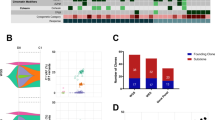
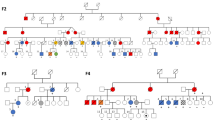
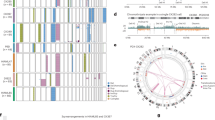
Abstract
Clinical next generation sequencing (NGS) typically relies on limited gene panels run on bulk marrow or blood. Current computational tools for inferring clonal relationships is generally limited by the use of a small panel of pathogenic mutations to define clones. We developed an online software (CloneTracker) that uses ‘incidentally-sequenced’ single nucleotide polymorphisms (SNPs) in the regions of recurrent somatic mutations in addition to conventional mutation data from bulk NGS gene panels to provide detailed visualizations of clonal evolution during cancer treatment, alongside clinical data. Tested on 29 patients who underwent non-myeloablative transplantation for AML, CloneTracker successfully reconstructed the evolutionary dynamics of donor engraftment from bulk NGS and rendered intuitive visualizations of residual patient-derived hematopoiesis and relapsing malignant clones. The software does not require sequencing donor samples, as donor-derived clones are identifiable from post-HCT SNP data. This manuscript aims to introduce CloneTracker to the BMT community and make it available for those who would ascertain its clinical utility, e.g, in BMT trials leveraging molecular minimal residual disease (MRD) monitoring and targeted interventions to pre-empt relapse.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
For original data and the code to reproduce all analyses, or to inquire about hosting CloneTracker on your own server or joining the CloneTracker development project, please contact Dr. Krakow.
References
Kröger N, Bishop M, Giralt S, Wayne A. Third International workshop on the biology, prevention, and treatment of relapse after stem cell transplantation. Bone Marrow Transpl. 2018;53:1–2. https://doi.org/10.1038/bmt.2017.218
Horowitz M, Schreiber H, Elder A, Heidenreich O, Vormoor J, Toffalori C, et al. Epidemiology and biology of relapse after stem cell transplantation. Bone Marrow Transpl. 2018;53:1379–89. https://doi.org/10.1038/s41409-018-0171-z
Hughes AE, Magrini V, Demeter R, Miller CA, Fulton R, Fulton LL, et al. Clonal architecture of secondary acute myeloid leukemia defined by single-cell sequencing. PLoS Genet. 2014;10:e1004462. https://doi.org/10.1371/journal.pgen.1004462
Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht T, O’Laughlin M, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25:379–92. https://doi.org/10.1016/j.ccr.2014.01.031
Potter N, Miraki-Moud F, Ermini L, Titley I, Vijaraghavan G, Papaemmanuil E, et al. Single cell analysis of clonal architecture in acute myeloid leukaemia. Leukemia. 2019;33:1113–23. https://doi.org/10.1038/s41375-018-0319-2
Ediriwickrema A, Aleshin A, Reiter JG, Corces MR, Kökhnke T, Stafford M, et al. Single-cell mutational profiling enhances the clinical evaluation of AML MRD. Blood Adv. 2020;4:943–52. https://doi.org/10.1182/bloodadvances.2019001181
Morita K, Wang F, Jahn K, Hu T, Tanaka T, Sasaki Y, et al. Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun. 2020;11:5327. https://doi.org/10.1038/s41467-020-19119-8
Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature. 2020;587:477–82. https://doi.org/10.1038/s41586-020-2864-x
García-Álvarez M, Yeguas A, Jiménez C, Medina-Herrera A, González-Calle V, Hernández-Ruano M, et al. Single-cell DNA sequencing and immunophenotypic profiling to track clonal evolution in an acute myeloid leukemia patient. Biomedicines. 2023;12:66. https://doi.org/10.3390/biomedicines12010066
Gillis S, Roth A. PyClone-VI: scalable inference of clonal population structures using whole genome data. BMC Bioinforma. 2020;21:571. https://doi.org/10.1186/s12859-020-03919-2
Deshwar AG, Vembu S, Yung CK, Jang GH, Stein L, Morris Q. PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 2015;16:35. https://doi.org/10.1186/s13059-015-0602-8
Grigoriadis K, Huebner A, Bunkum A, Colliver E, Frankell AM, Hill MS, et al. CONIPHER: a computational framework for scalable phylogenetic reconstruction with error correction. Nat Protoc. 2024;19:159–83. https://doi.org/10.1038/s41596-023-00913-9
Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol. 2014;10:e1003665. https://doi.org/10.1371/journal.pcbi.1003665
Cun Y, Yang TP, Achter V, Lang U, Peifer M. Copy-number analysis and inference of subclonal populations in cancer genomes using Sclust. Nat Protoc. 2018;13:1488–501. https://doi.org/10.1038/nprot.2018.033
Xiao Y, Wang X, Zhang H, Ulintz PJ, Li H, Guan Y. FastClone is a probabilistic tool for deconvoluting tumor heterogeneity in bulk-sequencing samples. Nat Commun. 2020;11:4469. https://doi.org/10.1038/s41467-020-18169-2
Fischer A, Vázquez-García I, Illingworth CJR, Mustonen V. High-definition reconstruction of clonal composition in cancer. Cell Rep. 2014;7:1740–52. https://doi.org/10.1016/j.celrep.2014.04.055
Popic V, Salari R, Hajirasouliha I, Kashef-Haghighi D, West RB, Batzoglou S. Fast and scalable inference of multi-sample cancer lineages. Genome Biol. 2015;16:91. https://doi.org/10.1186/s13059-015-0647-8
Zare H, Wang J, Hu A, Weber K, Smith J, Nickerson D, et al. Inferring clonal composition from multiple sections of a breast cancer. PLoS Comput Biol. 2014;10:e1003703. https://doi.org/10.1371/journal.pcbi.1003703
Jiang Y, Qiu Y, Minn AJ, Zhang NR. Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing. Proc Natl Acad Sci USA. 2016;113:E5528–37. https://doi.org/10.1073/pnas.1522203113
Caravagna G, Sanguinetti G, Graham TA, Sottoriva A. The MOBSTER R package for tumour subclonal deconvolution from bulk DNA whole-genome sequencing data. BMC Bioinforma. 2020;21:531. https://doi.org/10.1186/s12859-020-03863-1
Satas G, Raphael BJ. Tumor phylogeny inference using tree-constrained importance sampling. Bioinformatics. 2017;33:i152–60. https://doi.org/10.1093/bioinformatics/btx270
Deveau P, Colmet Daage L, Oldridge D, Bernard V, Bellini A, Chicard M, et al. QuantumClone: clonal assessment of functional mutations in cancer based on a genotype-aware method for clonal reconstruction. Bioinformatics. 2018;34:1808–16. https://doi.org/10.1093/bioinformatics/bty016
Myers MA, Satas G, Raphael BJ. CALDER: inferring phylogenetic trees from longitudinal tumor samples. Cell Syst. 2019;8:514–22.e5. https://doi.org/10.1016/j.cels.2019.05.010
Wintersinger JA, Dobson SM, Kulman E, Stein LD, Dick JE, Morris Q. Reconstructing complex cancer evolutionary histories from multiple bulk DNA samples using Pairtree. Blood Cancer Discov. 2022;3:208–19. https://doi.org/10.1158/2643-3230.BCD-21-0092
Malikic S, McPherson AW, Donmez N, Sahinalp CS. Clonality inference in multiple tumor samples using phylogeny. Bioinformatics. 2015;31:1349–56. https://doi.org/10.1093/bioinformatics/btv003
Zheng L, Niknafs N, Wood LD, Karchin R, Scharpf RB. Estimation of cancer cell fractions and clone trees from multi-region sequencing of tumors. Bioinformatics. 2022;38:3677–83. https://doi.org/10.1093/bioinformatics/btac367
El-Kebir M, Oesper L, Acheson-Field H, Raphael BJ. Reconstruction of clonal trees and tumor composition from multi-sample sequencing data. Bioinformatics. 2015;31:i62–70. https://doi.org/10.1093/bioinformatics/btv261
Jiao W, Vembu S, Deshwar AG, Stein L, Morris Q. Inferring clonal evolution of tumors from single nucleotide somatic mutations. BMC Bioinforma. 2014;15:35. https://doi.org/10.1186/1471-2105-15-35
Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars FG, Hinai AA, Zeilemaker A, et al. Molecular minimal residual disease in acute myeloid leukemia. N. Engl J Med. 2018;378:1189–99. https://doi.org/10.1056/NEJMoa1716863
Gaidzik VI, Weber D, Paschka P, Kaumanns A, Krieger S, Corbacioglu A, et al. DNMT3A mutant transcript levels persist in remission and do not predict outcome in patients with acute myeloid leukemia. Leukemia. 2018;32:30–7. https://doi.org/10.1038/leu.2017.200
Tettero JM, Freeman S, Buecklein V, Venditti A, Maurollo L, Kern W, et al. Technical aspects of flow cytometry-based measurable residual disease quantification in acute myeloid leukemia: experience of the European LeukemiaNet MRD Working Party. Hemasphere. 2021;6:e676. https://doi.org/10.1097/HS9.0000000000000676
Krakow EF, Gyurkocza B, Storer BE, Chauncey TR, McCune JS, Radich JP, et al. Phase I/II multisite trial of optimally dosed clofarabine and low-dose TBI for hematopoietic cell transplantation in acute myeloid leukemia. Am J Hematol. 2020;95:48–56. https://doi.org/10.1002/ajh.25665
Archer. Analysis 6.0 User Manual. https://cdn2.hubspot.net/hubfs/4445440/CS001.C%20Archer%20Analysis%206.0%20User%20Manual.pdf. Accessed 24 Mar 2025.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. https://doi.org/10.1038/nmeth0410-248
Tanner G, Westhead DR, Droop A, Stead LF. Benchmarking pipelines for subclonal deconvolution of bulk tumour sequencing data. Nat Commun. 2021;12:6396. https://doi.org/10.1038/s41467-021-26698-7
Chang WC, Joe, Allaire J, Sievert C, Schloerke B, Xie Y. R-Shiny. https://CRAN.R-project.org/package=shiny. Accessed 24 March 2025.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. https://doi.org/10.1038/gim.2015.30
Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. https://doi.org/10.1016/j.jmoldx.2016.10.002
Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–76. https://doi.org/10.1182/blood-2014-11-610543
Song J, Zhang H, Dong N, Zhang X, Hussaini M, Jain A, et al. The application of NextGen sequencing in the diagnosis of myeloid neoplasms in myeloma patients with cytopenia. Clin Lymphoma Myeloma Leuk. 2022;22:e414–26. https://doi.org/10.1016/j.clml.2021.12.008
Padella A, Ghelli Luserna Di Rorà A, Marconi G, Ghetti M, Martinelli G, Simonetti G. Targeting PARP proteins in acute leukemia: DNA damage response inhibition and therapeutic strategies. J Hematol Oncol. 2022;15:10. https://doi.org/10.1186/s13045-022-01228-0
Bischof L, Ussmann J, Grimm J, Bill M, Brauer D, Backhaus D, et al. Prognostic impact of measurable residual clonal hematopoiesis in acute myeloid leukemia patients after allogeneic hematopoietic stem cell transplantation. Leukemia. 2024;38:198–201. https://doi.org/10.1038/s41375-023-02072-y
Lamba R, Abella E, Kukuruga D, Savasan S, Abidi MH, Mohamed A, et al. Mixed hematopoietic chimerism at day 90 following allogenic myeloablative stem cell transplantation is a predictor of relapse and survival. Leukemia. 2004;18:1681–6. https://doi.org/10.1038/sj.leu.2403468
Craddock C, Jackson A, Loke J, Siddique S, Hodgkinson A, Mason J, et al. Augmented reduced-intensity regimen does not improve postallogeneic transplant outcomes in acute myeloid leukemia. J Clin Oncol. 2021;39:768–78. https://doi.org/10.1200/JCO.20.02308
Lindahl H, Vonlanthen S, Valentini D, Björklund AT, Sundin M, Mielke S, et al. Lineage-specific early complete donor chimerism and risk of relapse after allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia. Bone Marrow Transpl. 2022;57:753–9. https://doi.org/10.1038/s41409-022-01615-8
Bader P, Kreyenberg H, Hoelle W, Dueckers G, Handgretinger R, Lang P, et al. Increasing mixed chimerism is an important prognostic factor for unfavorable outcome in children with acute lymphoblastic leukemia after allogeneic stem-cell transplantation: possible role for pre-emptive immunotherapy? J Clin Oncol. 2004;22:1696–705. https://doi.org/10.1200/JCO.2004.05.198
Lindahl H, Valentini D, Vonlanthen S, Sundin M, Björklund AT, Mielke S, et al. Early relapse prediction after allogeneic hematopoietic stem cell transplantation for acute lymphoblastic leukemia (ALL) using lineage-specific chimerism analysis. EJHaem. 2022;3:1277–86. https://doi.org/10.1002/jha2.568
Saygin C, Cannova J, Stock W, Muffly L. Measurable residual disease in acute lymphoblastic leukemia: methods and clinical context in adult patients. Haematologica. 2022;107:2783–93. https://doi.org/10.3324/haematol.2022.280638
Lovisa F, Zecca M, Rossi B, Campeggio M, Magrin E, Giarin E, et al. Pre- and post-transplant minimal residual disease predicts relapse occurrence in children with acute lymphoblastic leukaemia. Br J Haematol. 2018;180:680–93. https://doi.org/10.1111/bjh.15086
Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604. https://doi.org/10.1038/ncomms7604
Ishida H, Iguchi A, Aoe M, Takahashi T, Tamefusa K, Kanamitsu K, et al. Panel-based next-generation sequencing identifies prognostic and actionable genes in childhood acute lymphoblastic leukemia and is suitable for clinical sequencing. Ann Hematol. 2019;98:657–68. https://doi.org/10.1007/s00277-018-3554-8
Qin G, Dai J, Chien S, Martin TJ, Loera B, Nguyen QH, et al. Mutation Patterns Predict Drug Sensitivity in Acute Myeloid Leukemia. Clin Cancer Res. 2024;30:2659–71. https://doi.org/10.1158/1078-0432.CCR-23-1674. 14
Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu J, et al. Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol Oncol. 2021;14:91. https://doi.org/10.1186/s13045-021-01105-2
Sarkar A, Stephens M. Separating measurement and expression models clarifies confusion in single-cell RNA sequencing analysis. Nat Genet. 2021;53:770–77. https://doi.org/10.1038/s41588-021-00873-4
Benard BA, Leak LB, Azizi A, Thomas D, Gentles AJ, Majeti R. Clonal architecture predicts clinical outcomes and drug sensitivity in acute myeloid leukemia. Nat Commun. 2021;12:7244. https://doi.org/10.1038/s41467-021-27472-5
Paguirigan AL, Smith J, Meshinchi S, Carroll M, Maley C, Radich JP. Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci Transl Med. 2015;7:281re2. https://doi.org/10.1126/scitranslmed.aaa0763
Hernandez-Boussard T, Macklin P, Greenspan EJ, Gryshuk AL, Stahlberg E, Syeda-Mahmood T, et al. Digital twins for predictive oncology will be a paradigm shift for precision cancer care. Nat Med. 2021;27:2065–66. https://doi.org/10.1038/s41591-021-01558-5
Acknowledgements
Sources of funding included the Archer Reagent Challenge Grant (CY), P30 CA015704, and P01 CA078902 (BS).
Author information
Authors and Affiliations
Contributions
EFK identified the clinical need and conceived the CloneTracker platform as detailed in Fig. 1 (that she created), assembled and led the research team, obtained IRB approval for donor sample sequencing, collected additional clinical data through chart review, and drafted the manuscript. CY and IB conceived of using incidentally-sequenced SNPs from the targeted gene panel sequencing output to identify clones. IB supervised PhD student NL, who evaluated, compared, selected and refined algorithms for clustering gene variants and inferring phylogenetic trees, and incorporated information from sex chromosomes in sex-matched and sex-mismatched transplant settings. NL performed statistical analysis comparing clonal prevalence in relapsed to non-relapsed patients, and created Fig. 2. EFK performed statistical analysis in Table 1 and created Fig. 3. LB and OST performed the Archer sequencing in the lab of JR. CY and OST evaluated the accuracy and pathogenicity of each putative gene variant, performed quality control, and established the filtering parameters with input from NL and IB, and they were implemented by NL. OST developed the harmonized vcf template with input from IB, CY, JR and EFK. IB provided simple criteria to assign donor origin to a clone. IJ developed the fishplot code and the visualization dashboard in R-Shiny. BF refined the dashboard, implemented the overall workflow (pipeline), and annotated the code in preparation for sharing it with future collaborators. BS designed, obtained IRB approval, and led the clinical trial in which serial patient samples were collected, and provided clinical data. OST, CY, NL and EFK drafted the eMethods supplement. OST prepared the supplemental Excel files. EFK, NL, CY, JR and IB interpreted the clonal dynamics in the context of the clinical treatment-and-response trajectories, patient-by-patient. All authors reviewed and edited the manuscript and supplemental files. Funding was provided by EFK, IB, CY, JR and BS.
Corresponding authors
Ethics declarations
Competing interests
Authors EFK, NL, IB, IJ, JR, OST and CY are inventors of patent applications filed by FHCC directed to the system for tracking cancer clones alongside clinical data as described in this manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Krakow, E.F., Lee, N., Jenkins, I. et al. A clinical solution for tracking clonal evolution of acute myeloid leukemia after allogeneic transplantation using bulk next generation sequencing. Bone Marrow Transplant 60, 1083–1091 (2025). https://doi.org/10.1038/s41409-025-02602-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41409-025-02602-5