NOD2, and PPARA, in the pharmacokinetics of tacrolimus was either inconsistent or insignificant3.
2. There was an error in the SUBJECTS and METHODS section, subsection “Determination of plasma concentrations of tacrolimus”, subsection “Pharmacokinetic analysis”, where “plasma concentration” should be “whole blood concentration”. A correction has been made, as well as their corresponding entries in Figure 1 and the caption on Table 3.
SUBJECTS AND METHODS
Determination of tacrolimus concentrations in whole blood
Whole-blood concentrations of tacrolimus were determined using a previously published LC/MS/MS method18 with some modifications. The blood sample preparation involved a liquid/liquid extraction with methyl tert-butyl ether.
Pharmacokinetic analysis
Tacrolimus concentrations from the reference formulation were used for the pharmacokinetic analysis in the present study. The maximum whole-blood concentration (Cmax) of tacrolimus was determined directly from the observed whole-blood concentration data.
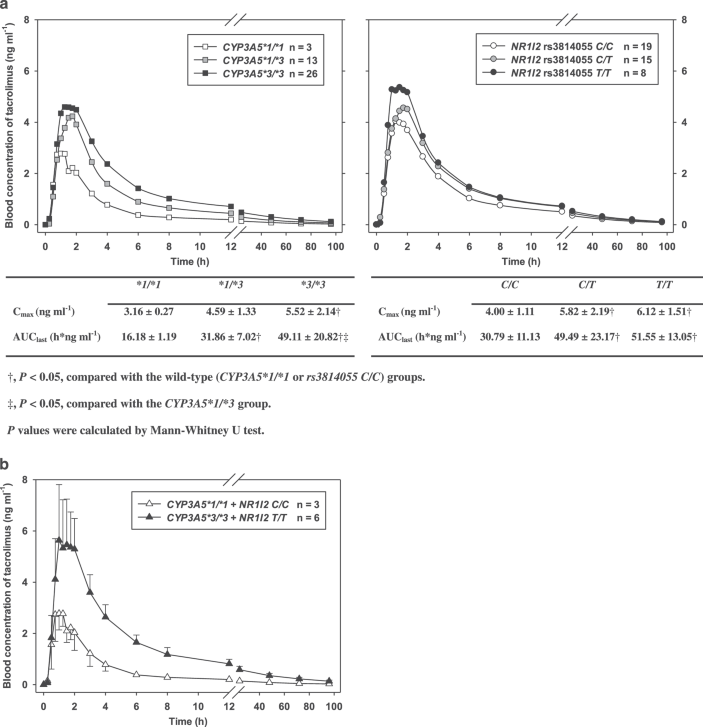
Figure 1. Mean concentration-time profiles of tacrolimus (a) by different CYP3A5 and NR1I2 genotypes (n = 42) and (b) by two different combined CYP3A5 and NR1I2 genotypes (n = 9) where the genotypes represented the highest (CYP3A5 *3/*3 and NR1I2 T/T) and the lowest (CYP3A5 *1/*1 and NR1I2 C/C) exposure to tacrolimus. The error bars represent the standard deviations.
3. There was an error in the RESULTS section, subsection “Genetic effects of CYP3A5 and NR1I2 on tacrolimus pharmacokinetics”, where “CYP2A5*3/*3” should be “CYP3A5*3/*3”.
Results
Genetic effects of CYP3A5 and NR1I2 on tacrolimus pharmacokinetics
The greater the number of nonfunctioning *3 alleles in the CYP3A5 gene, the greater the mean exposure to tacrolimus (Figure 1a). Consequently, the geometric mean AUClast and Cmax of tacrolimus was 2.78 (95% CI: 1.66–4.66) and 1.64 (95% CI: 1.04–2.60) times greater, respectively, in the CYP3A5*3/*3 homozygote than in the *1/*1 wild-type (P < 0.05; Figure 1a).
4. There was a typo in the DISCUSSION section. The corrected sentence appears below.
Discussion
Recently, a clinical trial in 32 kidney transplant patients showed that subjects with the rs3814055 C/C genotype had 1.2 and 1.5 times greater clearance of tacrolimus than the rs3814055 T carriers, C/T and T/T genotypes, respectively,27 which supports the findings in our study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Choi, Y., Jiang, F., An, H. et al. Correction to: A pharmacogenomic study on the pharmacokinetics of tacrolimus in healthy subjects using the DMETTM Plus platform. Pharmacogenomics J 24, 35 (2024). https://doi.org/10.1038/s41397-024-00354-x
Published:
Version of record:
DOI: https://doi.org/10.1038/s41397-024-00354-x
