Abstract
Background
Although high T cell density is a strong favourable prognostic factor in colorectal cancer, the significance of the spatial distribution of T cells is incompletely understood. We aimed to evaluate the prognostic significance of tumour cell-T cell co-localisation and T cell densities.
Methods
We analysed CD3 and CD8 immunohistochemistry in a study cohort of 983 colorectal cancer patients and a validation cohort (N = 246). Individual immune and tumour cells were identified to calculate T cell densities (to derive T cell density score) and G-cross function values, estimating the likelihood of tumour cells being co-located with T cells within 20 µm radius (to derive T cell proximity score).
Results
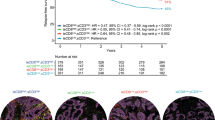
High T cell proximity score associated with longer cancer-specific survival in both the study cohort [adjusted HR for high (vs. low) 0.33, 95% CI 0.20–0.52, Ptrend < 0.0001] and the validation cohort [adjusted HR for high (vs. low) 0.15, 95% CI 0.05–0.45, Ptrend < 0.0001] and its prognostic value was independent of T cell density score.
Conclusions
The spatial point pattern analysis of tumour cell-T cell co-localisation could provide detailed information on colorectal cancer prognosis, supporting the value of spatial measurement of T cell infiltrates as a novel, robust tumour-immune biomarker.
Similar content being viewed by others
Background
Colorectal cancer is the third most common cancer, covering around 10% of all new cancer cases worldwide [1]. The tumour microenvironment is composed of neoplastic tumour cells and non-neoplastic cells, such as host immune cells, interacting through cell–cell contacts and inflammatory mediators [2]. The assessment of colorectal cancer prognosis and treatment is mainly based on evaluating neoplastic tumour cells and tumour spread rather than analysing the host immune response [3]. The most widely used clinical staging system is the American Joint Committee on Cancer/International Union Against Cancer (AJCC/UICC) TNM classification, which includes the extent of the primary tumour (T), presence of lymph node metastasis (N) and spread to distant organs (M), while World Health Organization (WHO) histologic grading categorises tumours according to their differentiation [4]. However, these methods do not entirely capture the characteristics of colorectal tumours and their prognoses.
Immunoscore® is a T cell scoring system based on computer-assisted quantification of CD3+ and CD8+ cell densities in the tumour centre and the invasive margin [5, 6]. T cell density varies within a single tumour and between different tumours. The density is generally higher in the invasive margin than in the centre of the tumour. In mismatch repair (MMR)-deficient colorectal cancers, T cell densities are also usually higher than in MMR proficient tumours [7, 8]. High Immunoscore® has been associated with better prognosis and has been internationally validated as an independent prognostic parameter in a cohort of more than 2600 disease stage I–III colorectal cancer cases [6]. However, most studies evaluating immune cell infiltrates in colorectal cancer have been limited to density-based analyses [9] and the significance of the co-localisation between tumour cells and T cells is not well-established.
In this study, we used immunohistochemistry and digital image analysis to identify CD3+ and CD8+ cells and tumour cells in 1229 colorectal cancer samples, including a study cohort of 983 patients and an independent validation cohort of 246 cases. We present the T cell proximity score as a novel prognostic parameter based on the evaluation of co-localisation of tumour cells with T cells. Our primary aim was to evaluate the prognostic value of the T cell proximity score and compare it to that of the T cell density score (based on the principles of Immunoscore®). We hypothesised that a high T cell proximity score (high likelihood of tumour cells being co-located with T cells) might be associated with favourable outcome. As secondary aims, we investigated the associations of T cell proximity score with tumour and patient characteristics and the prognostic significance of spatial T cell proximity measurements separately in the tumour centre and the invasive margin, in MMR proficient and MMR deficient tumour subgroups as well as in low and high disease stage tumours.
Methods
Patients
We identified 1343 patients who underwent resection for colorectal cancer at Central Finland Central Hospital between January 1, 2000 and December 31, 2015 and had adequate tumour samples available. The samples were retrospectively collected from the pathology registry of Central Finland Central Hospital, which covers all colorectal cancers diagnosed in Central Finland (the population of the area-averaged around 270,000 during the study period) [10]. Associated clinical data were collected from clinical patient records by study physicians. We excluded patients who died within 30 days after surgery (N = 40) or received any preoperative oncological treatments (radiotherapy, chemotherapy or chemoradiotherapy) (N = 243) due to their potential influences on tumour characteristics [11]. The final cohort with adequate samples in tissue microarrays and successful quantification of CD3+ and CD8+ cells included 983 patients. The median follow-up time for censored was 9.3 years (IQR 6.8–13.3 years). The main clinicopathologic features of the cases are shown in Table 1. Histological tumour parameters were re-evaluated by the study pathologist (J.P.V.), including tumour differentiation and lymphovascular invasion, using hematoxylin and eosin (H&E) stained whole slides. All the histological analyses were performed blinded to the clinical data.
Tissue microarrays
For tissue microarray construction, we selected one representative formalin-fixed paraffin-embedded tumour sample with the deepest cancer invasion for each patient. The arrays were constructed using a TMA Master II tissue microarrayer (3DHistech Ltd., Budapest, Hungary), and they included two 1 mm-diameter cores from representative areas of the tumour centre and the invasive margin (total: four cores). The core sites were annotated to best represent overall tumour morphology while avoiding necrosis. The invasive margin cores were targeted to span 500 µm into the healthy tissue and 500 µm into the tumour. In total, the cohort included 25 tissue microarray blocks, each containing two tonsil cores as staining controls. Tissue microarray blocks were tempered overnight at 60 °C and cut at 3.5 µm thickness.
Immunohistochemistry
The samples were screened for DNA mismatch repair (MMR) deficiency with MLH1, MSH2, MSH6 and PMS2 immunohistochemistry and for BRAF V600E mutation status with immunohistochemistry [12]. Immunohistochemistry for T cells and MMR genes were performed by BOND-III automated IHC stainer (Leica Biosystems, Buffalo Grove, IL, USA) with monoclonal antibodies and protocols shown in Table S1. All antibodies were in clinical use in the pathology laboratory of Central Finland Central Hospital, and appropriate staining was also confirmed by examining positive and negative controls. After bake, dewax and peroxide block, slides were processed with heat-induced antigen retrieval with EDTA-based buffer, pH 9.0 (BOND Epitope retrieval solution 2, Leica Biosystems, AR9640) at 100 °C. Antigen retrieval time was 30 min for MLH1, MSH2, MSH6 and PMS2 and 20 min for CD3 and CD8. Primary antibodies were incubated for 30 min. Visualisation was performed according to the manufacturer’s instructions using a BOND Polymer Refine Detection kit (Leica Biosystems, DS9800) with a horseradish peroxidase-conjugated secondary antibody (<25 µg/ml), 3.3-diaminobenzidine chromogen and hematoxylin (0.1%) counterstain. Stained slides were coverslipped with a Tissue-Tek Glas Automated Glass Coverslipper (Sakura) and digitalised with a NanoZoomer-XR (Hamamatsu Photonics, Hamamatsu City, Japan) slide scanner with a ×20 objective.
Immunohistochemistry to evaluate BRAF V600E mutation status was conducted using a BenchMark XT immunostainer (Ventana Medical Systems, Tucson, AZ) and a BRAF V600E mutation-specific mouse monoclonal antibody (clone: VE1, Spring Bioscience, Pleasonton, CA, US, dilution: 1:400). The amplification was done with OptiView Amplification (Ventana [13]).
Image analysis
T cell analyses were conducted with supervised machine learning approaches built-in QuPath (version 0.2.3), an open-source bioimage analysis software [14], using previously validated algorithms [15]. The software was trained to recognise tissue and cell types by manually annotating representative areas/cells. The identification of tissue from the background was done with the random forests pixel classifier. Cells were detected and classified into tumour cells, T cells, and other cells using the random forests object classifier. T cells were recognised by CD3 or CD8 expression and tumour cells were identified through their morphology. The remaining cells were classified as other. The workflow for image analysis is shown in Fig. S1. We confirmed the validity of the automated cell classifier by reviewing the classification result images. We further quantified the accuracy of the classifier by manually annotating each cell in 50 tumour regions (25 stained for CD3 and 25 stained for CD8; size 200 × 200 µm) and comparing the cell densities observed in these regions with those obtained with the automated classifier using the Spearman’s rank correlation test.
All tissue microarray cores were reviewed, and those with folding or detaching during processing, minimal amount or absence of tumour, or high amount of necrosis were excluded from the analyses. For the final analyses, we included only cases that at least one representative successfully analysed the tumour core from the tumour centre and the invasive margin for both CD3 and CD8. For each tissue microarray core, we calculated CD3+ and CD8+ cell densities by dividing the cell counts by the tumour core area in mm2. For tumours with multiple successfully analysed tumour centre cores or invasive margin cores, we calculated mean cell densities. As a result, each tumour had one density value for (1) CD3+ cells in the tumour centre, (2) CD3+ cells in the invasive margin, (3) CD8+ cells in the tumour centre and (4) CD8+ cells in the invasive margin.
To calculate the T cell density score, we followed the main principles of the Immunoscore assay [6]. The densities of CD3+ cells in the tumour centre, CD3+ cells in the invasive margin, CD8+ cells in the tumour centre and CD8+ cells in the invasive margin were converted to percentiles (0–100), which resulted in four separate percentile values for each tumour. T cell density score was determined by calculating the mean of the four percentiles and categorising it to low (0–25), intermediate (>25–70) or high (>70–100). The workflow for T cell density score analysis is shown in Fig. 1.
The figure shows analysis steps for one example tumour core from the tumour centre (CT) and the invasive margin (IM). Tumour cores stained with CD3 and CD8 (a) and corresponding phenotyping maps for T cells, tumour cells and other cells (b). G-cross (Gtumour:T cell) function curves, representing the likelihood of any tumour cell in the sample being co-located with at least one CD3+/CD8+ T cell within a radius r (c). Calculation charts for T cell proximity score (d) and for T cell density score (e). Respective example images for two remaining cores of the same sample are shown in Fig. S2.
In this study, we introduced the T cell proximity score as a new prognostic parameter based on tumour cell-T cell co-localisation. We estimated the empirical G-cross [Gtumour:T cell (r)] function for each sample, evaluating the likelihood of any tumour cell in the sample having at least one T cell at a specific radius r. The function is formed by measuring the distances from each tumour cell centroid to the closest immune cell centroid. Thus, higher G-cross function values result from a higher percentage of tumour cells harbouring T cells in their proximity and indicate greater co-localisation of tumour cells with T cells. We chose to examine the function values at 20 µm radius to identify T cell populations likely capable of effective, direct, cell-to-cell interaction with tumour cells, consistent with previous reports [15,16,17,18]. We applied the Kaplan–Meier correction for edge effects. T cell proximity score was calculated using a similar approach as in T cell density score. We calculated G-cross [Gtumour:T cell (20 µm)] function values for CD3+ and CD8+ cells in the tumour centre and in the invasive margin and converted these four values into percentiles. To determine the T cell proximity score for each tumour, we calculated the mean of the four percentiles and categorised it into low (0–25), intermediate (>25–70) or high (>70–100). In sensitivity analysis, we tested the T cell proximity score at G-cross function radii of 10, 30, 40, 50, 100 and 500 µm. The workflow for the proximity score analysis for two example cores of the same tumour is shown in Fig. 1. Figure S2 represents the respective analysis for the two remaining cores of the example tumour.
Validation cohort
For validation, we retrospectively analysed an independent, previously described colorectal cancer cohort operated at Oulu University Hospital from 2006 to 2014 [19]. Patients with preoperative treatment and unsuccessful CD3+ or CD8+ cell analysis were excluded, and the final data included 246 patients. The median follow-up time for censored cases was 6.2 years (IQR 5.0–7.6 years). Analyses were conducted for one to four 3 mm-diameter TMA cores per patient [19]. Antibodies and staining protocols are shown in Table S2. The densities and G-cross function values were compared with those obtained in the study cohort to convert them into percentiles.
Statistical analyses
Statistical analyses were performed using RStudio (version 1.3.1093) and R statistical programming (version 4.0.5, R Core Team) with packages gmodels (2.18.1), spatstat (2.1-0), survival (3.2-7), survminer (0.4.9) and tidyverse (1.3.0).
Categorical data were analysed by cross-tabulation of T cell scores and other variables and using Chi-square test to evaluate the statistical significance. Kaplan–Meier method was used for visualising the cumulative survival probabilities, and the comparison between categories was done with the Log-rank test. As our primary analyses, we utilised univariable and multivariable Cox proportion hazard regression to estimate mortality hazard ratio (HR) point estimates and their 95% confidence intervals (CIs). Cancer-specific survival was evaluated as the primary endpoint, and it was defined as the time from surgery to cancer death. Overall survival was evaluated as the secondary endpoint, and it was defined as the time between colorectal cancer surgery and death. We limited the follow-up to 10 years, considering that most colorectal cancer deaths occur within that period. Schoenfeld residual plots supported the proportionality of hazards during most of the follow-up period up to 10 years. Multivariable models included the following pre-determined indicator variables (with the reference category listed first): sex (male, female), age (<65, 65–75, >75), year of operation (2000–2005, 2006–2010, 2011–2015), tumour location (proximal colon, distal colon, rectum), disease stage (I–II, III, IV), tumour grade (well/moderately differentiated, poorly differentiated), lymphovascular invasion (negative, positive), MMR status (proficient, deficient), BRAF status (wild-type, mutant). Cases with missing data (validation cohort only) were included in the majority category of a given categorical covariate to limit the degrees of freedom. The following covariates had missing values in the validation cohort: disease stage (0.4% missing), differentiation (0.4% missing), lymphovascular invasion (1.2% missing), MMR status (0.4% missing). Excluding those missing cases in each covariate did not substantially alter results. We used a stringent alpha level of 0.005 according to the recommendation of an expert panel [20].
Results
Patient characteristics
We analysed T cell infiltrates in tumour samples of 983 colorectal cancer patients. Of the patients, 481 (49%) were women and the median age at the time of surgery was 72 years (range 36–100 years). The most prevalent site for the primary tumour was the proximal colon (cecum to transverse colon) with 478 (49%) cases. MMR deficiency was detected in 150 (15%) tumours (Table 1).
Cell analysis of tissue microarray cores
We successfully analysed a total of 2,351,513 CD3+ cells from 3632 tissue microarray cores and 1,105,424 CD8+ cells from 3608 tissue microarray cores. The average number of analysed cores was 3.7 per patient for both CD3 and CD8. Core-to-core correlations for G-cross (Gtumour:T cell) function values at 20 µm radius were 0.69 for CD3+ cells in the tumour centre 0.70 for CD3+ cells in the invasive margin, 0.69 for CD8+ cells in the tumour centre and 0.70 for CD8+ cells for the invasive margin (Fig. S3), being slightly higher than the respective core-to-core correlations for T cell densities (Fig. S4).
We tested the accuracy of machine-learning-based image analysis by manually annotating all cells (T cells, tumour cells and other cells) in 50 tumour images and then applying the optimised automated classifier to these images. The total number of detected cells was 11,312 in manual counting and 12,097 in automated cell counting. The Spearman’s rank correlations coefficient between automated and manual cell densities was 0.94 for T cells, 0.87 for tumour cells and 0.86 for other cells, indicating that the classifier had reached good accuracy (Fig. S5).
T cell proximity and density score
T cell proximity score was calculated based on tumour cell-T cell co-localisation measurements, using Gtumour:T cell function values at 20 µm (Fig. 1). Examples of tissue microarray cores with distinct T cell infiltration patterns and corresponding G-cross function curves are shown in Fig. S6. Of the patients, 545 (55%) cases were classified as intermediate for the proximity score, whereas low covered 20% and high covered 25% of the cases. High T cell proximity score was strongly associated with proximal tumour location (P = 0.0003), low disease stage, absence of lymphovascular invasion, MMR deficiency and BRAF mutation (all P < 0.0001; Table 1).
T cell density score was calculated based on the mean CD3+ and CD8+ cell densities in the tumour centre and the invasive margin according to the principles of Immunoscore® (Fig. 1). Like the proximity score, high density score was associated with proximal tumour location (P = 0.003), low disease stage (P = 0.0002), absence of lymphovascular invasion (P = 0.0004), MMR deficiency (P < 0.0001) and BRAF mutation (P = 0.001; Table S3).
Survival analyses
In total, there were 574 (58%) deaths including 278 (28%) colorectal cancer deaths. The 5-year and 10-year cancer-specific survival rates were 74% and 69% and overall survival rates were 61% and 46%, respectively.
Our primary aim was to evaluate the prognostic significance of the T cell proximity score and compare it to that of the T cell density score. High proximity and density scores predicted improved outcomes compared to low scores. The 10-year cancer-specific survival for patients with high and low proximity scores were 88% and 48%, respectively (Fig. 2, Table 2). High T cell proximity and density scores were associated with better cancer-specific and overall survival both in univariable and multivariable analyses. In cancer-specific survival analysis, the multivariable HR for high (vs. low) T cell proximity score was 0.33 (95% CI 0.20–0.52, Ptrend < 0.0001) and for high (vs. low) density score 0.47 (95% CI 0.31–0.73, Ptrend = 0.0007) (Table 2, Table S4).
To directly compare the prognostic value of T cell proximity and density scores for cancer-specific survival, we included these variables in the same multivariable Cox regression model for reciprocal adjustment (Table 3). This analysis indicated that the prognostic significance of the proximity score (Ptrend = 0.001) was independent of the density score (Ptrend = 0.75).
Of the proximity score high cases (N = 244), 179 (73%) cases were high and 65 (27%) were intermediate for the density score. Of the proximity score low cases (N = 196), 121 (62%) were low and 75 (38%) were intermediate for the density score. We categorised the tumours into four subgroups to evaluate the combined prognostic effect of the proximity and density scores (Fig. S7, Table S5). This analysis further indicated that a high T cell proximity score was associated with better survival regardless of the density score.
In secondary analyses, we evaluated the survival associations of the four components (CD3+ and CD8+ cells in the tumour centre and the invasive margin) of T cell proximity [(Gtumour:T cell) at a 20 µm radius] and density scores as ordinal quartile categories (Table 4, Fig. S8). In this analysis, higher values in all four components of both T cell proximity and T cell density score, except CD8+ cell density in the tumour centre, were statistically significantly associated with longer cancer-specific survival (all Ptrend < 0.005). The HR point estimates suggested stronger survival associations for the measurements based on the invasive margin, as compared to the tumour centre, and for the G-cross proximity estimates, as compared to the densities.
For sensitivity analysis, modified T cell proximity scores were derived from Gtumour:T cell function values at different radii (10–50, 100 and 500 µm) (Table S6). Univariable and multivariable Cox regression models for cancer-specific survival indicated strong prognostic associations for the proximity scores at 10–50 µm and 100 µm radii (all Ptrend < 0.0001), but not at 500 µm radius (Ptrend = 0.22). These results support the significance of tumour cell-T cell co-localisation within radii of 10–100 µm.
To further evaluate factors potentially influencing the prognostic significance of the T cell proximity score, we investigated the prognostic effect of the proximity score in MMR proficient and deficient tumour subgroups, as well as in different disease stages. The association between a higher T cell proximity score and longer cancer-specific survival did not significantly differ by MMR status (Pinteraction = 0.69) (Table S7), while a higher T cell proximity score was associated with longer cancer-specific survival in stages I–III but not in stage IV (Pinteraction < 0.0001) (Fig. S9, Tables S8, S9).
Validation cohort
We analysed an independent validation cohort of 246 patients. The clinicopathologic features for the validation cohort (N = 246) are shown in Table S10. High T cell proximity score was associated with low disease stage (P < 0.0001), absence of lymphovascular invasion (P = 0.0002), MMR deficiency (P < 0.0001) and BRAF mutation (P = 0.0002).
In total, there were 80 (33%) deaths including 58 (24%) colorectal cancer deaths. In this cohort, T cell proximity score was associated with longer cancer-specific survival [multivariable HR for high (vs. low) 0.15, 95% CI 0.05–0.45, Ptrend < 0.0001; Table S11]. As in the main cohort, the prognostic association of the proximity score was independent of the density score (Table S12).
Discussion
We investigated the spatial distribution and density of CD3+ and CD8+ cells in a large, population-based cohort of 983 colorectal cancer patients and an independent validation cohort of 246 colorectal cancer cases. T cell density is a well-established favourable prognostic parameter in colorectal cancer [9], but relatively few studies [15,16,17,18, 21,22,23,24] have investigated the role of the spatial organisation of immune cell infiltrates in cancer. G-cross function has been previously used for analyzing the spatial interactions of T cells and tumour cells in primary tumours [15, 17] and in liver metastases [22] of colorectal cancer, in non-small cell lung cancer [16] and in pancreatic cancer [18]. In this study, we utilised CD3 and CD8 immunohistochemistry and quantified G-cross (Gtumour:T cell) function values at 20 μm radius at the invasive margin and tumour centre and established T cell proximity score as a new, reproducible system for analyzing the co-localisation of tumour cells with T cells. Our main finding was that a high T cell proximity score was associated with favourable outcomes independent of potential confounding factors such as disease stage and MMR status, as well as T cell density score. We envision that the method would be applicable to the analysis of a variety of other solid tumours and could be used as a quantitative tumour-immune biomarker, evaluating not only the density but also the spatial patterns of T cell infiltrates in the tumour.
Our findings were consistent with a recent study in colorectal cancer, where the co-localisation of tumour cells with CD3+ T cells within 20 μm radius was a stronger prognostic factor than total T cell density in the tumour microenvironment [17]. Moreover, previous studies have demonstrated that strong engagement and mixing of CD8+ cytotoxic T cells with tumour cells in colorectal cancer liver metastases are associated with favourable outcomes [21, 22]. In our study of primary colorectal cancer, the prognostic value of the spatial measurements for CD3+ cells was as significant as those for CD8+ cells both in the tumour centre and in the invasive margin. The present study represents a comprehensive analysis of tumour cell-T cell co-localisation in two large colorectal cancer cohorts, with detailed clinicopathologic characterisation [17].
T cell proximity score, introduced in this study, specifically evaluates the co-localisation of tumour cells with T cells within 20 µm radius, ignoring T cells located further from tumour cells. In our analyses, the proximity score had a higher prognostic value than the density score. We hypothesise that this may be related to the proximity score focusing on T cells with the potential for direct cell–cell interactions with tumour cells, such as cytotoxicity [15, 16, 18, 24]. It is conceivable that distant T cells may have a reduced possibility for anti-tumoural activity compared with T cells in close tumour proximity. Moreover, a high stromal percentage predicts an unfavourable prognosis in colorectal cancer, while a low stromal percentage is associated with favourable outcomes [25, 26]. Considering that the majority of immune cells in the colorectal cancer microenvironment are located in tumour stromal rather than intraepithelial regions [15, 27, 28], the tumours with low stroma percentage might have low overall T cell densities as a result of low stromal content rather than a weak anti-tumour-immune response, while spatial point pattern analysis may still classify these tumours into higher T cell proximity score categories if T cells are located close to tumour cells. All four components of the proximity score (CD3+ and CD8+ cells in tumour centre and in invasive margin) had strong prognostic significance as separate variables. In addition, our sensitivity analyses showed that high G-cross (Gtumour:T cell) function values were associated with better prognosis within a range of radii between 10–50 µm and 100 µm, but not at 500 µm. These findings further highlight the robustness of the analysis, not dependent on a single, specific radius or component, and supports the potential of T cell proximity score as a relevant prognostic factor in colorectal cancer.
We identified the cell types with a machine-learning-based cell classifier using the QuPath software. We confirmed the adequacy of the classifier by manually viewing all result images and tested the accuracy of our cell classifier by comparing the densities of manually annotated cells and automatedly classified cells, supporting high concordance for all three cell types (T cells, tumour cells, and other cells). The machine-learning-based cell analysis for immunohistochemically stained tumour tissue samples using QuPath has also been validated in previous studies with high accuracy [15, 29, 30].
Some limitations should be considered. First, we used tissue microarrays, which may not totally represent the immunological milieu in the whole tumour [31, 32]. However, we successfully analysed on average 3.7 tumour cores for each patient, which should demonstrate relatively good concordance with the whole tumour [31]. We also observed reasonably good core-to-core correlation for both G-cross and density measurements, suggesting that T cell infiltrates can be evaluated using our tissue microarrays with reasonable accuracy. Moreover, measurement errors related to tissue microarrays would likely have a nearly random distribution, driving our findings towards the null hypothesis. Tissue microarrays also enabled staining of all samples at the same time, so the staining quality was uniform between the specimens. Second, the information on cancer treatment was lacking. Nevertheless, treatments have likely been principally based on the disease stage and MMR status rather than immune infiltrate, and we adjusted the multivariable survival models for several factors, including disease stage and MMR status. Third, we excluded all patients with preoperative treatment from analyses, which led to the under-representation of rectal cancers in these cohorts. The prognostic significance of the T cell proximity score should be interpedently evaluated in rectal cancer patients who have received neoadjuvant treatments. Fourth, although T cells play a critical role in anti-tumoural immunity, this study lacks the prognostic information of other immune cells in the tumour microenvironment. Fifth, most patients were non-Hispanic White, and the prognostic significance of the T cell proximity score should be confirmed in different populations.
There were several strengths in the study. This study included a large, thoroughly analysed study cohort [12, 33,34,35,36], as well as an independent validation cohort, which can forward the generalisability of the findings. The histological parameters were evaluated uniformly in accordance with the latest guidelines and the tumours were screened for two key molecular prognostic parameters (MMR status and BRAF mutation status). The machine learning assessment of immune cell infiltrates enabled uniform analysis throughout cases and spatial point pattern analyses based on positions of single cells [15]. This facilitated more detailed analyses of tumour cell-immune cell co-localisation than possible using traditional methods.
In conclusion, this study showed that the T cell proximity score, derived from G-cross measurements of co-localisation of tumour cells with T cells, was strongly associated with the survival of colorectal cancer patients, representing a new, quantitative prognostic parameter for colorectal cancer. Our results highlight the importance of the spatial context in the analysis of immune cell infiltrates in cancer and could be utilised to develop improved tumour-immune biomarkers for precision medicine.
Data availability
The datasets generated and/or analysed during this study are not publicly available. The sharing of data will require approval from relevant ethics committees and/or biobanks. Further information including the procedures to obtain and access data of Finnish Biobanks are described at https://finbb.fi/en/fingenious-service.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20:651–68.
Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20:662–80.
Argilés G, Tabernero J, Labianca R, Hochhauser D, Salazar R, Iveson T, et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31:1291–305.
Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, et al. Towards the introduction of the “Immunoscore” in the classification of malignant tumours. J Pathol. 2014;232:199–209.
Pagès F, Mlecnik B, Marliot F, Bindea G, Ou F-S, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet (Lond, Engl). 2018;391:2128–39.
Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17:79–92.
Dahlin AM, Henriksson ML, Van Guelpen B, Stenling R, Oberg A, Rutegård J, et al. Colorectal cancer prognosis depends on T-cell infiltration and molecular characteristics of the tumor. Mod Pathol. 2011;24:671–82.
Alexander PG, McMillan DC, Park JH. The local inflammatory response in colorectal cancer—type, location or density? A systematic review and meta-analysis. Cancer Treat Rev. 2020;83:101949.
Väyrynen V, Wirta E-V, Seppälä T, Sihvo E, Mecklin J-P, Vasala K, et al. Incidence and management of patients with colorectal cancer and synchronous and metachronous colorectal metastases: a population-based study. BJS Open. 2020;4:685–92.
Nagtegaal ID, Marijnen CAM, Kranenbarg EK, Mulder-Stapel A, Hermans J, van de Velde CJH, et al. Short-term preoperative radiotherapy interferes with the determination of pathological parameters in rectal cancer. J Pathol. 2002;197:20–7.
Seppälä TT, Böhm JP, Friman M, Lahtinen L, Väyrynen VMJ, Liipo TKE, et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer. 2015;112:1966–75.
Thiel A, Heinonen M, Kantonen J, Gylling A, Lahtinen L, Korhonen M, et al. BRAF mutation in sporadic colorectal cancer and Lynch syndrome. Virchows Arch. 2013;463:613–21.
Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878.
Väyrynen JP, Lau MC, Haruki K, Väyrynen SA, Dias Costa A, Borowsky J, et al. Prognostic significance of immune cell populations identified by machine learning in colorectal cancer using routine hematoxylin and eosin-stained sections. Clin Cancer Res. 2020;26:4326–38.
Barua S, Fang P, Sharma A, Fujimoto J, Wistuba I, Rao AUK, et al. Spatial interaction of tumor cells and regulatory T cells correlates with survival in non-small cell lung cancer. Lung Cancer. 2018;117:73–9.
Väyrynen JP, Haruki K, Lau MC, Väyrynen SA, Ugai T, Akimoto N, et al. Spatial organization and prognostic significance of NK and NKT-like cells via multimarker analysis of the colorectal cancer microenvironment. Cancer Immunol Res. 2021;10:215–28.
Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun. 2017;8:15095.
Väyrynen JP, Tuomisto A, Väyrynen SA, Klintrup K, Karhu T, Mäkelä J, et al. Preoperative anemia in colorectal cancer: relationships with tumor characteristics, systemic inflammation, and survival. Sci Rep. 2018;8:1–11.
Benjamin DJ, Berger JO, Johannesson M, Nosek BA, Wagenmakers E-J, Berk R, et al. Redefine statistical significance. Nat Hum Behav. 2018;2:6–10.
Lazarus J, Maj T, Smith JJ, Perusina Lanfranca M, Rao A, D’Angelica MI, et al. Spatial and phenotypic immune profiling of metastatic colon cancer. JCI insight. 2018;3:e121932.
Lazarus J, Oneka MD, Barua S, Maj T, Lanfranca MP, Delrosario L, et al. Mathematical modeling of the metastatic colorectal cancer microenvironment defines the importance of cytotoxic lymphocyte infiltration and presence of PD-L1 on antigen presenting cells. Ann Surg Oncol. 2019;26:2821–30.
Berthel A, Zoernig I, Valous NA, Kahlert C, Klupp F, Ulrich A, et al. Detailed resolution analysis reveals spatial T cell heterogeneity in the invasive margin of colorectal cancer liver metastases associated with improved survival. Oncoimmunology. 2017;6:e1286436.
Wang M, Huang Y-K, Kong JC, Sun Y, Tantalo DG, Yeang HXA, et al. High-dimensional analyses reveal a distinct role of T-cell subsets in the immune microenvironment of gastric cancer. Clin Transl Immunol. 2020;9:e1127.
van Pelt GW, Kjær-Frifeldt S, van Krieken JHJM, Al Dieri R, Morreau H, Tollenaar RAEM, et al. Scoring the tumor-stroma ratio in colon cancer: procedure and recommendations. Virchows Arch. 2018;473:405–12.
Eriksen AC, Sørensen FB, Lindebjerg J, Hager H, dePont Christensen R, Kjær-Frifeldt S, et al. The prognostic value of tumour stroma ratio and tumour budding in stage II colon cancer. A nationwide population-based study. Int J Colorectal Dis. 2018;33:1115–24.
Väyrynen JP, Haruki K, Väyrynen SA, Lau MC, Dias Costa A, Borowsky J, et al. Prognostic significance of myeloid immune cells and their spatial distribution in the colorectal cancer microenvironment. J Immunother Cancer. 2021;9:e002297.
Yoo S-Y, Park HE, Kim JH, Wen X, Jeong S, Cho N-Y, et al. Whole-slide image analysis reveals quantitative landscape of tumor-immune microenvironment in colorectal cancers. Clin Cancer Res. 2020;26:870–81.
Loughrey MB, Bankhead P, Coleman HG, Hagan RS, Craig S, McCorry AMB, et al. Validation of the systematic scoring of immunohistochemically stained tumour tissue microarrays using QuPath digital image analysis. Histopathology. 2018;73:327–38.
Bankhead P, Fernández JA, McArt DG, Boyle DP, Li G, Loughrey MB, et al. Integrated tumor identification and automated scoring minimizes pathologist involvement and provides new insights to key biomarkers in breast cancer. Lab Invest. 2018;98:15–26.
Voduc D, Kenney C, Nielsen TO. Tissue microarrays in clinical oncology. Semin Radiat Oncol. 2008;18:89–97.
Giltnane JM, Rimm DL. Technology insight: identification of biomarkers with tissue microarray technology. Nat Clin Pract Oncol. 2004;1:104–11.
Ahtiainen M, Wirta E-V, Kuopio T, Seppälä T, Rantala J, Mecklin J-P, et al. Combined prognostic value of CD274 (PD-L1)/PDCDI (PD-1) expression and immune cell infiltration in colorectal cancer as per mismatch repair status. Mod Pathol. 2019;32:866–83.
Porkka N, Lahtinen L, Ahtiainen M, Böhm JP, Kuopio T, Eldfors S, et al. Epidemiological, clinical and molecular characterization of Lynch-like syndrome: a population-based study. Int J Cancer. 2019;145:87–98.
Wirta E-V, Seppälä T, Friman M, Väyrynen J, Ahtiainen M, Kautiainen H, et al. Immunoscore in mismatch repair-proficient and -deficient colon cancer. J Pathol Clin Res. 2017;3:203–13.
Kellokumpu I, Kairaluoma M, Mecklin J-P, Kellokumpu H, Väyrynen V, Wirta E-V, et al. Impact of age and comorbidity on multimodal management and survival from colorectal cancer: a population-based study. J Clin Med. 2021;10:1751.
Funding
This study was funded by Cancer Foundation Finland (59-5619 to JPV). SO’s effort was supported in part by a U.S. National Institutes of Health grant (R35 CA197735). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Open Access funding provided by University of Oulu including Oulu University Hospital.
Author information
Authors and Affiliations
Contributions
Conceptualisation: HE, SO, JAN, JB, TK, JPV. Data curation: HE, MA, OH, E-VW, TTS, JB, MJM, JPM, JPV. Funding acquisition: MJM, JPM, JPV. Investigation: HE, MA, SAV, MF, OH, E-VW, TTS, JB, MJM, JPM, JPV. Methodology: HE, SAV, SO, JAN, MF, JPV. Formal analysis: HE, JPV. Resources: MJM, JB, JPM, TK, JPV. Supervision: TK, JPV. Visualisation: HE, SAV, JPV. Writing—original draft: HE, JPV. Writing—review and editing: All authors.
Corresponding author
Ethics declarations
Competing interests
JAN reports grants from NanoString, Akoya Biosciences and Illumina outside the submitted work. TTS is the CEO and co-owner of Healthfund Finland Oy and reports interview honoraria from Boehringer Ingelheim Finland. The remaining authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the hospital administration and the ethics board (Dnro13U/2011, 1/2016 and 8/2020) and the National Supervisory Authority for Welfare and Health (Valvira). The need to obtain informed consent from the study patients was waived (Valvira Dnro 3916/06.01.03.01/2016). The validation study was conducted under permissions form the Ethics Committee of Oulu University Hospital (25/2002, 42/2005, 122/2009, 37/2020) and Biobank Borealis (BB-2017_1012).
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Elomaa, H., Ahtiainen, M., Väyrynen, S.A. et al. Prognostic significance of spatial and density analysis of T lymphocytes in colorectal cancer. Br J Cancer 127, 514–523 (2022). https://doi.org/10.1038/s41416-022-01822-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41416-022-01822-6
This article is cited by
-
Urothelium marker UPK2 identifies aggressive colorectal cancers with distinct molecular and histological features
British Journal of Cancer (2026)
-
Automated deep learning-based assessment of tumour-infiltrating lymphocyte density determines prognosis in colorectal cancer
Journal of Translational Medicine (2025)
-
Associations of serum and tissue TIMP1 with host response and survival in colorectal cancer
Scientific Reports (2025)
-
Stroma AReactive Invasion Front Areas (SARIFA), tumour immune microenvironment, and survival in colorectal cancer
British Journal of Cancer (2025)
-
Associations of mucinous differentiation and mucin expression with immune cell infiltration and prognosis in colorectal adenocarcinoma
British Journal of Cancer (2025)