Abstract
Biopsychosocial factors substantially increase the risk of depression in adults, and DNA methylation has been implicated as a mechanism through which these factors influence this risk. This study determined whether methylation levels moderate the association between biopsychosocial factors and depression. Using cross-sectional data from the Taiwan Biobank 2016–2017, the study examined the effects of biopsychosocial factors and DNA methylation on depression. The sample consisted of 96 participants aged 30 to 68 years. Depression and biopsychosocial factors were evaluated using self-reported questionnaires. Biopsychosocial factors included biological factors (age, sex, physical illness, body mass index [BMI]), psychological factors (alcohol experience, smoking experience, and exercise habit), and social factors (education, marriage, and dependency). To obtain methylated gene data, the Taiwan Biobank 2016–2017, 65 biological risk genes, and the GSE113725 dataset were intersected, revealing 5 genes and 14 CpG sites potentially associated with depression (IL2RB-cg02238178, IL2RB-cg11558856, IL15RA-cg03108606, IL15RA-cg07796897, IL15RA-cg08676905, IL6R-cg25853020, IL6R-cg09257526, IL6R-cg04715245, FTL-cg04385818, FTL-cg03039974, ZNF614-cg09503196, ZNF614-cg25776555, ZNF614-cg03293882, and ZNF614-cg15684917). The results indicated that BMI was negatively associated with depression risk (adjusted odds ratio [aOR] = 0.320, 95% confidence interval [CI] [0.117–0.876]), whereas the methylation of IL6R_cg09257526 increased the risk of depression (aOR = 2.535, 95% CI [1.006–6.391]) and significantly moderated the association between BMI and depression (aOR = 4.687, 95% CI [1.185–18.542]). BMI plays a crucial role in biological factors and together with DNA methylation of the IL6R_cg09257526 gene contributes to the occurrence of depression in the Taiwanese population.
Similar content being viewed by others
Introduction
Depression affects cognitive processes, emotional wellbeing, and physical health [1]. Depression is a major concern among adults and affects individuals of any age, sex, or lifestyle [2]. In 2023, the World Health Organization reported that depression affects approximately 3.8% of the global population, 5% of adults, and 5.7% of individuals aged 60 years or older; and that an expected 280 million individuals worldwide have depression [3]. In particular, adults aged 30 years or older have an increased risk of depression and risk factors include social isolation, financial stress, and chronic health conditions [4]. Over a 10-year period, the incidence of depression in Taiwan increased by 12%. The prevalence of treated depression increased from 1.6% in 2007 to 1.92% in 2016 [5]. These findings indicate the importance of investigating depression in the Taiwanese population. Although various factors contribute to depression, many remain unidentified. Regulation of biopsychosocial dynamics is hypothesized to reduce the prevalence of depression. Thus, the impact of biopsychosocial factors warrants further investigation [6].
Depression is associated with numerous factors, among which biopsychosocial factors play a essential role [7]. Biopsychosocial models serve as comprehensive frameworks that can be used to examine biological, psychological, and social factors contributing to the development and manifestation of both physical and psychological health conditions [8]. The “biological” component includes an individual’s biological, genetic, and physical characteristics [9], such as age [10,11,12], sex [11,12,13,14,15], physical health status [16, 17], and body mass index (BMI) [18,19,20,21]. Recent studies have reported that genetic factors, such as DNA methylation, can affect the onset of depression through epigenetic mechanisms [22, 23]. The “psychological” aspect involves developmental, psychological, and psychopathological factors [24]. Psychological risk factors, such as smoking [25, 26], alcohol consumption [27], and exercise habits [28, 29], are associated with an increased risk of depression. The “social” component focuses on broader environmental factors, including social and cultural surroundings [8]. Social factors, such as education [30,31,32], marital status [2, 33], and dependency [34], are also associated with depression. Studies have explored how biological factors, such as BMI, may interact with genetic factors, such as DNA methylation, to affect depression [22, 23]. For instance, DNA methylation may modulate the relationship between various biopsychosocial factors and depression, suggesting the presence of a more complex interplay than has been understood to date. Thus, investigating DNA methylation as a potential moderator of biopsychosocial factors can provide new insights into mechanisms underlying depression.
DNA methylation plays a critical role in the development of depression. Global DNA methylation, which regulates gene expression across the genome, is associated with key neurobiological processes, such as stress response, immune function, and inflammation, all of which are implicated in the development of depression [35]. In particular, gene-specific methylation, such as at IL6R and IL15RA, affects immune and stress responses, thereby influencing mental health outcomes [36, 37]. The methylation of IL6R is particularly important because it moderates inflammation associated with depression [38].
DNA methylation is an essential process that inhibits the transcription of repetitive DNA sequences. DNA methylation is involved in various cellular processes, such as X-chromosome inactivation, genomic imprinting, transposon suppression, chromatin structure regulation, retroviral gene silencing, and epigenetic memory maintenance [22]. Changes in DNA methylation might occur in various brain areas, especially the hippocampus and amygdala [39]. Several studies have suggested an association between DNA methylation and depression [22, 40,41,42]. Hypomethylation, particularly in the hippocampus, can lead to decreased expression of glucocorticoid receptors, a process that is further exacerbated by depression and suicide [39]. A study observed considerably higher hypomethylation in individuals with depression than in those without depression [43]. Another study indicated that epigenetic alterations, including DNA methylation, may contribute to neuropsychiatric disorders. These changes are potentially reversible and could be as important as genetic defects in the pathogenesis of depression [23]. Therefore, understanding DNA methylation is essential because it can either increase or reduce the risk of depression by acting as a moderating factor.
In statistical analysis, moderating effects refer to the role of DNA methylation in modifying the strength or direction of the association between BMI and depression. Specifically, DNA methylation may amplify, attenuate, or even reverse the nature of this association, highlighting its potential regulatory influence on the interplay between biological and psychological factors [44]. Multiple studies have explored the association between biopsychosocial factors and depression [7, 24, 45] but few studies on this topic have focused on the Taiwanese population [46]. Furthermore, some studies have examined DNA methylation in the Taiwanese population, but only few studies have investigated its association with depression [47,48,49,50]. A study examined DNA methylation as a moderator in the relationship between sleep quality and depression in young Chinese adults [51]. Another study explored the link between childhood maltreatment and depression during adolescence [52]. However, no studies have investigated DNA methylation as a moderator in the relationship between biopsychosocial factors and depression. Thus, the current study explored DNA methylation as a moderating variable in the relationship between biopsychosocial factors and depression in Taiwan. In particular, this study examined whether biopsychosocial factors can predict depression in this population. In addition, this study determined the extent to which DNA methylation moderates the relationship between biopsychosocial factors and depression. We hypothesized that DNA methylation moderates this relationship, such that individuals with specific methylation profiles would exhibit depression outcomes.
Methods
Study design
This cross-sectional study was conducted using data from the Taiwan Biobank collected between 2016 and 2017. This database contains information on biological and lifestyle factors, thus serving as a valuable resource for identifying health risk factors in Taiwan. Recruitment for the Taiwan Biobank is conducted through multiple channels, including media outlets, posters, pamphlets, and websites. Data collection is facilitated through the collaboration of 29 centers across Taiwan. The Taiwan Biobank strictly adheres to regulations governing data protection and privacy [2].
Sample
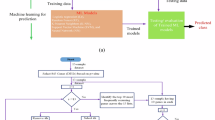
Initially, 4888 participants were recruited from the Taiwan Biobank. The Taiwan Biobank only contains data on individuals aged between 30 and 68 years. Participants were included if they had complete data regarding age, sex, weight, height, personal physical illness history, smoking history, alcohol consumption history, exercise habits, education level, marital status, dependency, depression, and DNA methylation. Participants were excluded if they had incomplete data, were undergoing treatment for mental health, or were receiving medication. Finally, 310 eligible participants were identified.
Among the eligible participants, 252 did not have depression, and 58 had depression. Because of the unequal distribution of participants between the two groups [53, 54], a matching analysis was performed. Matching analysis improves study efficiency by reducing the sample size without losing essential information, thereby enhancing the precision of statistical estimates [53]. By minimizing selection bias, matching ensures more accurate and reliable comparisons, increases the study’s validity, and reduces bias [53]. In this study, we conducted matching analysis by manually pairing participants with the same sex and age from both the depression and no depression groups. This process resulted in 48 participants in the depression group and 48 participants from the no depression group. The total number of participants included was 96 (Fig. 1). This study was approved by the Joint Institutional Review Board of Taipei Medical University (approval number: N202306111).
Measures
Biopsychosocial
Biopsychosocial factors were categorized into biological, psychological, and social dimensions on the basis of participants’ characteristics. Biological factors included age and BMI, both recorded as continuous variables, along with sex and physical illness status. BMI was calculated using the following formula: BMI = weight (kg)/height² (m²). This calculation provided a continuous measurement of BMI for each participant [55]. Psychological factors included alcohol experience, smoking experience, and exercise habits. Alcohol experience status was assessed using the question, “Do you currently have a habit of drinking?” Smoking experience status was determined using the question, “Do you currently smoke?” Exercise habits were assessed using the question, “Do you have a habit of exercising regularly?” Social factors included the highest level of education attained, current marital status, and whether the participant lives alone. The variables were categorized as follows: sex (male/female), physical illness (no physical illness/at least one physical illness), alcohol experience (yes/no), smoking experience (yes/no), exercise habits (yes/no), education level (below bachelor’s degree/bachelor’s degree or above), marital status (not currently married/currently married), and dependency (not living alone/living alone).
Depression
Depression was measured using a self-reported item from the Taiwan Biobank 2016–2017 questionnaire. Participants were asked the following question: ‘Do you or your family have depression?’ with response options of ‘Yes’ and ‘No.’ This question was presented alongside another item inquiring about the year they were diagnosed with depression by a physician.
DNA methylation
DNA methylation data were processed using established quality control protocols to ensure data integrity. Specifically, methylation intensity values were normalized using the normal-exponential out-of-band (noob) method to correct for background fluorescence and ensure the accurate quantification of methylation levels [56]. Batch effect correction was applied using the ComBat method to adjust for technical variations across different sample plates, rows, columns, and chip array positions [49]. These standard procedures have been widely used in DNA methylation studies to ensure high-quality data free from technical biases. To obtain DNA methylation data, several steps were followed. First, data from the Taiwan Biobank (2016–2017) were analyzed using the Limma (Linear Models for Microarray and RNA-Seq data) package in R/Bioconductor framework. In total, 2942 genes in 2016 and 2761 genes in 2017 related to DNA methylation were identified. Second, the Gene Expression Omnibus (GEO) dataset (GSE113725) was used to identify genes related to depression in humans [40]. Analysis was conducted using GEO2R, and 7595 genes were identified. Third, from other study, 65 genes related to biological factors in depressed participants were identified from the Taiwan Biobank 2016–2017 dataset [57].
All the genes were intersected, and the results revealed 5 genes and 14 CpG sites potentially associated with depression. These identified genes and CpG sites were included in the current study: IL2RB (cg02238178 and cg11558856), IL15RA (cg03108606, cg07796897, and cg08676905), IL6R (cg25853020, cg09257526, and cg04715245), FTL (cg04385818 and cg03039974), and ZNF614 (cg09503196, cg25776555, cg03293882, and cg15684917).
Statistical analyses
Descriptive statistics are presented as means and standard deviations (SDs) for continuous data which are analyzed using the independent-samples t-test. Categorical data were compared and statistically analyzed using the chi-square (X²) test and presented frequency and percentage. To standardize numerical data, including age, BMI, and DNA methylation, z-scores were calculated. This step mitigated problems arising from differing variable ranges and minimized the effects of outliers [58, 59]. Collinearity was further assessed by evaluating the variance inflation factor (VIF) within the regression model. The analysis indicated that multicollinearity among the independent variables was unlikely, as all VIF values adhered to the commonly accepted threshold of being below 10 [60]. This approach balances the need for model precision with the inclusion of meaningful variables, ensuring that the findings are robust and interpretable. Furthermore, adjusted odd ratio (aOR) and 95% confidence intervals (CIs) were obtained by performing a conditional logistic regression for depression in association with significant variables including BMI, physical illness, IL15RA_cg03108606, IL15RA_cg08676905, IL6R_cg09257526, FTL_cg04385818, and ZNF614_cg09503196. Moderation analysis conducted using conditional logistic regression with interaction terms to evaluate whether DNA methylation moderated the relationship between BMI, physical illness, and depression, which was considered statistically significant at P ≤ 0.05. Therefore, model reliability was assessed using the conditional logistic regression to identify the best-fitting model, balancing goodness-of-fit and model complexity [61]. In addition, the likelihood ratio test was used to determine whether adding predictors significantly improved model fit. Pseudo R² was calculated as a measure of the model’s explanatory power. Effect sizes were reported as aORs with 95% CIs to ensure the precision and robustness of the estimates [62]. Statistical analyses were performed using STATA with a p value of ≤0.05 considered statistically significant.
The forest plot was used to illustrate the odds ratios (ORs) and 95% confidence intervals (CIs) for the associations between five methylated genes and depression across BMI subgroups (categorized based on lower mean BMI values and a mean BMI greater or equal to the threshold) [63, 64]. This visualization provides a clear representation of effect sizes across different BMI categories, indicating the moderating role of methylated genes in depression.
Results
Table 1 summarizes the characteristics of 96 adult Taiwanese participants across biological, psychological, social, and methylation factors. Regarding biological factors, the mean age of participants was 51.02 (SD = 11.65) years in both the depression and no depression groups. The majority of participants in this study were female (56.25%); however, the proportions of males and females were balanced between the depression and no-depression groups. This balance indicates that the manual matching process, which paired participants by sex and age, was effectively implemented, ensuring comparability between the two groups. Additionally, 80.21% of the participants reported having at least one physical illness. The mean BMI was 24.55 kg/m² (SD = 3.32). Physical illness and BMI were significantly associated with depression (P ≤ 0.05), whereas age and sex were not significantly associated with depression. Regarding psychological factors, most participants were no alcohol drinking (91.67%) and no smoking (63.54%). In addition, 57.29% of the participants reported irregular exercise habits. Alcohol experience, smoking experience, and exercise habits were not significantly associated with depression (P > 0.05). Regarding social factors, 52.08% of the participants had a bachelor’s degree or above, and 64.58% of the participants were currently married. Most participants (86.46%) were not living alone. Marital status, education level, and dependency were not associated with depression (P > 0.05). The mean ± SD DNA methylation levels at significant CpG sites were as follows: IL15RA_cg03108606 (0.062 ± 0.053), IL15RA_cg08676905 (0.128 ± 0.019), IL6R_cg09257526 (0.310 ± 0.024), FTL_cg04385818 (0.270 ± 0.158) and ZNF614_cg09503196 (0.050 ± 0.010). The association between BMI and physical illness variables for Taiwanese and depression severity were shown to present relevant covariates. The results revealed that individuals who have high BMI and at least one has physical illness showed significantly more depression.
Table 2 presents the results of the conditional logistic regression analysis, investigating the association between BMI, 4 methylated genes, depression, and their interactions. The variables that exhibited low multicollinearity, with variance inflation factor (VIF) values below 5 and condition indices within acceptable thresholds were physical illness, BMI, IL15RA_cg03108606, IL15RA_cg08676905, IL6R_cg09257526, FTL_cg04385818, and ZNF614_cg09503196. These factors included relevant covariates in the moderation analysis. However, IL15RA_cg03108606 and FTL_cg04385818 showed severe multicollinearity (VIF > 10), which could potentially destabilize parameter estimates and reduce interpretability in the conditional logistic regression model. Finally, model incorporated only one of the collinear genes (Table 2). The findings reveal that a higher BMI was significantly associated with a reduced likelihood of depression (aOR = 0.320, 95% CI [0.117–0.748], p = 0.027), after controlling for physical illness and methylation levels at IL15RA_cg08676905, IL6R_cg09257526, FTL_cg04385818, and ZNF614_cg09503196. Higher methylation levels at IL6R_cg09257526 were significantly associated with an increased risk of depression (aOR = 2.535, 95% CI [1.006–6.391], p = 0.049). Furthermore, methylation at IL6R_cg09257526 significantly moderated the relationship between BMI and depression, as demonstrated by the significant interaction term (aOR = 4.687, 95% CI [1.185–18.542], p = 0.028). Physical illness was also analyzed; however, none of the results showed significant associations with depression when methylated genes were included as moderators.
Figure 2 illustrates the association between DNA methylation at specific CpG sites and depression risk, classified by BMI categories. BMI was categorized based on the mean value of 24.55 (Table 1), with participants grouped into lower (BMI < 24.55; Fig. 2A) and greater or equal (BMI ≥ 24.55; Fig. 2B) categories. Among participants with a BMI < 24.55, significant associations were identified between the methylation of IL15RA_cg08676905 (OR = 0.337, 95% CI [0.120–0.947]) and ZNF614_cg09503196 (OR = 0.296, 95% CI [0.105–0.837]) and depression, while the other 3 genes analyzed did not show significant associations (p > 0.05). Among participants with a BMI ≥ 24.55, all genes significant associations were observed. Methylation at IL15RA_cg03108606 was linked to a notably high odds ratio (OR = 3.603, 95% CI [1.146–11.333]), suggesting a significant increased risk of depression. Methylation at IL6R_cg09257526 showed a strong positive association with depression, with a markedly high odds ratio (OR = 10.138, 95% CI [3.009–34.156]). In contrast, methylation at IL15RA_cg08676905 was associated with a significantly lower odds ratio (OR = 0.311, 95% CI [0.099–0.972]), also indicating a protective effect. Furthermore, methylation at FTL_cg04385818 (OR = 0.295, 95% CI [0.094–0.925]) and ZNF614_cg09503196 (OR = 0.272, 95% CI [0.086–0.856]) were both associated with reduced odds of depression, supporting their potential protective roles. These findings suggest that the relationship between DNA methylation and depression may be influenced by BMI, with stronger and more significant associations observed in individuals with a higher BMI.
Discussion
This is the first study to examine DNA methylation as a potential moderating factor in the relationships between biological, psychological, and social factors and depression by performing matching analysis in a Taiwanese population. The results of the current study revealed that the DNA methylation gene of IL6R_cg09257526 moderates the strength of the correlation between BMI and depression in the Taiwan population.
Our findings indicated that an increase in BMI was associated with a 0.320-fold reduction in the risk of depression (95% CI [0.117–0.876]; P = 0.027; Table 2). Thus, a higher BMI was consistently correlated with a decreased risk of depression, even after controlling for the effect of biopsychosocial factors. In another study, depression was less prevalent among overweight individuals than among underweight individuals [55]. These finding challenges traditional understandings, especially for women in Korea, who experience considerable weight-related pressures and stress due to the high level of societal concern about physical size and weight [55, 65]. BMI does not always correlate with the risk of depression because weight loss efforts can occasionally contribute to an increase in depression [66]. BMI is related to C-reactive protein, and a high level of C-reactive protein is associated with depression, confirming the assumption that other causes of peripheral inflammation (e.g., the interleukin 6 receptor [IL6R]) may affect the development of depression [67].
The methylated gene IL6R_cg09257526 was significantly correlated with depression (P ≤ 0.05; Table 2). The mean depression score for this gene (0.318; Table 1) was higher than that of the control group (0.301; Table 1). IL6R acts as a receptor for the proinflammatory cytokine interleukin-6 (IL-6), enabling it to transmit signals. IL6R is a type 1 cytokine receptor consisting of an IL-6 binding protein and a signal transducing protein [68, 69]. Hypermethylation of IL6R was associated with low expression in the no depression group and high expression in the depression group [70]. Other studies have reported that individuals with depression have higher hypomethylation than do those without depression [39, 43]. This finding suggests that increased methylation of IL6R_cg09257526 can exacerbate depression, supporting its negative effect. This finding is in line with that of a study that identified IL-6 as a potent biomarker for depression, with increased plasma levels of IL-6 genetically predicted to correlate with major depression [71]. Furthermore, another study found that an IL6R functional variant could reduce inflammation, and high serum levels of IL-6/IL6R can increase the severity of depression [67]. The current study indicated that an increase in the methylation of IL6R_cg09257526 significantly increased the risk of depression by 2.535 times (95% CI [1.006–6.391]; P = 0.049; Table 2). This finding is consistent with that of a study indicating the correlation of an increased IL-6 level with depression [72]. The pathogenesis of depression is correlated with inflammation, specifically the IL-6/IL-6R pathways; peripheral blood levels of inflammatory markers increased during the acute episodes of depression [73]. Thus, managing IL-6/IL-6R levels in the circulatory system could control both inflammation and depression.
The present study found that the DNA methylation of the IL6R_cg09257526 gene significantly moderated the relationship between BMI and depression (OR = 4.687; 95% CI [1.185–18.542]; P = 0.028; Table 2). A study reported that biopsychosocial factors such as lifestyle are associated with depression and strongly affect DNA methylation patterns [74]. Another study demonstrated that IL-6 moderated the relationships among physical activity, fatigue, and depressed mood [75]. When IL-6 levels are low, the association between physical activity and fatigue is weaker. However, when IL-6 levels are high, physical activity increases fatigue, which is a common symptom of depressed mood [75]. IL-6 can affect the balance of neurotransmitters in the brain (e.g., serotonin, dopamine, and norepinephrine) that are closely involved in mood regulation. Alterations in the level of these neurotransmitters are related to depression [76]. Norepinephrine stimulates the release of IL-6, and IL-6 inhibits the secretion of dopamine and serotonin [77, 78]. Both excessively high and excessively low levels of IL-6 may contribute to mental health problems, such as depression, anxiety, and schizophrenia [79, 80]. IL-6 may cause alterations in the central nervous system [81] and contribute to the development and severity of depression [82]. Furthermore, vagal afferents, circumventricular organs, and brain regions outside the blood–brain barrier can transmit cytokine signals from the periphery to the brain [82], potentially resulting in neurotransmission alterations and subsequent depression [83]. Moreover, in the brain, an increased IL-6 level may cause neuroinflammation, which disrupts normal brain function and affects mood regulation [84, 85]. Higher levels of inflammatory markers and altered neural functioning in several brain regions have been correlated with depression [86]. These genomic and neurobiological pathways might explain why IL-6 is the only inflammatory modifier with sufficient sensitivity to predict the early signs of depression [86].
The strength of the current study lies in its novelty. To the best of our knowledge, no study has examined the role of DNA methylation in Taiwanese individuals with depression considering biopsychosocial factors and investigated DNA methylation as a moderator. The current study performed matching analysis to enhance the robustness of the findings. This technique has been effectively used as an analytical approach in social and community studies [53].
The present study has several limitations. First, the sample size was small, however, another study also included a small number [87], which might affect the generalizability of the results. Thus, caution is advised when interpreting these initial findings, particularly at the CpG level. Second, depression was assessed using self-reported measures that captured participants’ perception of their depression status, based on a prior diagnosis by a healthcare professional. However, self-reported data may be subject to recall bias and could lead to either underreporting or overreporting of depressive symptoms [88, 89]. Third, because the present study had a cross-sectional design, the findings were correlational, and causality cannot be inferred [52].
The findings of this study offer vital implications. Protective factors against depression related to gene methylation were identified. Interventions should focus on helping individuals improve their physical wellbeing. Gene methylation can undergo a reversal process, leading to gene demethylation in the presence of various environmental conditions [22, 23]. Gene methylation is reversible, indicating the importance of adopting a healthy lifestyle [23]. To maintain physical wellbeing, individuals should focus on consuming nutritious food, engaging in regular physical exercise, and ensuring adequate sleep quality [90]. Maintaining a BMI within the normal range is essential for monitoring nutrition status, which can assist in the management of depression [91].
Conclusion
The findings of this study suggest that biopsychosocial factors, particularly BMI, contribute to the development of depression, with methylated gene of IL6R_cg09257526 acting as a moderator that exacerbates the condition. In addition, the study highlights the importance of maintaining physical health and achieving an ideal weight as a preventive measure against depression. These findings can assist health-care providers in designing and implementing effective interventions that integrate biopsychosocial factors into mental health treatments. Future research should focus on exploring how depression can be improved through interventions or new strategies, particularly within public health policies addressing BMI status.
References
Cui R. Editorial: a systematic review of depression. Curr. Neuropharmacol. 2015;13:480.
Hsu MY, Huang SC, Liu PL, Yeung KT, Wang YM, Yang HJ. The interaction between exercise and marital status on depression: a cross-sectional study of the taiwan biobank. Int. J. Env. Res. Public. Health. 2022;19:1876.
WHO. Depressive disorder (depression) 2023. 2023. https://www.who.int/news-room/fact-sheets/detail/depression.
Mykyta L. Living alone and feelings of depression among adults age 18 and older. Natl. Health Stat. Rep. 2024:1–11, https://doi.org/10.15620/cdc:136451.
Wang H-H, Chang C-M, Chang S-S, Yang AC, Liu Y-H, Liao S-C, et al. Ten-year trends in depression care in Taiwan. J. Formos. Med. Assoc. 2022;121:2001–11.
Schneider FW, Gruman JA, Coutts LM. Applied social psychology: Understanding and addressing social and practical problems. Thousand Oaks, CA, US: Sage Publications, Inc; 2005. p. 449.
Amsah N, Md Isa Z, Ahmad N. Biopsychosocial and nutritional factors of depression among type 2 diabetes mellitus patients: a systematic review. Int. J. Env. Res. Public. Health. 2022;19:4888.
Saasa S, Miller S. Biopsychosocial predictors of depression and anxiety symptoms among first-generation black african immigrants. Soc. Work. 2021;66:329–38.
Habtewold TD, Islam MA, Radie YT, Tegegne BS. Comorbidity of depression and diabetes: an application of biopsychosocial model. Int. J. Ment. Health Syst. 2016;10:74.
Almeida OP. Prevention of depression in older age. Maturitas. 2014;79:136–41.
Salk RH, Hyde JS, Abramson LY. Gender differences in depression in representative national samples: meta-analyses of diagnoses and symptoms. Psychol. Bull. 2017;143:783–822.
Tseng H, Lee JI, Geng JH, Chen SC. Sex difference in the associations among risk factors with depression in a large Taiwanese population study. Front. Public. Health. 2023;11:1070827.
Kuehner C. Why is depression more common among women than among men? Lancet Psychiatry. 2017;4:146–58.
Noble RE. Depression in women. Metabolism. 2005;54(5 Suppl 1):49–52.
Sassarini DJ. Depression in midlife women. Maturitas. 2016;94:149–54.
Li A, Rosella LC, Kurdyak P, Wodchis WP. Depression as a risk factor for physical illness and multimorbidity in a cohort with no prior comorbidity. Can. J. Psychiatry. 2021;66:726–36.
Lin SC, Tyus N, Maloney M, Ohri B, Sripipatana A. Mental health status among women of reproductive age from underserved communities in the United States and the associations between depression and physical health. A cross-sectional study. PLoS One. 2020;15:e0231243.
Cui N, Cui J, Xu X, Aslam B, Bai L, Li D, et al. Health conditions, lifestyle factors and depression in adults in qingdao, china: a cross-sectional study. Front. Psychiatry. 2021;12:508810.
Jantaratnotai N, Mosikanon K, Lee Y, McIntyre RS. The interface of depression and obesity. Obes. Res. Clin. Pract. 2017;11:1–10.
Woo YS, McIntyre RS, Kim JB, Lee MS, Kim JM, Yim HW, et al. Association of treatment response with obesity and other metabolic risk factors in adults with depressive disorders: results from a national depression cohort study in Korea (the CRESCEND study). J. Affect. Disord. 2016;203:190–8.
Alkharji TM, Alharbi RS, Bakhsh EA, Alghalibi M, Alraddadi RA. The association between depression and obesity among adults in jeddah, Saudi Arabia, in 2022. Cureus. 2023;15:e35428.
Chen D, Meng L, Pei F, Zheng Y, Leng J. A review of DNA methylation in depression. J. Clin. Neurosci. 2017;43:39–46.
Chmielewska N, Szyndler J, Maciejak P, Plaznik A. Epigenetic mechanisms of stress and depression. Psychiatr. Pol. 2019;53:1413–28.
Santos HP Jr., Adynski H, Harris R, Bhattacharya A, Incollingo Rodriguez AC, Cali R, et al. Biopsychosocial correlates of psychological distress in Latina mothers. J. Affect. Disord. 2021;282:617–26.
Fluharty M, Taylor AE, Grabski M, Munafo MR. The association of cigarette smoking with depression and anxiety: a systematic review. Nicotine Tob. Res. 2017;19:3–13.
Luger TM, Suls J, Vander Weg MW. How robust is the association between smoking and depression in adults? A meta-analysis using linear mixed-effects models. Addict. Behav. 2014;39:1418–29.
Jia H, Zack MM, Gottesman II, Thompson WW. Associations of smoking, physical inactivity, heavy drinking, and obesity with quality-adjusted life expectancy among us adults with depression. Value Health. 2018;21:364–71.
Gordon BR, McDowell CP, Hallgren M, Meyer JD, Lyons M, Herring MP. Association of efficacy of resistance exercise training with depressive symptoms: meta-analysis and meta-regression analysis of randomized clinical trials. JAMA Psychiatry. 2018;75:566–76.
Imboden C, Gerber M, Beck J, Holsboer-Trachsler E, Puhse U, Hatzinger M. Aerobic exercise or stretching as add-on to inpatient treatment of depression: similar antidepressant effects on depressive symptoms and larger effects on working memory for aerobic exercise alone. J. Affect. Disord. 2020;276:866–76.
Patel JS, Oh Y, Rand KL, Wu W, Cyders MA, Kroenke K, et al. Measurement invariance of the Patient Health Questionnaire-9 (PHQ-9) depression screener in U.S. adults across sex, race/ethnicity, and education level: NHANES 2005-2016. Depress. Anxiety. 2019;36:813–23.
Nguyen AW, Walton QL, Thomas C, Mouzon DM, Taylor HO. Social support from friends and depression among african americans: the moderating influence of education. J. Affect. Disord. 2019;253:1–7.
Wickersham A, Sugg HVR, Epstein S, Stewart R, Ford T, Downs J. Systematic review and meta-analysis: the association between child and adolescent depression and later educational attainment. J. Am. Acad. Child. Adolesc. Psychiatry. 2021;60:105–18.
Rehman US, Gollan J, Mortimer AR. The marital context of depression: research, limitations, and new directions. Clin. Psychol. Rev. 2008;28:179–98.
Srivastava S, Debnath P, Shri N, Muhammad T. The association of widowhood and living alone with depression among older adults in India. Sci. Rep. 2021;11:21641.
Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8:355–67.
Lorente-Sorolla C, Garcia-Gomez A, Català-Moll F, Toledano V, Ciudad L, Avendaño-Ortiz J, et al. Inflammatory cytokines and organ dysfunction associate with the aberrant DNA methylome of monocytes in sepsis. Genome Med. 2019;11:66.
Ali MM, Naquiallah D, Qureshi M, Mirza MI, Hassan C, Masrur M, et al. DNA methylation profile of genes involved in inflammation and autoimmunity correlates with vascular function in morbidly obese adults. Epigenetics. 2022;17:93–109.
Roohi E, Jaafari N, Hashemian F. On inflammatory hypothesis of depression: what is the role of IL-6 in the middle of the chaos? J. Neuroinflammation. 2021;18:45.
Lin E, Tsai SJ. Epigenetics and depression: an update. Psychiatry Investig. 2019;16:654–61.
Crawford B, Craig Z, Mansell G, White I, Smith A, Spaull S, et al. DNA methylation and inflammation marker profiles associated with a history of depression. Hum. Mol. Genet. 2018;27:2840–50.
Zhu JH, Bo HH, Liu BP, Jia CX. The associations between DNA methylation and depression: a systematic review and meta-analysis. J. Affect. Disord. 2023;327:439–50.
Penner-Goeke S, Binder EB. Epigenetics and depression. Dialogues Clin. Neurosci. 2019;21:397–405.
Ryan J, Pilkington L, Neuhaus K, Ritchie K, Ancelin M-L, Saffery R. Investigating the epigenetic profile of the inflammatory gene IL-6 in late-life depression. BMC Psychiatry. 2017;17:354.
Nussbeck FW, Fuchs P Moderation (Statistical). In: Zeigler-Hill V, Shackelford TK, editors. Encyclopedia of personality and individual differences. Cham: Springer International Publishing; 2017. pp. 1-5.
Robinson MA, Kim I, Mowbray O, Disney L. African Americans, caribbean blacks and depression: which biopsychosocial factors should social workers focus on? Results from the National Survey of American Life (NSAL). Community Ment. Health J. 2022;58:366–75.
Chiu CJ, Li ML, Chou CY. Trends and biopsychosocial correlates of physical disabilities among older men and women in Taiwan: examination based on ADL, IADL, mobility, and frailty. BMC Geriatr. 2022;22:148.
Weng JT, Wu LS, Lee CS, Hsu PW, Cheng AT. Integrative epigenetic profiling analysis identifies DNA methylation changes associated with chronic alcohol consumption. Comput. Biol. Med. 2015;64:299–306.
Chen YC, Huang RL, Huang YK, Liao YP, Su PH, Wang HC, et al. Methylomics analysis identifies epigenetically silenced genes and implies an activation of beta-catenin signaling in cervical cancer. Int. J. Cancer. 2014;135:117–27.
Chiu K-C, Hsieh M-S, Huang Y-T, Liu C-Y. Exposure to ambient temperature and heat index in relation to DNA methylation age: a population-based study in Taiwan. Environ. Int. 2024;186:1–14.
Chou YH, Tantoh DM, Wu MC, Tyan YS, Chen PH, Nfor ON, et al. PM(2.5) exposure and DLEC1 promoter methylation in Taiwan biobank participants. Env. Health Prev. Med. 2020;25:68.
Li T, Xie Y, Tao S, Zou L, Yang Y, Mou X, et al. Moderating effects of PER3 gene DNA methylation on the association of sleep quality with mental health in Chinese young adults. J. Affect. Disord. 2023;323:716–22.
Comtois-Cabana M, Barr E, Provencal N, Ouellet-Morin I. Association between child maltreatment and depressive symptoms in emerging adulthood: the mediating and moderating roles of DNA methylation. PLoS One. 2023;18:e0280203.
Iwagami M, Shinozaki T. Introduction to matching in case-control and control studies. Ann. Clin. Epidemiol. 2022;4:33–40.
Pearce N. Analysis of matched case-control studies. BMJ. 2016;352:i969.
Noh JW, Kwon YD, Park J, Kim J. Body mass index and depressive symptoms in middle aged and older adults. BMC Public. Health. 2015;15:310.
Lin W-Y, Wang Y-C, Teng I-H, Liu C, Lou X-Y. Associations of five obesity metrics with epigenetic age acceleration: Evidence from 2474 Taiwan biobank participants. Obes. Soc. 2021;29:1731–8.
Lesmana MHS, Le NQK, Chiu WC, Chung KH, Wang CY, Irham LM, et al. Genomic-analysis-oriented drug repurposing in the search for novel antidepressants. Biomedicines. 2022;10:1947.
DeVore GR. Computing the Z score and centiles for cross-sectional analysis: a practical approach. J. Ultrasound Med. 2017;36:459–73.
Plichta SB, Kelvin E. Munro’s statistical methods for health care research. New York: Wolter Kluwer; 2013.
García CB, García J, López Martín MM, Salmerón R. Collinearity: revisiting the variance inflation factor in ridge regression. J. Appl. Stat. 2015;42:648–61.
Archer KJ, Lemeshow S, Hosmer DW. Goodness-of-fit tests for logistic regression models when data are collected using a complex sampling design. Comput Stat. Data Anal. 2007;51:4450–64.
Jané MB, Ben-Shachar MS, Dunleavy DJ, Moreau D, Steele J, Xiao Q, et al. Guide to effect sizes and confidence intervals. James Steele; 2024.
Wallace C, Vandevijvere S, Lee A, Jaacks LM, Schachner M, Swinburn B. Dimensions of national culture associated with different trajectories of male and female mean body mass index in countries over 25 years. Obes. Rev. 2019;20(Suppl 2):20–9.
Gabbrielli R, Pugno NM. The impact of mean body mass index on reported mortality from COVID-19 across 181 countries. Front. Public. Health. 2023;11:1106313.
Lovejoy JC, Sainsbury A. Stock Conference Working G. Sex differences in obesity and the regulation of energy homeostasis. Obes. Rev. 2009;10:154–67.
Ross CE. Overweight and Depression. J. Health Soc. Behav. 1994;35:63–78.
Khandaker GM, Zammit S, Burgess S, Lewis G, Jones PB. Association between a functional interleukin 6 receptor genetic variant and risk of depression and psychosis in a population-based birth cohort. Brain Behav. Immun. 2018;69:264–72.
Singh SS, Jois SD Chapter One - Homo- and Heterodimerization of Proteins in Cell Signaling: Inhibition and Drug Design. In: Donev R, editor. Advances in protein chemistry and structural biology. 111: Academic Press; London, 2018. p. 1-59.
Srinivasan L, Harris MC, Kilpatrick LE Cytokines and Inflammatory Response in the Fetus and Neonate. In: Polin RA, Abman SH, Rowitch DH, Benitz WE, Fox WW, editors. Fetal and neonatal physiology (Fifth Edition): Elsevier; Philadelphia, 2017. pp. 1241-54.e4.
Alipour S, Sakhinia E, Khabbazi A, Samadi N, Babaloo Z, Azad M, et al. Methylation status of interleukin-6 gene promoter in patients with Behçet’s disease. Reumatol. Clin. 2020;16:229–34.
Mikhalitskaya EV, Vyalova NM, Ermakov EA, Levchuk LA, Simutkin GG, Bokhan NA, et al. Association of Single Nucleotide Polymorphisms of Cytokine Genes with Depression, Schizophrenia and Bipolar Disorder. Genes. 2023;14:1460.
Flouri E, Francesconi M, Midouhas E, Lewis G. Prenatal and childhood adverse life events, inflammation and depressive symptoms across adolescence. J. Affect. Disord. 2020;260:577–82.
Khandaker GM, Zuber V, Rees JMB, Carvalho L, Mason AM, Foley CN, et al. Shared mechanisms between coronary heart disease and depression: findings from a large UK general population-based cohort. Mol. Psychiatry. 2020;25:1477–86.
Barbu MC, Shen X, Walker RM, Howard DM, Evans KL, Whalley HC, et al. Epigenetic prediction of major depressive disorder. Mol. Psychiatry. 2021;26:5112–23.
Cohen M, Levkovich I, Katz R, Fried G, Pollack S. Low physical activity, fatigue and depression in breast cancer survivors: moderation by levels of IL-6 and IL-8. Int. J. Psychophysiol. 2020;158:96–102.
Kummer KK, Zeidler M, Kalpachidou T, Kress M. Role of IL-6 in the regulation of neuronal development, survival and function. Cytokine. 2021;144:155582.
Levandovski R, Pfaffenseller B, Carissimi A, Gama CS, Hidalgo MPL. The effect of sunlight exposure on interleukin-6 levels in depressive and non-depressive subjects. BMC Psychiatry. 2013;13:75.
Hodes GE, Menard C, Russo SJ. Integrating Interleukin-6 into depression diagnosis and treatment. Neurobiol. Stress. 2016;4:15–22.
Frommberger UH, Bauer J, Haselbauer P, Fräulin A, Riemann D, Berger M. Interleukin-6-(IL-6) plasma levels in depression and schizophrenia: comparison between the acute state and after remission. Eur. Arch. Psychiatry Clin. Neurosci. 1997;247:228–33.
Ting EY, Yang AC, Tsai SJ. Role of interleukin-6 in depressive disorder. Int. J. Mol. Sci. 2020;21:2194.
Ricciotti E, FitzGerald GA. Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 2011;31:986–1000.
Maier SF. Bi-directional immune-brain communication: implications for understanding stress, pain, and cognition. Brain Behav. Immun. 2003;17:69–85.
Accortt EE, Schetter CD, Peters RM, Cassidy-Bushrow AE. Lower prenatal vitamin D status and postpartum depressive symptomatology in African American women: preliminary evidence for moderation by inflammatory cytokines. Arch. Womens Ment. Health. 2016;19:373–83.
Han KM, Ham BJ. How inflammation affects the brain in depression: a review of functional and structural MRI studies. J. Clin. Neurol. 2021;17:503–15.
Jeon SW, Kim YK. Neuroinflammation and cytokine abnormality in major depression: cause or consequence in that illness? World J. Psychiatry. 2016;6:283–93.
Kautz MM, Coe CL, McArthur BA, Mac Giollabhui N, Ellman LM, Abramson LY, et al. Longitudinal changes of inflammatory biomarkers moderate the relationship between recent stressful life events and prospective symptoms of depression in a diverse sample of urban adolescents. Brain Behav. Immun. 2020;86:43–52.
Fuchikami M, Morinobu S, Segawa M, Okamoto Y, Yamawaki S, Ozaki N, et al. DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS One. 2011;6:e23881.
Arias-de la Torre J, Vilagut G, Serrano-Blanco A, Martin V, Molina AJ, Valderas JM, et al. Accuracy of self-reported items for the screening of depression in the general population. Int. J. Env. Res. Public. Health. 2020;17:7955.
Hunt M, Auriemma J, Cashaw AC. Self-report bias and underreporting of depression on the BDI-II. J. Pers. Assess. 2003;80:26–30.
Martinez-de-Quel O, Suarez-Iglesias D, Lopez-Flores M, Perez CA. Physical activity, dietary habits and sleep quality before and during COVID-19 lockdown: a longitudinal study. Appetite. 2021;158:105019.
Herhaus B, Kersting A, Brahler E, Petrowski K. Depression, anxiety and health status across different BMI classes: a representative study in Germany. J. Affect. Disord. 2020;276:45–52.
Acknowledgements
This manuscript was edited by Wallace Academic Editing.
Funding
This study was supported by Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (DP2-TMU-112-N-07).
Author information
Authors and Affiliations
Contributions
RMK, MHL, MHC designed the study. BSW, RMK, and MS performed the statistical analyses. RMK. and VLA wrote the manuscript. MHC assisted with the preparation and proofreading of the manuscript. RMK and MA constructed result and discussion of manuscript. All authors have approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kusuma, R.M., Lesmana, M.H.S., Amelia, V.L. et al. DNA methylation of the IL6R gene moderates the association between biopsychosocial factors and depression. Transl Psychiatry 15, 499 (2025). https://doi.org/10.1038/s41398-025-03596-w
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41398-025-03596-w