Abstract
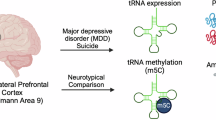
A moderating hub between resting state networks (RSNs) and the medial temporal lobe (MTL) is the parahippocampal cortex (PHC). Abnormal activity has been reported in depressed patients and suicide attempters in this region. Alterations in neuronal mitochondrial function may contribute to depression and suicidal behavior. However, little is known about the underlying molecular level changes in relevant structures. Specifically, expressional changes related to suicide have not been reported in the PHC. In this study, we compared the protein expression levels of genes encoding tricarboxylic acid (TCA) cycle enzymes in the PHC of adult individuals who died by suicide by reverse phase protein array (RPPA), which was corroborated by qRT-PCR at the mRNA level. Postmortem human brain samples were collected from 12 control and 10 suicidal individuals. The entorhinal cortex, which is topographically anterior to the PHC in the parahippocampal gyrus, and some other cortical brain regions were utilized for comparison. The results of the RPPA analysis revealed that the protein levels of DLD, OGDH, SDHB, SUCLA2, and SUCLG2 subunits were significantly elevated in the PHC but not in other cortical brain regions. In accordance with these findings, the mRNA levels of the respective subunits were also increased in the PHC. The subunits with altered levels are implicated in enzyme complexes involved in the oxidative decarboxylation branch of glutamine catabolism. These data suggest a potential role of glutaminolysis in the pathophysiology of suicidal behavior in the PHC.

Similar content being viewed by others
Introduction
There is evidence to suggest that alterations in the functioning and connectivity of resting state networks (RSNs), including the default mode, salience, and central executive networks, may contribute to suicidal behavior by increasing negative rumination, impairing emotional regulation, and reducing cognitive control and decision-making abilities [1]. The default mode network (DMN) is considered a task-negative network of brain regions that deactivates during cognitive goal-directed tasks [2] while exhibiting increased activation during internally focused processes such as autobiographical memory retrieval and emotional and reward processing [3,4,5]. A growing number of studies consider the DMN to encompass functionally distinct subdivisions, including the dorsal medial prefrontal cortex (DMPFC), the core DMN, and the medial temporal lobe (MTL) subsystems [6,7,8]. The MTL comprises heterogeneous brain areas, including the hippocampus and the parahippocampal gyrus, which play an essential role in memory and cognitive processes [9,10,11]. The anterior part of the parahippocampal gyrus is comprised of the entorhinal and perirhinal cortices, while the posterior part of the parahippocampal gyrus is constituted by the parahippocampal cortex (PHC) [12]. In-depth studies have revealed that the PHC functions are considerably more intricate than previously assumed, encompassing a multitude of processes beyond mere spatial and visual information processing as originally suggested [13, 14]. Recent studies employing resting-state functional magnetic resonance imaging (fMRI) have demonstrated that the MTL subsystem co-activates with the default mode subnetworks during rest [15,16,17] and self-referential processing [18, 19] with the parahippocampal cortex (PHC) serving as a key mediator of this interaction.
Structural and functional abnormalities have been identified in brain circuits involved in emotional processing in patients with major depression [20,21,22] and suicidal ideation [1, 23,24,25]. These include regions belonging to the MTL, such as the hippocampus, PHC, and amygdala, and prefrontal cortical regions, such as the orbitofrontal cortex (OFC), medial prefrontal cortex (mPFC), and anterior cingulate cortex (ACC), and parietal cortical areas such as the posterior cingulate cortex (PCC), precuneus, and retrosplenial cortex. The PHC is also considered a key region of the limbic system and, together with the amygdala and hippocampus, is important in the operation of the reward system [26]. It also exhibits essential roles in emotion regulation, motivation, and cognitive control [13, 27, 28]. Emerging evidence suggests that dysregulation in the MTL is increasingly associated with functional impairments in individuals with mood disorders [29,30,31]. Recall of the past suicide attempt was associated with increased activation in the ACC, mPFC, and hippocampus, while recall of mental pain when committing suicide was associated with the deactivation of frontal cortical regions and activation of the PHC [32]. Other neuroimaging studies have demonstrated that depressed subjects looking at negative pictures evoked an increase in the activity of the PHC [4]. In contrast, the absence or delay of rewards was correlated with diminished PHC activation in adult suicide attempters with major depressive disorder (MDD) [33]. Zamoscik et al. have reported that remitted depressed individuals had stronger connectivity between the PCC and the PHC during the recollection of negative life events compared to healthy controls [34], which supports the assumption that connectivity between the DMN and MTL is dynamic and influenced by changes in emotional states [30].
In recent years, molecular investigations using high-throughput omics technologies have increased significantly. However, relatively few have examined the molecular alterations in individual regions of the human brain networks involved in psychiatric disorders. Images of brain metabolism and gene expression studies have reported altered mitochondrial functioning associated with psychiatric disorders [35,36,37,38,39,40,41]. Mitochondria are pivotal in the brain’s energy metabolism, supporting essential processes such as dendritic reconstruction, neurotransmitter release, neurite extension, and neuronal differentiation, which are crucial for neuroplasticity [42]. Patients with MDD frequently exhibit mitochondrial energy metabolism obstacles and energy imbalances in the brain, suggesting a correlation between depression and energy metabolism [43]. Disturbances in mitochondrial energy metabolism can contribute to mitochondrial dysfunction, which has been identified as a crucial vulnerability factor for the development of depression [44,45,46] and suicide completion by violent means [35, 47]. Recent studies have suggested that the activity of enzymes involved in the tricarboxylic acid (TCA) cycle is associated with MDD and depression-like behavior [48, 49]. The TCA cycle intermediates also play a role in the synthesis of neurotransmitters that are critical for brain function and mood regulation [50,51,52]. It has been demonstrated that depressed individuals may exhibit disturbances in neurotransmitter metabolism related to the TCA pathway [53]. Emerging evidence suggests a complex interplay between mitochondrial dysfunction and neuroinflammation, both of which contribute significantly to the pathogenesis of depression [54]. Mitochondrial damage releases damage-associated molecular patterns (DAMPs), which are recognized by microglial immune receptors, amplifying neuroinflammation [55]. In turn, activated glial cells release inflammatory factors that disrupt mitochondrial metabolism and function, creating a vicious cycle exacerbating depressive symptoms [56]. A recent study linked post-inflammatory behavioral despair in mice with reduced cortical glutamate-glutamine ratios, indicating that their aberrant metabolism may play a pivotal role in the pathogenesis of depression [57]. A proteomic enrichment analysis of affective disorders also revealed an alteration in glutamate metabolism [58]. However, systematic studies assessing changes in TCA cycle-related genes in brain regions relevant to suicide symptomatology are scarce. Consequently, further investigation of the gene expression changes associated with this phenotype is required to gain a deeper understanding of the underlying pathological mechanisms. Previous studies have identified proteomic and genomic differences in the PHC in neurodegenerative disorders [59,60,61,62], and mass spectrometry-based proteomics studies have identified alterations in TCA cycle enzymes in MDD and in its animal models [63,64,65]. However, proteomic alterations related to depression and suicide have not been reported in the PHC of the human brain.
To better understand the connection between mitochondrial dysfunction and suicidal behavior, in the present study, we measured the protein expression of TCA cycle enzyme subunits in the parahippocampal cortex of individuals who died by suicide and compared the alterations to those in other brain regions using microarray-based reverse phase protein array (RPPA) technology. This technology enables the measurement of protein levels in a large number of samples simultaneously using antibodies against the targets of interest. In contrast to traditional immunoassay techniques, RPPA is reversed and requires monospecific antibodies for quantification [66]. The generated signal is proportional to the amount of the selected antibody bound to the spotted proteins and can be quantified to estimate relative protein concentrations. We identified five differentially expressed subunits of TCA cycle enzymes between suicide and control groups in the parahippocampal, but not in the adjacent entorhinal cortex.
Methods and materials
Human brain tissue samples
Human brain samples were collected in accordance with the Ethical Rules for Using Human Tissues for Medical Research in Hungary (HM 34/1999) and the Code of Ethics of the World Medical Association (Declaration of Helsinki). Tissue samples were obtained during brain autopsy at the Department of Pathology, Forensic and Insurance Medicine of Semmelweis University and the Clinical Centre of Pathology, University of Debrecen, in the framework of the Human Brain Tissue Bank (HBTB), Budapest. The activity of the HBTB has been authorized by the Committee of Science and Research Ethics of the Ministry of Health Hungary (ETT TUKEB: 189/KO/02.6008/2002/ETT) and the Semmelweis University Regional Committee of Science and Research Ethics (No. 32/1992/TUKEB). The study was conducted in accordance with a protocol approved by the Committee of Science and Research Ethics, Semmelweis University (TUKEB 189/2015). The subjects’ medical histories were obtained from clinical records, interviews with family members and relatives, as well as pathological and neuropathological reports. Informed consent was obtained from all subjects. All personal data is stored in accordance with the highest ethical standards, and samples were coded before the analysis of tissues.
Tissue preparation
Postmortem human brain tissue samples were acquired by the Human Brain Tissue Bank (Semmelweis University, Budapest, Hungary). Tissue samples from the parahippocampal cortex and comparison brain regions such as the entorhinal, dorsomedial prefrontal, anterior cingulate, and insular cortices were collected from 22 individuals. The control group includes 12 subjects (4 females and 8 males, mean age of 50.2 ± 3.0), while the suicide group consists of 10 subjects who died by suicide (3 females and 7 males, mean age of 49.7 ± 4.3). The ages of the subjects ranged from 26 to 71 years (Table 1, Supplementary Table 1). The selected control subjects were not diagnosed with any psychiatric disorder, while 9 out of 10 of the suicide individuals were without evidence of any clinical care or known drug treatment.
The brains were removed from the skull with a postmortem delay of 1–10 h, frozen rapidly on dry ice, and stored at −80 °C until microdissection. Serial coronal sections were made from frozen brain slices. During sample collection, we obtained representative cortical samples from areas corresponding to previously reported MNI scores of the activated parahippocampal regions in depressed and suicidal individuals [24, 32, 67,68,69]. Therefore, sections between −11.0 and −35.0 mm from the origin of the MNI standard space were excised (see ref. [70]). The cortical samples were punched out using specialized microdissection needles with 8 and 15 mm internal diameters [71] (Fig. 1). The entorhinal cortical samples were obtained from Brodmann area 34, anterior to Brodmann area 35 and 36 (which include the perirhinal and parahippocampal cortices) [72]. Accordingly, the EC samples were derived from the medial intermediate subfield of the entorhinal cortex (EMI) [73]. The dissected cortical tissue pellets included both gray and white matter portions within the gyrus. Samples were collected in 1.5 mL Eppendorf tubes and stored at −80 °C until use. Throughout the microdissection procedure, the tissue samples were kept frozen. In subsequent experiments, the investigator was blinded to the group allocation during the experiment and when assessing the outcome.
A and B The location and topographical extension of the entorhinal (EC, MNI anteroposterior coordinate [y] = −12) (A) and parahippocampal (PHC, MNI anteroposterior coordinate [y] = −27) (B) cortices at the level of anterior (A) and posterior (B) hippocampus in the human temporal lobe [60]. C and D The coronal section of the human brain represent the dissected regions, namely the entorhinal (C) and parahippocampal (D) cortices. The dissected areas are demarcated by dashed circles. The section was stained using the Levanol-Fast Cyanine 5RN method. The scale bar is 1 cm. Amg amygdala, EC entorhinal cortex, FuG fusiform gyrus, HPC hippocampus, ITG inferior temporal gyrus, MTG middle temporal gyrus, PHC parahippocampal cortex, sfs superior frontal sulcus, STG superior temporal gyrus.
Sample handling and reverse phase protein array (RPPA) measurement
RPPA was performed on postmortem human brain samples to determine the protein expression changes related to suicidal behavior in the parahippocampal and entorhinal cortices. For the measurements of PHC samples, 10 control subjects (3 females and 7 males, mean age of 50.8 ± 3.6) and 10 subjects who died by suicide (3 females and 7 males, mean age of 49.7 ± 4.3) were used. For the measurements of entorhinal cortical samples, 7 control subjects (2 females and 5 males, mean age of 44.7 ± 3.4) and 5 subjects who died by suicide (1 female and 4 males, mean age of 39.8 ± 3.1) were used. For the measurements of additional comparison brain regions as the dorsomedial prefrontal cortical samples, 6 control subjects (2 females and 4 males, mean age of 50.8 ± 3.6) and 10 subjects who died by suicide (3 females and 7 males, mean age of 49.7 ± 4.3), for the anterior cingulate cortical samples 10 control subjects (3 females and 7 males, mean age of 49.6 ± 3.6) and 8 subjects who died by suicide (3 females and 5 males, mean age of 50.4 ± 5.8), and for the insular cortical samples 10 control subjects (2 females and 8 males, mean age of 50.9 ± 2.0) and 9 subjects who died by suicide (3 females and 6 males, mean age of 47.9 ± 4.3) were used, respectively (see Supplementary Table 1). We used the above-described number of individuals in the groups to ensure a balanced yet feasible sample size for detecting significant differences in protein expression between the groups. This sample size was determined based on practical considerations of availability and ethical constraints while aiming for sufficient statistical power to detect differences in proteins of interest. Additionally, prior studies suggested that similar sample sizes were adequate for detecting proteomic changes in brain tissue studies.
RPPA allows for the semi-quantitative measurement of protein levels in a large number of samples simultaneously using antibodies against the targets of interest [66]. The generated signal is proportional to the amount of the selected antibody bound to the spotted proteins and can be quantified to estimate relative protein concentrations. The procedure was carried out as previously described [74]. Briefly, the delipidation of the samples and the purification of proteins were carried out in a buffer containing benzonase and protease inhibitors. Following purification, the protein concentration was adjusted to 1.0 mg/ml for each sample. Aliquots were then stored at −80 °C until use. Before quantitative measurement, samples were serially diluted in lysis buffer to produce four serial two-fold dilutions (1, 0.5, 0.25, and 0.125 mg/ml concentrations). Subsequently, the samples were printed onto ONCYTE nitrocellulose slides (Sigma-Aldrich, cat. no. 705170) under conditions of constant 70% relative humidity using an Aushon BioSystems 2470 microarrayer (Quanterix, Billerica, MA, USA). Two technical replicates were printed per sample dilution. Following the printing of the samples, the slides were hydrated in deionized water, washed with Tris-buffered saline (TBS) containing 0.1% (w/v) Tween 20 (TBS-T), and blocked with TBS-T containing 5% (w/v) bovine serum albumin (TBS-T-BSA). Following the washing procedures, the slides were incubated with avidin-biotin and peroxidase blocking reagents (VECTOR) and incubated with primary antibodies (primary antibodies are listed in Supplementary Table 3). It should be noted that the specificity and selectivity of the antibodies included in the array have been pre-validated (see Supplementary Table 3 and Dobolyi et al. [75]). Bound antibodies were detected by incubation with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Europe, Ely, UK). To amplify the signals, slides were incubated with Bio-Rad Amplification Reagent (Bio-Rad, Watford, UK, cat. no. 1708230). After signal amplification, arrays were washed and probed with IRDye800CW-coupled streptavidin fluorophore, then incubated with TBS-T-BSA and washed in TBS-T. In order to normalize against total protein, an additional slide was prepared, and total protein was stained with Fast Green FCF (Sigma-Aldrich, cat. no. F7258). Subsequently, the slides were washed with deionized water, dried, and stored in the dark. The slides were then read using the InnoScan 710-IR slide scanner (Innopsys) at an excitation wavelength of 785 nm for IRDye800CW and an excitation wavelength of 670 nm for Fast Green FCF. The images were acquired as tagged image file format (TIFF) files. The relative fluorescence intensity of each sample spot was quantified using Mapix software (Innopsys). To determine nonspecific signals, the primary antibody incubation step was omitted. Processed signal intensities were normalized using the corresponding total protein stain signal intensity. Means of technical replicate sample spots were calculated, and data were written to a comma-separated values (CSV) file. Further details on all RPPA protocols can be found at http://rppa.hu/Protocols.html.
RPPA data analysis
Normalized RPPA data were expressed as log2-transformed values for each biological sample, based on the serial dilution curve and technical replicates. The transformed normalized data were stored as matrices arranged in rows of different samples and columns of different antibodies. The median of all samples was calculated, and this was subtracted from each element in that column. Matrices of antibody-wise median-centered data were horizontally integrated into a single matrix. The normalized and log2-transformed data were subsequently exported to a CSV file. The investigator was blinded to the group allocation during the experiment until the CSV files were exported.
Gene expression analysis with qRT-PCR
The procedure was carried out as described previously [76]. In brief, total RNA was isolated from approximately 20 mg of frozen postmortem brain tissue using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as lysis buffer combined with the RNeasy Mini kit (Qiagen, Germany) in accordance with the manufacturer’s instructions. The quality and quantity of extracted RNA were determined using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and only those with an A260/A280 ratio between 1.8 and 2.1 were used in subsequent experiments. The concentration of RNA was adjusted to 1000 ng/µL, and it was treated with Amplification Grade DNase I (Invitrogen, Carlsbad, CA, USA). The isolated RNA concentration was calculated and normalized with RNase‐free water and reverse transcribed into cDNA using SuperScript II Reverse Transcriptase Kit (Invitrogen, Carlsbad, CA, USA). After a 10‐fold dilution, 2.5 μl of the resulting cDNA was used as a template in PCR performed in duplicates using SYBR Green dye (Sigma, St. Louis, MO, USA). The PCR reactions were conducted on the CFX-96 C1000 Touch Real-Time System (Bio-Rad Laboratories, Hercules, CA, USA) with iTaq DNA polymerase (Bio‐Rad Laboratories, Hercules, CA, USA) in total volumes of 12.5 μl under the following conditions: 95 °C for 3 min, followed by 35 cycles of 95 °C for 0.5 min, 60 °C for 0.5 min, and 72 °C for 1 min using specific primers employed. A melting curve was performed at the conclusion of the amplification cycles to verify the specificity of the PCR products. The primers utilized for qRT‐PCR were synthesized by Integrated DNA Technologies, Inc., (IDT, Coralville, IA, USA) and employed at a final concentration of 300 nM. Housekeeping genes, including ACTB, GAPDH, and LDHA, were used as internal controls. The applied primers are listed in Supplementary Table 4. The relative gene expression values were calculated from the ratio of genes and the averages of housekeeping genes using the 2 −△△Ct method.
Preparation of in situ hybridization probes
The PCR products using primer pairs for OGDH isoforms 1 and 2 (CTGTGCTTGGCTGGAAAACC and AGCATCCAAAATCCCCAGGG) were purified from gel to obtain non-overlapping probes to demonstrate specific labeling. The purified PCR products were applied as templates in a PCR reaction, with the primer pairs specific for the probe, and the T7 RNA polymerase recognition site (GTAATACGACTCACTATAGGGCGAATTGGGTA) was added to the reverse primers. Finally, the identities of the cDNA probes were verified by sequencing them with T7 primers.
In situ hybridization histochemistry
A fixed medial temporal lobe brain block of a 48-year-old male control subject (cause of death: acute myocardial infarction; PMI: 24 h) was used for in situ hybridization (ISH). Using a cryostat, serial coronal sections (20 μm thick) were cut and mounted on positively charged slides (SuperfrostPlus®, Fisher Scientific), dried, and stored at −80 °C until use. Further steps were performed according to the procedure described previously by Dobolyi et al. [77]. Briefly, both sense and antisense [35S]UTP-labeled riboprobes were generated from the above-described DNA probes using T3 and T7 RNA polymerases of the MAXIscript Transcription Kit (Ambion, Austin, TX, USA) and used for hybridization at 1 million DPM (discharges per minute) activity per slide. Washing procedures included a 30 min incubation in RNase A, followed by decreasing concentrations of sodium-citrate buffer (pH 7.4) at room temperature and subsequently at 65 °C. Following hybridization and washes, slides were dipped in NTB nuclear track emulsion (Eastman Kodak) and stored at 4 °C for 3 weeks for autoradiography. Then, the slides were developed and fixed with Kodak Dektol developer and Kodak fixer, respectively, counterstained with Giemsa, and coverslipped with Cytoseal 60 (Stephens Scientific, Kalamazoo, MI, USA). A cell was considered to express OGDH if the number of autoradiography grains accumulated in a seemingly Gaussian distribution around a center was at least three times higher than the background level in an area corresponding to an average cell size (a circle with a diameter of 25 μm) in the same section.
Combination of in situ hybridization histochemistry with S100 and Iba1 immunohistochemistry
Slide attached sections of fixed brain tissue block from the medial temporal lobe were first processed for ISH, as described above. Thus, tightly bound RNA–RNA pairs were already formed by the time immunohistochemistry was performed, immediately before dipping the slides into an autoradiographic emulsion. In addition, the solutions used for perfusion and immunohistochemistry were prepared with DEPC-treated RNAse-free water, which ensured that the labeling intensity of the ISH did not decrease significantly. The immunolabeling protocol for mouse anti-S100b (1:250 dilution, Sigma-Aldrich, Cat. No. S2532) and goat anti-ionized calcium-binding adapter molecule 1 (Iba1) (1:100 dilution, Abcam, Cat. No. ab107159) was the same as that described above for double labeling immunohistochemistry. Immunoreactivity was visualized using DAB reactions, after which the ISH procedure was continued by dipping the slides into the emulsion. Each double-labeling experiment included controls, which were carried out through the double-labeling procedure without the application of radioactive ISH probes. These controls demonstrated that the DAB signal did not induce an autoradiography signal. An OGDH-expressing cell was considered S100- or Iba-immunopositive if at least 70% of the area of the circle containing the accumulation of autoradiography grains contained immunoreactivity for S100 or Iba1 and the center of the autoradiography grains was within the immunolabeled cell. To define the distribution of ISH signals over the cells, we used ImageJ software [78]. After uploading the images, the channels were separated, and the red channel was selected. Using the “Threshold” function, the darkest pixels were selected by setting the slider between 0 and 128, ensuring that only the ISH signal was selected, excluding the DAB-stained glial cells. A critical step in our quantitative analysis was to first determine the average area of glial cells, which was then used as a baseline to measure the ISH signal intensity across both glial and non-glial cells. We outlined five randomly selected glial cells per image across four different images to establish this average area. This standardized area was then applied to each instance of accumulated OGDH signals to ensure uniformity in the measurement scale across all cells analyzed (see Supplementary Fig. 3). The “Analyze Particles” Image J function was used to measure the number and area of the dark pixels, representing the OGDH mRNA within each predefined area. Parameters were set to include all sizes (0-infinity) and circularity (0.00–1.00) to include all relevant signals. For each cell, we quantified the intensity of the OGDH signals by measuring the dark pixels within the marked areas and adjusted these values by subtracting the dark pixel area observed in the background. The corrected values were then used to calculate the percentage of distribution relative to each cell’s area.
Microscopy and photography
Sections were examined using an Olympus BX60 light microscope, also equipped with fluorescent epi-illumination and a dark-field condenser. Images were captured at 2048 × 2048 pixel resolution with a SPOT Xplorer digital CCD camera (Diagnostic Instruments, Sterling Heights, MI, USA) using ×10–×100 objectives for bright-field images. The contrast and sharpness of the images were adjusted using the ‘levels’ and ‘sharpness’ commands in Adobe Photoshop CS 8.0. Full resolution was maintained until the photomicrographs were cropped, at which point the images were adjusted to a resolution of at least 300 dpi.
Statistical analyses
A quantitative analysis of protein expression by reverse phase protein array (RPPA) and the mRNA expression patterns of TCA cycle enzyme subunits was conducted in the PHC and the neighboring EC of people who died by suicide. We used postmortem brain tissue samples from 12 controls and 10 suicidal individuals. The numbers of samples were chosen based on a power analysis to ensure adequate power to detect the pre-specified effect size in mitochondrial protein levels. This analysis involved estimating the expected effect size and variability in the data and setting a target power level of 80% and a significance level of 0.05. These factors allow the selection of the sample size to balance sensitivity in detecting meaningful differences while considering practical constraints such as sample availability. The demographic data of the control and suicidal individuals were compared using the Chi-square test or Welch’s unequal variances t-test (p < 0.05). A statistical analysis of RPPA data was performed using a Python Statistical Analysis Tool (Python 3.8.5) to identify differentially expressed proteins between control and suicide groups. The signal intensities of <1% of undiluted samples were excluded. A principal component analysis (PCA) plot was generated for PHC samples across all the measured proteins using the ggfortify package in R Studio [79]. Levene’s test indicated that the variances are not significantly different for RPPA data. Therefore, two-way ANOVA was employed for the statistical analysis of altered protein expression for each brain region, using the protein expression levels of the 11 examined mitochondrial genes and the groups (suicide vs. control) as factors, followed by Sidak’s multiple comparison test. A Welch’s unequal variances t-test was carried out to compare mRNA expression between groups (GraphPad Prism 8.0.1). To examine the relationship between protein expression levels and postmortem interval (PMI), we calculated the Pearson correlation coefficients for each protein. To further explore the relationship between protein expression and other covariates, linear regression analyses were conducted using the RStudio software. Statistical significance was determined at p < 0.05, and the results were plotted as mean ± SEM (standard error of the mean).
Results
Comparing suicide and control samples
A reverse phase protein array (RPPA) was used to examine mitochondrial protein expression changes in individuals who had died of suicide. A total of 11 TCA cycle enzyme subunits were profiled in the parahippocampal (PHC) and comparison brain regions. The demographic information for the control and suicide groups is listed in Suppl. Table 1. The age, sex, and postmortem interval (PMI) covariates were compared between the control and the suicidal individuals using Welch’s unequal variances t-test or Chi-Square test. No significant differences were found between the two groups for the age and sex covariates. However, a significant difference was observed between the controls and suicide samples regarding the PMI covariate (Supplementary Table 1).
Characterizing the expression levels of selected TCA cycle enzymes in people who died by suicide
The expression values for the 11 TCA cycle enzyme subunits were compared across 22 cortical samples to identify differentially expressed proteins between the control and suicide groups. Two of the validated antibodies can recognize transcript variants. The anti-oxoglutarate dehydrogenase (OGDH) antibody (Proteintech, cat. no. 15212-1-AP) recognizes two transcripts, OGDH isoform 1 and isoform 2. Another anti-OGDH antibody (Sigma-Aldrich, cat. no. HPA020347) recognizes the OGDH isoform 3. Significant changes were observed between groups as the expression levels of dihydrolipoamide dehydrogenase (DLD), OGDH isoforms 1 and 2, succinate dehydrogenase B (SDHB), succinyl-CoA ligase ATP-forming subunit beta (SUCLA2), and succinyl-CoA ligase GTP-forming subunit beta (SUCLG2) enzymes were upregulated in the PHC of suicidal individuals (Fig. 2A). In contrast, no significant differences were observed between the suicide and control groups in the EC for any of the examined proteins (Fig. 2B). The mRNA levels for each of the 5 significantly altered proteins were analyzed in the PHC of both groups. The quantitative PCR analyses identified three significantly altered genes, thus, we verified an increased expression of the OGDH isoforms 1 and 2, SDHB, and SUCLG2, whose expression level alterations confirmed their protein expression (Fig. 2C). The mRNA expression of SDHC was also significantly increased in the PHC, despite the protein level not changing significantly. However, the p-value of its expression level was less than 0.1 (p = 0.056) (Fig. 2C). Although the mRNA expression of DLD and SUCLA2 was higher in people who died by suicide than in control subjects, the observed differences were not statistically significant (not shown; SUCLA2: p = 0.109, DLD: p = 0.233).
A Bar graphs show representative examples of 5 differentially expressed subunits of the TCA cycle in the PHC. B At the same time, there were no significant changes in the protein expression of these subunits in the EC. Data are based on 10 control and 10 suicide individuals for PHC, and 7 control and 5 suicide individuals for EC as indicated in Table 3. Protein expression was analyzed using two-way ANOVA followed by Sidak’s multiple comparisons tests. C The mRNA expression analysis of the altered TCA enzyme subunit DLD, OGDH isoforms 1 and 2, SDHB, SUCLA2, and SUCLG2 showed that gene expression of OGDH isoforms 1 and 2, SDHB and SUCLG2 were significantly elevated in the PHC as expected. Data are based on 9 control and 9 suicide individuals for PHC, and the gene expression was analyzed using Welch’s unequal variances t-test. Bar graphs represent mean ± SEM (*p < 0.05, **p < 0.01, ***p < 0.001). EC entorhinal cortex, PHC parahippocampal cortex.
Protein expression changes of non-mitochondrial proteins in people who died by suicide
Based on the previous results, changes in both mRNA and protein levels of the selected TCA cycle enzyme subunits are prominent in the PHC. Therefore, we sought to determine whether changes in the levels of some non-mitochondrial proteins, which do not play a role in mitochondrial processes, occur in this region. To this end, three proteins were selected for analysis: the epidermal growth factor receptor (EGFR), the Notch receptor 1 (NOTCH1), and the G protein subunit alpha q (GNAQ). These proteins have been demonstrated to play a role in cell signaling processes [80,81,82]. The protein levels of these three genes did not differ between the control and suicide groups, either in the PHC or the EC (Fig. 3A, B).
A and B For these proteins, statistical analyses showed no differences between the control and suicide groups in the PHC (A) and EC (B) (p < 0.05). Data are based on 10 control and 10 suicide individuals for PHC, and 7 control and 5 suicide individuals for EC. Protein expression was analyzed using two-way ANOVA followed by Sidak’s multiple comparisons tests. Error bars represent the SEM. EC entorhinal cortex, EGFR epidermal growth factor receptor, GNAQ G protein subunit alpha q, NOTCH1 Notch receptor 1, PHC parahippocampal cortex.
Characterization of the protein expression of selected TCA cycle enzyme subunits in regions belonging to the default mode and salience networks
Studies have shown that individuals with depression exhibit alterations in DMN and salience network (SN) function, which may contribute to negative rumination and difficulties in regulating emotions, and an increased sensitivity to stressors [83, 84]. Similarly, alterations in the interaction between the DMN and the SN have been observed in individuals who have attempted suicide [68, 85]. To ascertain whether the altered expression of TCA cycle enzymes occurs in cortical brain regions belonging to the DMN and SN, we conducted a comparative analysis of the expression levels of all 11 examined subunits in the DMPFC, ACC, and insular cortices between the control and suicide groups. Our findings revealed no significant alteration between the two groups for the examined proteins (Fig. 4).
A Bar graphs show that the expression levels of the investigated proteins did not change in the DMPFC of suicide individuals compared to control subjects. B and C Similarly, there were no differences in the expression levels of the examined enzyme subunits in the ACC (B) and insular cortex (C) between the suicide and control groups. Protein expression was analyzed using two-way ANOVA followed by Sidak’s multiple comparisons tests. Error bars represent the SEM (control group for DMPFC: n = 6; suicide group for DMPFC: n = 10; control group for ACC: n = 10; suicide group for ACC: n = 8; control group for insular cortex: n = 10; suicide group for insular cortex: n = 9).
Analysis of the PMI-dependent protein expression of the PHC and EC samples
To ascertain whether the difference in PMI could influence protein expression, principal component analysis (PCA) was performed (Supplementary Fig. 1). This method enables the identification of clusters comprising samples exhibiting similar expressional features. The majority of the variance observed (58%, PC1) across the PHC samples can be attributed to the differences between the control and suicide groups, as demonstrated by their distinct clustering (Supplementary Fig. 1A). However, there were minor overlaps, with one control and one suicide sample showing similarities to the opposite cluster. Based on the plot, it can be concluded that PMI does not significantly affect protein expression in the PHC samples. We also performed PCA on the EC samples (Supplementary Fig. 1B), which further supports the same conclusion.
To further validate the impact of PMI on our findings, we performed Pearson correlation and linear regression analyses on PMI-dependent protein expression for all the examined TCA cycle enzyme subunits, as well as on the three examined non-mitochondrial proteins in the PHC and EC samples. The results indicate that PMI did not affect the expression levels of the examined proteins and genes (Supplementary Table 2).
The distribution of mRNA expression of OGDH in the human parahippocampal cortex
To investigate the cell-type-specific expression of one of the significantly upregulated TCA cycle enzymes, OGDH isoforms 1 and 2, we performed ISH combined with immunolabeling for glia markers in the PHC. ISH histochemistry revealed the distribution of the mRNA levels of OGDH (Fig. 5A–C). In the PHC, all layers contained OGDH mRNA. In sections labeled for OGDH mRNA, both S100-immunoreactive and Iba1-immunoreactive cells were visualized (Fig. 5D, E). Cells expressing high levels of OGDH mRNA did not co-localize with S100b and Iba1 immunoreactivity. Conversely, cells containing S100 and Iba1 exhibited only low levels of OGDH mRNA. The average background-corrected distribution of OGDH mRNA over non-glial cells was 16.69 ± 0.67%, indicating strong labeling. In contrast, the average corrected distribution of OGDH mRNA over S100-immunoreactive cells was 3.83 ± 0.39%, and over Iba1-immunoreactive cells it was 2.29 ± 0.4%. These values suggest that the ISH signals in glial cells are slightly higher than in the background, indicating weak OGDH expression. Details of the validation of ISH specificity and control for DAB staining are shown in Supplementary Fig. 2, and a quantitative analysis of OGDH ISH signal intensity across different cell types is shown in Supplementary Fig. 3.
OGDH mRNA expression in the PHC of a control individual was visualized by ISH histochemistry. A and B Bright-field photomicrographs show the high intensity of OGDH isoforms 1 and 2 hybridization signals in the PHC. C A high-magnification bright-field microphotograph shows the expression of OGDH. D and E High-magnification pictures demonstrate the location of autoradiography grains representing OGDH mRNA in relation to S100- (D) and Iba1-immunoreactive glial cells (E). White arrowheads indicate the OGDH mRNA-expressing cells. In contrast, S100-immunoreactive astrocytes indicated by black arrowheads in D are not labeled for OGDH mRNA. Also, mRNA expression of OGDH is absent in Iba1-immunoreactive microglia (black arrowheads) in (E). Scale bars are 500 μm for A, 100 μm for B, D, E, and 12.5 μm for (C).
Discussion
The primary finding of the study is the increase in the level of mitochondrial proteins in the parahippocampal cortex (PHC) of people who died by suicide, which participate in the oxidative decarboxylation branch of glutamine catabolism. Samples from the postmortem brains of 12 control and 10 suicidal individuals were used to establish this finding (Supplementary Table 1). However, not all brain regions were available from all patients throughout. It is noteworthy that RPPA data in the PHC were obtained from 10 control and 10 suicidal individuals. The number of samples in comparison brain regions contained a smaller number of brains, which, however, consistently exceeded a minimal brain number of n = 5 per group. In these brain regions, no changes or marked differences were observed in the level of the same mitochondrial proteins, which suggests that the alterations are specific to the PHC. Given that the neighboring PHC and EC samples were derived from the same side of identical brains, it is reasonable to exclude any potential methodological bias due to differences in brain tissue or substance preservation or local effects (e.g. acidification, freezing time, etc.).
Another methodological consideration is whether differences between patients, aside from suicidal behavior, play a role in the observed changes. In that regard, it should be noted that clinical and pathological diagnosis was available for 9 control subjects and 9 people who died by suicide. In the control group, acute cardiac insufficiency was the most frequent cause of death. While cardiac as well as respiratory insufficiency can lead to hypoxia, brain tissue hypoperfusion also occurred in 7 of the suicidal individuals whose cause of death was by hanging (Suppl. Table 1), even though in the latter cases, the degree and extent of the hypoxia could not be determined. Despite the fact that the number of samples belonging to these subgroups is insufficient for statistical analysis, it is unlikely that the observed differences in protein levels were influenced by these factors instead of suicidal behavior. It is also noteworthy that none of the suicidal individuals in our study had a previous diagnosis of depression. While this statement does not exclude the possibility that some of them suffered from depressive disorder, it is rather unlikely that they had received antidepressant medication, an important factor in altering gene and protein expression.
It is argued that the results and data interpretation were not affected by sex and age factors. Males and females were included in both control and suicidal groups with almost the same ratio, and the mean age of both groups was similar. However, the postmortem delay was higher in the suicide group. While previous analysis using postmortem samples stored at 4 °C did not identify protein decay within 24 h [86], the maximum PMI in the present study reached 10 h. Therefore, we addressed this question statistically. A principal component analysis showed clusters of the PHC samples based on which group they belong to but not based on PMI (Supplementary Fig. 1). In addition, a linear regression analysis of PHC and EC samples did not show a correlation with PMI, thereby refuting the hypothesis that PMI plays a role in protein level differences between the 2 groups in the PHC. Finally, the lack of difference between the protein levels in other than parahippocampal brain areas also implies that PMI is not responsible for the observed differences in the PHC. Protein levels of 5 subunits differed among the 11 examined mitochondrial TCA cycle enzyme subunits in the PHC (Fig. 2A). Protein levels of the examined subunits were compared in the adjacent EC samples derived from the intermediate subfield of the EC. Although the entorhinal and parahippocampal cortices are two adjacent and anatomically and functionally interconnected brain regions [87, 88], we found no alteration in protein expression of EC samples for the examined proteins between the control and suicide groups (Fig. 2B). Mitochondrial alterations were also not found in other brain regions belonging to the DMN and SN, which are known to be involved in depression and suicide, such as the DMPFC, ACC, and insular cortex. It is noteworthy that other researchers have observed altered functional connectivity between the DMN and PHC in chronic depression and suicidal behavior [34, 89,90,91]. In light of these observations, our protein expression results suggest a specific role of the PHC in suicide.
In order to ascertain whether transcriptional events could be responsible for the increased protein levels, the mRNA levels of these proteins were also investigated (Fig. 2C). Three out of five altered proteins demonstrated elevated mRNA levels suggesting the involvement of transcriptional regulation. Thus, mRNA measurements and RRPA data analysis, as independent techniques, highlight and validate the identified changes in the PHC. While a relatively large percentage of mitochondrial protein expression was changed in the PHC, none of the examined non-mitochondrial proteins, EGFR, NOTCH1, and GNAQ, exhibited any differences among groups for the same brain area (Fig. 3A, B). Given that the non-mitochondrial proteins under investigation are involved in common metabolic signaling processes [80,81,82], their unaltered levels between the control and suicide groups suggest that altered processes involving mitochondrial rather than cytosolic metabolism are more prominent in the PHC in relation to the pathophysiology of suicidal behavior.
Studies have shown that depression is associated with mitochondrial damage, disrupted electron transport, increased oxidative stress, neuroinflammation, and apoptosis, leading to impaired neuroplasticity and neurogenesis [44, 92,93,94,95,96]. Maintaining mitochondrial quality control is essential for cellular function, and mitochondrial homeostasis is emerging as a potential therapeutic target for mood disorders. More recently, Punzi et al. [47] considered that suicidal completion by violent methods may be associated with elevated mitochondrial metabolism in the dorsolateral prefrontal cortex (DLPFC). Given that the majority of people who died by suicide included in our study (8 out of 10) died by violent methods, in line with the observations presented by Punzi and colleagues, our findings confirm the activation of mitochondrial metabolism in the PHC. Research has consistently demonstrated that gender plays a significant role in determining the mode of death among people who die by suicide. Specifically, males are more likely to choose violent methods such as firearms or hanging, whereas females tend to opt for less violent means such as drug overdose or self-poisoning [97]. These sex-specific patterns in suicide methods may stem from various factors, including biological differences, societal norms, and access to means [98, 99]. However, it is important to acknowledge the potential influence of the predominantly male composition of our study cohort. While our findings align with existing literature, future research should consider including a more balanced representation of both sexes to ensure robust conclusions regarding the influence of gender on suicide behavior.
Our data allowed more specific conclusions to be drawn since the altered mitochondrial proteins catalyze specific reactions belonging to the TCA cycle. Among these, the ketoglutarate dehydrogenase complex (KGDHC), which contains OGDH and DLD subunits, catalyzing the irreversible decarboxylation of α-ketoglutarate to succinyl-CoA while the SUCLA2 and the SUCLG2 are necessary for converting succinyl-CoA to succinate (Fig. 6). Currently, there is a paucity of data regarding the impact of alterations in these enzymes on psychiatric disorders. Mass spectrometric proteomic studies have demonstrated that OGDH is downregulated in the prefrontal cortex in a rat model of depression [100] and in the DLPFC of MDD patients [26]. Furthermore, expression studies have identified reduced gene expression levels of SUCLA2 in the prefrontal cortex of depressed individuals [101] and increased protein expression of SDHB in rats exhibiting high-anxiety-like behavior [102]. Recently, Linghu et al. reported that depressive rats caused by chronic, unpredictable mild stress show a significantly decreased TCA cycle based on a stable isotope-resolved metabolomics study [97]. In turn, a genome-wide association study (GWAS) has reported that a specific variation of DLD was associated with MDD [37]. Another GWAS study has indicated that genetic variation in SUCLA2 is associated with an increased likelihood of fatal suicide behavior [103]. Our study builds on the examination of individuals who died by suicide but had no diagnosed history of mental illness. The upregulation of certain enzymes in our study contrasts with the downregulation observed in MDD patients, suggesting different or compensatory mechanisms might be at play in the absence of a diagnosed mental illness. Understanding these differences could provide insight into the metabolic dysregulation associated with suicidal behavior, independent of psychiatric diagnoses. It highlights the need for further research to delineate whether these enzymatic changes are a cause or consequence of suicide and how they interact with other biological and environmental factors. Within the TCA cycle, genes with altered expression levels in people who died by suicide are all involved in the conversion of α-ketoglutarate to fumarate through the concerted action of KGDHC, SUCL, and SDH (Fig. 6), which is part of the oxidative decarboxylation pathway of glutaminolysis. Recent research has shown that fumarate can affect cellular function in neurons through a variety of mechanisms [104, 105]. The evidence currently available suggests that fumarate can directly release mitochondrial DNA (mtDNA) through the activation of the TCA cycle in cells [106, 107] and influence neuronal functions [108]. However, the precise mechanisms by which fumarate may impact mtDNA release and neuronal function remain to be discovered. What is known is, that disruptions in mitochondrial function and mtDNA integrity can contribute to the development of neurological disorders [109], and fumarate has been shown to play a role in various cellular processes related to neuronal function, such as oxidative stress and inflammation [110, 111]. Although fumarate has been demonstrated to exert a pivotal role in cellular metabolism and signaling, further research is necessary to elucidate the intricate interactions between fumarate, the TCA cycle, mtDNA, and neuronal functioning.
The figure was adapted from Fig. 1 of Dhami et al. [124] and modified. The genes in colors under boxes are either isozymes part of the enzyme complex or are subunits catalyzing the TCA reaction. The significantly upregulated TCA cycle enzyme subunits are indicated with red color, while the significantly not altered subunits are indicated with blue color. ACO2 aconitase 2, CS citrate synthase, DLAT dihydrolipoamide S-acetyl transferase, DLD dihydrolipoamide dehydrogenase, DLST dihydrolipoamide succinyltransferase, FH fumarate hydratase, GLS glutaminase, GLUD glutamate dehydrogenase, IDH isocitrate dehydrogenase, FADH2 flavin adenine dinucleotide (hydroquinone form), MDH malate dehydrogenase, NADH nicotinamide adenine dinucleotide reduced form, OGDH oxoglutarate dehydrogenase, PDC pyruvate dehydrogenase complex, PDHA1 pyruvate dehydrogenase alpha, SCS succinate CoA-synthase and SDH succinyl dehydrogenase.
Glutaminolysis is an adjuvant/alternative pathway for energy production that converts glutamine to glutamate and then to α-ketoglutarate [112], which eventually follows mitochondrial substrate-level phosphorylation [113, 114]. It has been demonstrated that energy production by the TCA cycle is altered in depression [115]. The present data suggest that subunits of all major TCA enzymes belonging to glutaminolysis were upregulated in the PHC of people who died by suicide, implying the potentiation of glutaminolysis in suicidal behavior. It is plausible that increased glutaminolysis occurs predominantly in neurons rather than glial cells in the PHC. This is because the TCA cycle is less active in glial cells than in neurons, considering that some of the enzyme subunits exhibiting increased expression in people who died by suicide, such as OGDH, SUCLA2, SUCLG2, SDHB, and SDHC, are primarily expressed in neurons rather than glial cells, especially in the human cerebral cortex [75, 77, 116]. To confirm this hypothesis, we conducted ISH in the PHC targeting OGDH mRNA combined with immunolabeling for astrocyte marker S100 and microglia marker Iba1. Based on our findings, double mRNA expression was detected more in glial cells than in neurons, which is consistent with our previous double immunolabeling study [75]. It should be noted that our study did not specifically quantify OGDH mRNA expression or use neuronal markers to explicitly demonstrate neuronal localization. Nevertheless, the observed pattern of OGDH labeling in the parahippocampal cortex aligns with findings from a recent quantitative study in the human neocortex and hippocampal formation, which demonstrated that mitochondrial enzymes involved in oxidative phosphorylation are predominantly localized in neurons [116]. This alignment supports the inference that OGDH, which is essential for metabolic processes in neurons, exhibits a similar distribution in the PHC. Since the ISH was performed in a control subject, we do not know if changes in the cellular distribution of OGDH and other mitochondrial enzymes changed in people who died by suicide. Glutaminolysis is not active in glial cells because they transport glutamine to the neurons rather than using it as an energy source. In turn, the major source of glutamine in neurons arrives from astrocytes via the glutamate-glutamine shuttle [117]. The astrocyte-derived glutamine can be used by the neurons to produce energy via the TCA cycle or to produce glutamate for neurotransmission. The data do not provide information regarding the amount of glutamate used as a neurotransmitter in suicidal individuals. However, the increased glutaminolysis implies increased energy production, which typically serves to maintain increased neuronal activity. Previous studies have reported that a positive BOLD response reflects increased energy production during activation [118,119,120]. Consequently, the augmented TCA cycle activity in the PHC of suicidal individuals is consistent with the findings of fMRI studies reporting elevated BOLD responses to the PHC of suicide attempters [121,122,123].
Conclusion
The data point towards altered glutaminolysis in the PHC of people who died by suicide. Thereby, a molecular component potentially fueling increased neuronal activity in this brain area was identified. Additionally, the observed molecular changes in the PHC also suggest that the cause of suicidal behavior may originate in this brain region through molecular dysfunctions. The PHC may then potentially affect other regions within the default mode network through its increased activity, which could contribute to suicidal behavior.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
References
Schmaal L, van Harmelen AL, Chatzi V, Lippard ETC, Toenders YJ, Averill LA, et al. Imaging suicidal thoughts and behaviors: a comprehensive review of 2 decades of neuroimaging studies. Mol Psychiatry. 2020;25:408–27.
Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. PNAS. 2001;98:676–82.
Buckner RL, Andrews-Hanna JR, Schacter DL. The brain’s default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38.
Sheline YI, Barch DM, Price JL, Rundle MM, Vaishnavi SN, Snyder AZ, et al. The default mode network and self-referential processes in depression. Proc Natl Acad Sci USA. 2009;106:1942–7.
Maresh EL, Allen JP, Coan JA. Increased default mode network activity in socially anxious individuals during reward processing. Biol Mood Anxiety Disord. 2014;4:7.
Andrews-Hanna JR. The brain’s default network and its adaptive role in internal mentation. Neuroscientist. 2012;18:251–70.
Dixon ML, Andrews-Hanna JR, Spreng RN, Irving ZC, Mills C, Girn M, et al. Interactions between the default network and dorsal attention network vary across default subsystems, time, and cognitive states. NeuroImage. 2017;147:632–49.
Figley TD, Bhullar N, Courtney SM, Figley CR. Probabilistic atlases of default mode, executive control and salience network white matter tracts: an fMRI-guided diffusion tensor imaging and tractography study. Front Hum Neurosci. 2015;9:585.
Andrews-Hanna JR, Smallwood J, Spreng RN. The default network and self-generated thought: component processes, dynamic control, and clinical relevance. Ann N Y Acad Sci. 2014;1316:29–52.
Squire LR, Stark CE, Clark RE. The medial temporal lobe. Annu Rev Neurosci 2004;27:279–306.
Bird CM, Burgess N. The hippocampus and memory: insights from spatial processing. Nat Rev Neurosci. 2008;9:182–94.
Raslau FD, Mark IT, Klein AP, Ulmer JL, Mathews V, Mark LP. Memory part 2: the role of the medial temporal lobe. AJNR Am J Neuroradiol. 2015;36:846–9.
Aminoff EM, Kveraga K, Bar M. The role of the parahippocampal cortex in cognition. Trends Cogn Sci. 2013;17:379–90.
Price G, Cercignani M, Parker GJ, Altmann DR, Barnes TR, Barker GJ, et al. White matter tracts in first-episode psychosis: a DTI tractography study of the uncinate fasciculus. Neuroimage. 2008;39:949–55.
Qin S, Duan X, Supekar K, Chen H, Chen T, Menon V. Large-scale intrinsic functional network organization along the long axis of the human medial temporal lobe. Brain Struct Funct. 2016;221:3237–58.
Seoane S, Modroño C, González-Mora JL, Janssen N. Medial temporal lobe contributions to resting-state networks. Brain Struct Funct. 2022;227:995–1012.
Ward AM, Schultz AP, Huijbers W, Van Dijk KR, Hedden T, Sperling RA. The parahippocampal gyrus links the default-mode cortical network with the medial temporal lobe memory system. Hum Brain Mapp. 2014;35:1061–73.
Andrews-Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL. Functional-anatomic fractionation of the brain’s default network. Neuron. 2010;65:550–62.
Xu X, Yuan H, Lei X. Activation and connectivity within the Default Mode Network contribute independently to future-oriented thought. Sci Rep. 2016;6:21001.
Drevets WC. Functional anatomical abnormalities in limbic and prefrontal cortical structures in major depression. Prog Brain Res. 2000;126:413–31.
Sheline YI. Neuroimaging studies of mood disorder effects on the brain. Biol Psychiatry. 2003;54:338–52.
Sheline YI, Price JL, Yan Z, Mintun MA. Resting-state functional MRI in depression unmasks increased connectivity between networks via the dorsal nexus. Proc Natl Acad Sci USA. 2010;107:11020–5.
Cao J, Chen J-m, Kuang L, Ai M, Fang W-d, Gan Y, et al. Abnormal regional homogeneity in young adult suicide attempters with no diagnosable psychiatric disorder: a resting state functional magnetic imaging study. Psychiatry Res: Neuroimaging. 2015;231:95–102.
Kang SG, Na KS, Choi JW, Kim JH, Son YD, Lee YJ. Resting-state functional connectivity of the amygdala in suicide attempters with major depressive disorder. Prog Neuro-Psychopharmacol Biol Psychiatry. 2017;77:222–7.
Zhang S, Chen J-m, Kuang L, Cao J, Zhang H, Ai M, et al. Association between abnormal default mode network activity and suicidality in depressed adolescents. BMC Psychiatry. 2016;16:337.
Jin W, Jie F, Wenwei Z, Bin Z, Chen S, Wei S, et al. The medial temporal lobe structure and function support positive affect. Neuropsychologia. 2022;176:108373.
Tyng CM, Amin HU, Saad MN, Malik AS. The influences of emotion on learning and memory. Front Psychol. 2017;8:1454.
Gosselin N, Samson S, Adolphs R, Noulhiane M, Roy M, Hasboun D, et al. Emotional responses to unpleasant music correlates with damage to the parahippocampal cortex. Brain. 2006;129:2585–92.
Bobilev AM, Perez JM, Tamminga CA. Molecular alterations in the medial temporal lobe in schizophrenia. Schizophr Res. 2020;217:71–85.
Chen X, Chen N-X, Shen Y-Q, Li H-X, Li L, Lu B, et al. The subsystem mechanism of default mode network underlying rumination: a reproducible neuroimaging study. NeuroImage. 2020;221:117185.
Just MA, Pan L, Cherkassky VL, McMakin DL, Cha C, Nock MK, et al. Machine learning of neural representations of suicide and emotion concepts identifies suicidal youth. Nat Hum Behav. 2017;1:911–9.
Reisch T, Seifritz E, Esposito F, Wiest R, Valach L, Michel K. An fMRI study on mental pain and suicidal behavior. J Affect Disord. 2010;126:321–5.
Vanyukov PM, Szanto K, Hallquist MN, Siegle GJ, Reynolds CF 3rd, Forman SD, et al. Paralimbic and lateral prefrontal encoding of reward value during intertemporal choice in attempted suicide. Psychol Med. 2016;46:381–91.
Zamoscik V, Huffziger S, Ebner-Priemer U, Kuehner C, Kirsch P. Increased involvement of the parahippocampal gyri in a sad mood predicts future depressive symptoms. Soc Cogn Affect Neurosci. 2014;9:2034–40.
Cabrera B, Monroy-Jaramillo N, Fries GR, Mendoza-Morales RC, García-Dolores F, Mendoza-Larios A, et al. Brain gene expression pattern of subjects with completed suicide and comorbid substance use disorder. Complex Psychiatry. 2019;5:60–73.
Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2004;14:241–53.
Giannakopoulou O, Lin K, Meng X, Su M-H, Kuo P-H, Peterson RE, et al. The genetic architecture of depression in individuals of East Asian ancestry: a Genome-Wide Association Study. JAMA Psychiatry. 2021;78:1258–69.
Pantazatos SP, Huang Y, Rosoklija GB, Dwork AJ, Arango V, Mann JJ. Whole-transcriptome brain expression and exon-usage profiling in major depression and suicide: evidence for altered glial, endothelial and ATPase activity. Mol Psychiatry. 2017;22:760–73.
Romero-Pimentel AL, Almeida D, Muñoz-Montero S, Rangel C, Mendoza-Morales R, Gonzalez-Saenz EE, et al. Integrative DNA methylation and gene expression analysis in the prefrontal cortex of Mexicans who died by suicide. Int J Neuropsychopharmacol. 2021;24:935–47.
Rothman DL, de Graaf RA, Hyder F, Mason GF, Behar KL, De Feyter HM. In vivo (13)C and (1) H-[(13)C] MRS studies of neuroenergetics and neurotransmitter cycling, applications to neurological and psychiatric disease and brain cancer. NMR Biomed. 2019;32:e4172.
Henkel ND, Wu X, O’Donovan SM, Devine EA, Jiron JM, Rowland LM, et al. Schizophrenia: a disorder of broken brain bioenergetics. Mol Psychiatry. 2022;27:2393–404.
Cheng A, Hou Y, Mattson MP. Mitochondria and neuroplasticity. ASN Neuro. 2010;2:e00045.
Rezin GT, Amboni G, Zugno AI, Quevedo J, Streck EL. Mitochondrial dysfunction and psychiatric disorders. Neurochem Res. 2009;34:1021–9.
Bansal Y, Kuhad A. Mitochondrial dysfunction in depression. Current Neuropharmacol. 2016;14:610–8.
Allen J, Romay-Tallon R, Brymer KJ, Caruncho HJ, Kalynchuk LE. Mitochondria and mood: mitochondrial dysfunction as a key player in the manifestation of depression. Front Neurosci. 2018;12:386.
Czarny P, Wigner P, Galecki P, Sliwinski T. The interplay between inflammation, oxidative stress, DNA damage, DNA repair and mitochondrial dysfunction in depression. Prog Neuro-Psychopharmacol Biol Psychiatry. 2018;80:309–21.
Punzi G, Ursini G, Chen Q, Radulescu E, Tao R, Huuki LA, et al. Genetics and brain transcriptomics of completed suicide. Am J Psychiatry. 2022;179:226–41.
Gu X, Ke S, Wang Q, Zhuang T, Xia C, Xu Y, et al. Energy metabolism in major depressive disorder: Recent advances from omics technologies and imaging. Biomed Pharmacother. 2021;141:111869.
Xie X, Shen Q, Yu C, Xiao Q, Zhou J, Xiong Z, et al. Depression-like behaviors are accompanied by disrupted mitochondrial energy metabolism in chronic corticosterone-induced mice. J Steroid Biochem Mol Biol. 2020;200:105607.
Sarawagi A, Soni ND, Patel AB. Glutamate and GABA homeostasis and neurometabolism in major depressive disorder. Front Psychiatry. 2021;12:637863.
Karnecki K, Świerczyński J, Steiner J, Krzyżanowska M, Kaliszan M, Gos T, The left-lateralisation of citrate synthase activity in the anterior cingulate cortex of male violent suicide victims. Eur Arch Psychiatry Clin Neurosci 2022.
Rose J, Brian C, Pappa A, Panayiotidis MI, Franco R. Mitochondrial metabolism in astrocytes regulates brain bioenergetics, neurotransmission and redox balance. Front Neurosci. 2020;14:536682.
Clay HB, Sillivan S, Konradi C. Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neurosci. 2011;29:311–24.
Lin MM, Liu N, Qin ZH, Wang Y. Mitochondrial-derived damage-associated molecular patterns amplify neuroinflammation in neurodegenerative diseases. Acta Pharmacol Sin. 2022;43:2439–47.
Dantzer R. Neuroimmune interactions: from the brain to the immune system and vice versa. Physiol Rev. 2018;98:477–504.
Song Y, Cao H, Zuo C, Gu Z, Huang Y, Miao J, et al. Mitochondrial dysfunction: a fatal blow in depression. Biomed Pharmacother. 2023;167:115652.
Chan SY, Probert F, Radford-Smith DE, Hebert JC, Claridge TDW, Anthony DC, et al. Post-inflammatory behavioural despair in male mice is associated with reduced cortical glutamate-glutamine ratios, and circulating lipid and energy metabolites. Sci Rep. 2020;10:16857.
Gottschalk MG, Wesseling H, Guest PC, Bahn S. Proteomic enrichment analysis of psychotic and affective disorders reveals common signatures in presynaptic glutamatergic signaling and energy metabolism. Int J Neuropsychopharmacol. 2014;18:pyu019.
Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, et al. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–85.
Jia Y, Wang X, Chen Y, Qiu W, Ge W, Ma C. Proteomic and transcriptomic analyses reveal pathological changes in the entorhinal cortex region that correlate well with dysregulation of ion transport in patients with Alzheimer’s disease. Mol Neurobiol. 2021;58:4007–27.
Mendonça CF, Kuras M, Nogueira FCS, Plá I, Hortobágyi T, Csiba L, et al. Proteomic signatures of brain regions affected by tau pathology in early and late stages of Alzheimer’s disease. Neurobiol Disease. 2019;130:104509.
Wang W-Y, Liu Y, Wang H-F, Tan L, Sun F-R, Tan M-S, et al. Impacts of CD33 genetic variations on the atrophy rates of hippocampus and parahippocampal gyrus in normal aging and mild cognitive impairment. Mol Neurobiol. 2017;54:1111–8.
Wang Z, Li W, Chen J, Shi H, Zhao M, You H, et al. Proteomic analysis reveals energy metabolic dysfunction and neurogenesis in the prefrontal cortex of a lipopolysaccharide-induced mouse model of depression. Mol Med Rep. 2016;13:1813–20.
Palmfeldt J, Henningsen K, Eriksen SA, Müller HK, Wiborg O. Protein biomarkers of susceptibility and resilience to stress in a rat model of depression. Mol Cell Neurosci. 2016;74:87–95.
Rahman SU, Hao Q, He K, Li Y, Yang X, Ye T, et al. Proteomic study reveals the involvement of energy metabolism in the fast antidepressant effect of (2R, 6R)-hydroxy norketamine. Proteomics Clin Appl. 2020;14:e1900094.
Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–21.
Fan T, Wu X, Yao L, Dong J. Abnormal baseline brain activity in suicidal and non-suicidal patients with major depressive disorder. Neurosci Lett. 2013;534:35–40.
Ho TC, Walker JC, Teresi GI, Kulla A, Kirshenbaum JS, Gifuni AJ, et al. Default mode and salience network alterations in suicidal and non-suicidal self-injurious thoughts and behaviors in adolescents with depression. Transl Psychiatry. 2021;11:1–14.
Sun J, Du Z, Ma Y, Chen L, Wang Z, Guo C, et al. Altered functional connectivity in first-episode and recurrent depression: a resting-state functional magnetic resonance imaging study. Front Neurol. 2022;13:922207.
Mai JK, Majtanik M, and Paxinos G, Atlas of the Human Brain. 4th ed. 2016: Elsevier.
Palkovits M. Microdissection of individual brain nuclei and areas. In: Boulton AA, Baker GB, editors. General neurochemical techniques. Totowa, NJ: Humana Press; 1985. p. 1–17.
Insausti R, Córcoles-Parada M, Ubero MM, Rodado A, Insausti AM, Muñoz-López M. Cytoarchitectonic areas of the gyrus ambiens in the human brain. Front Neuroanat. 2019;13:21.
Insausti R, Muñoz-López M, Insausti AM, Artacho-Pérula E. The human periallocortex: layer pattern in presubiculum, parasubiculum and entorhinal cortex. A review. Front Neuroanat. 2017;11:84.
Byron A, Bernhardt S, Ouine B, Cartier A, Macleod KG, Carragher NO, et al. Integrative analysis of multi-platform reverse-phase protein array data for the pharmacodynamic assessment of response to targeted therapies. Sci Rep. 2020;10:21985.
Dobolyi A, Bago A, Palkovits M, Nemeria NS, Jordan F, Doczi J, et al. Exclusive neuronal detection of KGDHC-specific subunits in the adult human brain cortex despite pancellular protein lysine succinylation. Brain Struct Funct. 2020;225:639–67.
Dora F, Renner E, Keller D, Palkovits M, Dobolyi A. Transcriptome profiling of the dorsomedial prefrontal cortex in suicide victims. Int J Mol Sci. 2022;23:7067.
Dobolyi A, Ostergaard E, Bagó AG, Dóczi T, Palkovits M, Gál A, et al. Exclusive neuronal expression of SUCLA2 in the human brain. Brain Struct Funct. 2015;220:135–51.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5.
Tang Y, Horikoshi M, Li W. ggfortify: unified interface to visualize statistical results of popular R packages. R J. 2016;8:474.
Maner BS, Dupuis L, Su A, Jueng JJ, Harding TP, Meisenheimer J, et al. Overview of genetic signaling pathway interactions within cutaneous malignancies. J Cancer Metastasis Treatment. 2020;6:37.
Porcelli L, Mazzotta A, Garofoli M, Di Fonte R, Guida G, Guida M, et al. Active notch protects MAPK activated melanoma cell lines from MEK inhibitor cobimetinib. Biomed Pharmacother. 2021;133:111006.
Amaro A, Mirisola V, Angelini G, Musso A, Tosetti F, Esposito AI, et al. Evidence of epidermal growth factor receptor expression in uveal melanoma: inhibition of epidermal growth factor-mediated signalling by Gefitinib and Cetuximab triggered antibody-dependent cellular cytotoxicity. Eur J Cancer. 2013;49:3353–65.
Scult MA, Fresco DM, Gunning FM, Liston C, Seeley SH, García E, et al. Changes in functional connectivity following treatment with emotion regulation therapy. Front Behav Neurosci. 2019;13:10.
Ho TC, Connolly CG, Henje Blom E, LeWinn KZ, Strigo IA, Paulus MP, et al. Emotion-dependent functional connectivity of the default mode network in adolescent depression. Biol Psychiatry. 2015;78:635–46.
Dai Z, Shao J, Zhou H, Chen Z, Zhang S, Wang H, et al. Disrupted fronto-parietal network and default-mode network gamma interactions distinguishing suicidal ideation and suicide attempt in depression. Prog Neuro-Psychopharmacol Biol Psychiatry. 2022;113:110475.
Ferrer I, Santpere G, Arzberger T, Bell J, Blanco R, Boluda S, et al. Brain protein preservation largely depends on the postmortem storage temperature: implications for study of proteins in human neurologic diseases and management of brain banks: a BrainNet Europe Study. J Neuropathol Exp Neurol. 2007;66:35–46.
Clark RE, Squire LR. Similarity in form and function of the hippocampus in rodents, monkeys, and humans. Proc Natl Acad Sci USA. 2013;110:10365–70.
Eichenbaum H, Lipton PA. Towards a functional organization of the medial temporal lobe memory system: role of the parahippocampal and medial entorhinal cortical areas. Hippocampus. 2008;18:1314–24.
Figueroa CA, Mocking RJT, van Wingen G, Martens S, Ruhé HG, Schene AH. Aberrant default-mode network-hippocampus connectivity after sad memory-recall in remitted-depression. Soc Cogn Affect Neurosci. 2017;12:1803–13.
Renner F, Siep N, Arntz A, van de Ven V, Peeters F, Quaedflieg C, et al. Negative mood-induction modulates default mode network resting-state functional connectivity in chronic depression. J Affect Disord. 2017;208:590–6.
Hwang J, Legarreta M, Bueler CE, DiMuzio J, McGlade E, Lyoo IK, et al. Increased efficiency of brain connectivity networks in veterans with suicide attempts. NeuroImage: Clinical. 2018;20:318–26.
Manji H, Kato T, Di Prospero NA, Ness S, Beal MF, Krams M, et al. Impaired mitochondrial function in psychiatric disorders. Nat Rev Neurosci. 2012;13:293–307.
Einat H, Yuan P, Manji HK. Increased anxiety-like behaviors and mitochondrial dysfunction in mice with targeted mutation of the Bcl-2 gene: further support for the involvement of mitochondrial function in anxiety disorders. Behav Brain Res. 2005;165:172–80.
Shao L, Martin MV, Watson SJ, Schatzberg A, Akil H, Myers RM, et al. Mitochondrial involvement in psychiatric disorders. Ann Med. 2008;40:281–95.
Javani G, Babri S, Farajdokht F, Ghaffari-Nasab A, Mohaddes G. Mitochondrial transplantation improves anxiety-and depression-like behaviors in aged stress-exposed rats. Mech Ageing Dev. 2022;202:111632.
Giménez-Palomo A, Dodd S, Anmella G, Carvalho AF, Scaini G, Quevedo J, et al. The role of mitochondria in mood disorders: from physiology to pathophysiology and to treatment. Front Psychiatry. 2021;12:977.
Linghu T, Zhao Y, Wu W, Gao Y, Tian J, Qin X. Novel targets for ameliorating energy metabolism disorders in depression through stable isotope-resolved metabolomics. Biochim Biophys Acta Bioenerg. 2022;1863:148578.
Milner A, Witt K, Maheen H, LaMontagne AD. Access to means of suicide, occupation and the risk of suicide: a national study over 12 years of coronial data. BMC Psychiatry. 2017;17:125.
McDermott R. Sex and death: gender differences in aggression and motivations for violence. Int Organ. 2015;69:753–75.
Yang Y, Yang D, Tang G, Zhou C, Cheng K, Zhou J, et al. Proteomics reveals energy and glutathione metabolic dysregulation in the prefrontal cortex of a rat model of depression. Neuroscience. 2013;247:191–200.
Kim S, Webster M. Correlation analysis between genome-wide expression profiles and cytoarchitectural abnormalities in the prefrontal cortex of psychiatric disorders. Mol Psychiatry. 2010;15:326–36.
Filiou MD, Zhang Y, Teplytska L, Reckow S, Gormanns P, Maccarrone G, et al. Proteomics and metabolomics analysis of a trait anxiety mouse model reveals divergent mitochondrial pathways. Biol Psychiatry. 2011;70:1074–82.
Coon H, Darlington TM, DiBlasi E, Callor WB, Ferris E, Fraser A, et al. Genome-wide significant regions in 43 Utah high-risk families implicate multiple genes involved in risk for completed suicide. Mol Psychiatry. 2020;25:3077–90.
Jové M, Pradas I, Mota-Martorell N, Cabré R, Ayala V, Ferrer I, et al. Succination of protein thiols in human brain aging. Front Aging Neurosci. 2020;12:52.
Riechers S-P, Mojsilovic-Petrovic J, Belton TB, Chakrabarty RP, Garjani M, Medvedeva V, et al. Neurons undergo pathogenic metabolic reprogramming in models of familial ALS. Mol Metab. 2022;60:101468.
Zecchini V, Paupe V, Herranz-Montoya I, Janssen J, Wortel IMN, Morris JL, et al. Fumarate induces vesicular release of mtDNA to drive innate immunity. Nature. 2023;615:499–506.
Hooftman A, Peace CG, Ryan DG, Day EA, Yang M, McGettrick AF, et al. Macrophage fumarate hydratase restrains mtRNA-mediated interferon production. Nature. 2023;615:490–8.
Nakamya MF, Sil S, Buch S, Hakami RM. Mitochondrial extracellular vesicles in CNS disorders: new frontiers in understanding the neurological disorders of the brain. Front Mol Biosci. 2022;9:840364.
Zhao Y, Liu B, Xu L, Yu S, Fu J, Wang J, et al. ROS-Induced mtDNA Release: the emerging messenger for communication between neurons and innate immune cells during neurodegenerative disorder progression. Antioxidants. 2021;10:1917.
Scuderi SA, Ardizzone A, Paterniti I, Esposito E, Campolo M. Antioxidant and anti-inflammatory effect of Nrf2 inducer dimethyl fumarate in neurodegenerative diseases. Antioxidants. 2020;9:630.
Campolo M, Casili G, Biundo F, Crupi R, Cordaro M, Cuzzocrea S, et al. The neuroprotective effect of dimethyl fumarate in an MPTP-mouse model of Parkinson’s disease: involvement of reactive oxygen species/nuclear factor-κB/nuclear transcription factor related to NF-E2. Antioxid Redox Signal. 2017;27:453–71.
Mohamed A, Deng X, Khuri FR, Owonikoko TK. Altered glutamine metabolism and therapeutic opportunities for lung cancer. Clin Lung Cancer. 2014;15:7–15.
Ravasz D, Bui D, Nazarian S, Pallag G, Karnok N, Roberts J, et al. Residual Complex I activity and amphidirectional Complex II operation support glutamate catabolism through mtSLP in anoxia. Sci Rep. 2024;14:1729.
Seyfried TN, Arismendi-Morillo G, Mukherjee P, Chinopoulos C. On the origin of ATP synthesis in cancer. iScience. 2020;23:101761.
Morris G, Berk M. The many roads to mitochondrial dysfunction in neuroimmune and neuropsychiatric disorders. BMC Med. 2015;13:68.
Dobolyi A, Cservenák M, Bagó AG, Chen C, Stepanova A, Paal K, et al. Cell-specific expression of key mitochondrial enzymes limits OXPHOS in astrocytes of the adult human neocortex and hippocampal formation. Commun Biol. 2024;7:1045.
Siracusa R, Fusco R, Cuzzocrea S. Astrocytes: role and functions in brain pathologies. Front Pharmacol. 2019;10:1114.
Mangia S, Giove F, DiNuzzo M. Metabolic pathways and activity-dependent modulation of glutamate concentration in the human brain. Neurochem Res. 2012;37:2554–61.
Bednařík P, Tkáč I, Giove F, DiNuzzo M, Deelchand DK, Emir UE, et al. Neurochemical and BOLD responses during neuronal activation measured in the human visual cortex at 7 Tesla. J Cereb Blood Flow Metab. 2015;35:601–10.
Martínez-Maestro M, Labadie C, Möller HE. Dynamic metabolic changes in human visual cortex in regions with positive and negative blood oxygenation level-dependent response. J Cerebr Blood Flow Metab. 2019;39:2295–307.
Ballard ED, Reed JL, Szczepanik J, Evans JW, Yarrington JS, Dickstein DP, et al. Functional imaging of the implicit association of the self with life and death. Suicide Life‐Threat Behav. 2019;49:1600–8.
Gosnell S, Fowler JC, Salas R. Classifying suicidal behavior with resting‐state functional connectivity and structural neuroimaging. Acta Psychiatr Scand. 2019;140:20–29.
Ballard ED, Gilbert JR, Fields JS, Nugent AC, Zarate CA Jr. Network changes in insula and amygdala connectivity accompany implicit suicidal associations. Front Psychiatry. 2020;11:577628.
Dhami N, Trivedi DK, Goodacre R, Mainwaring D, Humphreys DP. Mitochondrial aconitase is a key regulator of energy production for growth and protein expression in Chinese hamster ovary cells. Metabolomics. 2018;14:136.
Acknowledgements
We thank Magdolna Toronyay-Kasztner for managing the HBTB database. We appreciate the technical assistance of Nikolett Hanák, Szilvia Deák and Viktória Dellaszéga-Lábas in the project.
Funding
Grant support was provided by NAP2022-I-3/2022 and NAP2022-I-4/2022 (National Brain Program 3.0 of the Hungarian Academy of Sciences), NKFIH OTKA K146077, and Excellence 151425, and 2024-1.2.3-HU-RIZONT research grants of the National Research, Developmental and Innovation Office, CELSA/24/020 to AD, the NKFIH ([TKP2021-EGA-25], VEKOP 2.3.3-15-2016-00012, and K135027) grants to CC, EFOP-3.6.3-VEKOP-16-2017-00009 grant from the Higher Education Institutional Excellence Program of the Semmelweis University, the Higher Education Institutional Excellence Program of the Ministry of Human Capacities in Hungary, within the framework of the Neurology Thematic Program of Semmelweis University (TKP 2021 EGA-25) for AA and AD, NAP2022-I-1/2022 of the Hungarian Academy of Sciences for AA, the New National Excellence Program of the Ministry for Culture and Innovation from the source of the National Research, Development and Innovation Fund (ÚNKP-23-4), the Gedeon Richter Plc. Centenary Foundation (Gyömrői út 19-21 Budapest, 1103), and the NKFIH EKÖP-2024 fellowship to FD.
Author information
Authors and Affiliations
Contributions
Fanni Dóra: Conceptualization, methodology, validation, formal analysis, investigation, resources, data curation, writing—original draft, writing—review and editing, visualization, funding acquisition. Tamara Hajdu: Methodology and investigation. Éva Renner: Methodology, investigation, and project administration. Krisztina Paál: Methodology and investigation. Alán Alpár: Investigation, writing—review and editing, project administration, and funding acquisition. Miklós Palkovits: Conceptualization, writing—original draft, writing—review and editing, supervision, project administration, funding acquisition. Christos Chinopoulos: Conceptualization, methodology, resources, writing—original draft, writing—review and editing, supervision, project administration, funding acquisition. Árpád Dobolyi: Conceptualization, methodology, resources, writing—original draft, writing—review and editing, supervision, project administration, funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
The study was conducted according to the guidelines of the Declaration of Helsinki and the Ethical Rules for Using Human Tissues for Medical Research in Budapest, Hungary (HM 34/1999). Approval to collect and store brain samples was granted by the Committee of Science and Research Ethics of the Ministry of Health of Hungary (6008/8/2002/ETT) and the Semmelweis University Regional Committee of Science & Research Ethics (31/1992/TUKEB).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dóra, F., Hajdu, T., Renner, É. et al. Reverse phase protein array-based investigation of mitochondrial genes reveals alteration of glutaminolysis in the parahippocampal cortex of people who died by suicide. Transl Psychiatry 14, 479 (2024). https://doi.org/10.1038/s41398-024-03137-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-024-03137-x