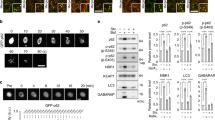
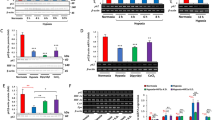
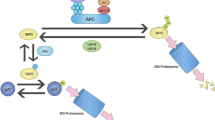
Abstract
p62/SQSTM1 is a selective autophagy receptor that drives ubiquitinated cargos towards autophagic degradation. This receptor is also a stress-induced scaffold protein that helps cells to cope with oxidative stress through activation of the Nrf2 pathway. Functional disorders of p62 are closely associated with multiple neurodegenerative diseases and cancers. The gene encoding the E3 ubiquitin ligase substrate-binding adapter SPOP is frequently mutated in prostate cancer (PCa), but the molecular mechanisms underlying how SPOP mutations contribute to PCa tumorigenesis remain poorly understood. Here, we report that cytoplasmic SPOP binds and induces the non-degradative ubiquitination of p62 at residue K420 within the UBA domain. This protein modification decreases p62 puncta formation, liquid phase condensation, dimerization, and ubiquitin-binding capacity, thereby suppressing p62-dependent autophagy. Moreover, we show that SPOP relieves p62-mediated Keap1 sequestration, which ultimately decreases Nrf2-mediated transcriptional activation of antioxidant genes. We further show that PCa-associated SPOP mutants lose the capacity to ubiquitinate p62 and instead promote autophagy and the redox response in a dominant-negative manner. Thus, our findings indicate oncogenic roles of autophagy and Nrf2 activation in the tumorigenesis of SPOP-mutated PCa.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Feng YC, He D, Yao ZY, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41.
Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Sign 2014;20:460–73.
Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–69.
Kirkin V, Rogov VV. A diversity of selective au tophagy receptors determines the specificity of the autophagy pathway. Mol Cell. 2019;76:268–85.
Sanchez-Martin P, Komatsu M. p62/SQSTM1-steering the cell through health and disease. J Cell Sci. 2018;131:jcs222836.
Lamark T, Svenning S, Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Signal Mechanisms Autophagy. 2017;61:609–24.
Sanchez-Martin P, Saito T, Komatsu M. p62/SQSTM1: ‘Jack of all trades’ in health and cancer. Febs J. 2019;286:8–23.
Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50.
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–U107.
Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–5.
Wang ZW, Song YZ, Ye MM, Dai XM, Zhu XQ, Wei WY. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat Rev Urol. 2020;17:339–50.
Geng CD, He B, Xu LM, Barbieri CE, Eedunuri VK, Chew SA, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci USA. 2019;116:14386–7.
Blattner M, Liu DL, Robinson BD, Huang D, Poliakov A, Gao D, et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer cell. 2017;31:436–51.
Yang J, Peng H, Xu YM, Xie XD, Hu RG. SQSTM1/p62 (sequestosome 1) senses cellular ubiquitin stress through E2-mediated ubiquitination. Autophagy 2018;14:1072–3.
Pan JA, Sun Y, Jiang YP, Bott AJ, Jaber N, Dou ZX, et al. TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol Cell. 2016;62:149–51.
Heath RJ, Goel G, Baxt LA, Rush JS, Mohanan V, Paulus GLC, et al. RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell Rep. 2016;17:2183–94.
Jongsma MLM, Berlin I, Wijdeven RHM, Janssen L, Janssen GMC, Garstka MA, et al. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell 2016;166:152–66.
Lin Q, Dai Q, Meng HX, Sun AQ, Wei J, Peng K, et al. The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J Cell Sci. 2017;130:3839–50.
Song PP, Li SS, Wu H, Gao RZ, Rao GH, Wang DM, et al. Parkin promotes proteasomal degradation of p62: implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson’s disease. Protein Cell. 2016;7:114–29.
Lee Y, Chou TF, Pittman SK, Keith AL, Razani B, Weihl CC. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell Rep. 2017;20:1994.
Jin XF, Wang J, Gao K, Zhang PZ, Yao LF, Tang Y, et?al. Dysregulation of INF2-mediated mitochondrial fission in SPOP-mutated prostate cancer. Plos Genet. 2017;13:e1006748.
Jin X, Shi Q, Li Q, Zhou L, Wang J, Jiang L, et al. CRL3-SPOP ubiquitin ligase complex suppresses the growth of diffuse large B-cell lymphoma by negatively regulating the MyD88/NF-kappaB signaling. Leukemia 2020;34:1305–14.
Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat cell Biol. 2010;12:213–U17.
Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu TD, Jiang T, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–85.
Sun DX, Wu RB, Zheng JX, Li PL, Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28:405–15.
Yang Y, Willis TL, Button RW, Strang CJ, Fu YH, Wen X, et?al. Cytoplasmic DAXX drives SQSTM1/p62 phase condensation to activate Nrf2-mediated stress response. Nature Communications. 2019;10:3759.
Sun DX, Wu RB, Li PL, Yu L. Phase separation in regulation of aggrephagy. J Mol Biol. 2020;432:160–9.
Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51:283–96.
Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer cell. 2008;13:343–54.
Dao TP, Kolaitis RM, Kim HJ, O’Donovan K, Martyniak B, Colicino E, et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol Cell. 2018;69:965–78. e6
An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657–69.
Acknowledgements
We thank the members of Wang and Gao labs for their technical support. This work was in part supported by the National Natural Science Foundation of China (No. 91954106, 81872109 to K.G.; No. 91957125, 81972396 to C.W.; No. 81872260, 82172938 to P.Z.; No. 31801165 to X.J.), the Natural Science Foundation of Shanghai (No. 18ZR1430100 to K.G.), the Open Research Fund of State Key Laboratory of Genetic Engineering (No. SKLGE-2111 to K.G.). Science and Technology Research Program of Shanghai (19DZ2282100).
Author information
Authors and Affiliations
Contributions
CJW and KG conceived the study. KG, QS, PZZ, QL, ZHL, YD, YJW, YLH, HYH, and XYZ performed the experiments and data analyses. SMZ, YL, KG, and CJW analyzed and interpreted the data. CJW and KG wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by E Baehrecke
Supplementary information
Rights and permissions
About this article
Cite this article
Shi, Q., Jin, X., Zhang, P. et al. SPOP mutations promote p62/SQSTM1-dependent autophagy and Nrf2 activation in prostate cancer. Cell Death Differ 29, 1228–1239 (2022). https://doi.org/10.1038/s41418-021-00913-w
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-021-00913-w
This article is cited by
-
TRIM24-mediated K27-linked ubiquitination of ULK1 alleviates energy stress-induced autophagy and promote prostate cancer growth in the context of SPOP mutation
Cell Death & Differentiation (2025)
-
Comprehensive analysis of m7G-related genes METTL1 and WDR4 for predicting prognosis and oncogenic functions in prostate cancer
Discover Oncology (2025)
-
Integrative analysis of mitochondrial-related gene profiling identifies prognostic clusters and drug resistance mechanisms in low-grade glioma
Discover Oncology (2025)
-
The protective role of baicalin regulation of autophagy in cancers
Cytotechnology (2025)
-
Fisetin Attenuates Mutant SOD1 Aggregation in Amyotrophic Lateral Sclerosis via Nrf2-Mediated Autophagy Activation
Journal of Molecular Neuroscience (2025)