Abstract
The cytoskeleton, composed of microfilaments, intermediate filaments, and microtubules, provides the structural basis for cellular functions such as motility and adhesion. Equally crucial, phosphoinositide (PIPn) signaling is a critical regulator of these processes and other biological activities, though its precise impact on cytoskeletal dynamics has yet to be systematically investigated. This review explores the complex interplay between PIPn signaling and the cytoskeleton, detailing how PIPn modulates the dynamics of actin, intermediate filaments, and microtubules to shape cellular behavior. Dysregulation of PIPn signaling is implicated in various diseases, including cancer, highlighting promising therapeutic opportunities through targeted modulation of these pathways. Future research should aim to elucidate the intricate molecular interactions and broader cellular responses to PIPn signaling perturbations, particularly in disease contexts, to devise effective strategies for restoring cytoskeletal integrity.
Similar content being viewed by others
Facts
-
The molecular mechanisms underlying the direct interaction between PIPns and intermediate filaments remain poorly defined despite evidence of their role in regulating IF dynamics.
-
PIPn signaling exhibits spatial and temporal regulation across cytoskeletal components, raising key questions about signal coordination during processes like migration and division.
-
The therapeutic potential of targeting PIPn-cytoskeleton interactions remains largely unexplored despite their crucial roles in cellular dynamics and disease pathogenesis.
Open questions
-
What mechanisms enable PIPns to specifically recognize and target distinct cytoskeletal components, and how is this selectivity determined?
-
What are the full sets of PIPn-binding proteins involved in cytoskeletal regulation, and how do their functions vary in different cellular contexts?
-
How do PIPn signaling pathways integrate with other networks to orchestrate cytoskeletal dynamics during development, homeostasis, and disease?
Introduction
The cell cytoskeleton is a complex network of protein filaments that extends throughout the cell. It provides structural support and plays a dynamic role in cellular processes such as motility, division, and intracellular transport. The cytoskeleton comprises three main types of protein polymers: microfilaments, intermediate filaments (IFs), and microtubules, each with distinct roles and properties [1].
Phosphoinositide (PIPn) signaling is a pivotal regulatory mechanism that modulates cytoskeletal dynamics. Phosphatidylinositol (PI/PtdIns), which consists of a glycerol backbone, two fatty acid acyl chains, and a myo-inositol ring, undergoes reversible phosphorylation at the 3rd, 4th, and 5th positions of the myo-inositol ring by specific kinases and phosphatases (Fig. 1) [2]. This phosphorylation produces seven unique PIPn isomers, including three monophosphorylated forms (PtdIns3P, PtdIns4P, and PtdIns5P), three bisphosphorylated forms (PtdIns(3,4)P2, PtdIns(3,5)P2, and PtdIns(4,5)P2), and one triphosphorylated form (PtdIns(3,4,5)P3). These PIPns serve as key signaling molecules that influence a plethora of cellular processes.
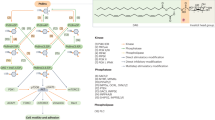
This diagram provides a comprehensive overview of PI metabolism within the cellular environment, highlighting the structure of the inositol ring with the 3rd, 4th, and 5th hydroxyl groups capable of phosphorylation. This process generates seven distinct PIPn isomers, whose interconversion is facilitated by PIPn kinases, phosphatases, and phospholipases—critical enzymes in this metabolic pathway. The diagram also highlights the PI3K/AKT signaling pathway, a key downstream effector of PIPn metabolism that regulates various cellular processes, particularly cell dynamics such as motility and adhesion. Notably, PtdIns(4,5)P2 4-phosphatase type I/II (PIP4P1/2), also known as TMEM55B/TMEM55A, has been proposed as PtdIns(4,5)P2 4-phosphatase [172]. However, the follow-up study found PIP4P1/TMEM55B to be catalytically inactive and lacking the typical catalytic CX5R motif [173]. To reflect this controversy, PIP4P1/2 is accompanied by a question mark, denoting the ongoing debate in the field. This diagram is generated using BioRender.
Discovered in the 1950s by Lowell and Mabel Hokin, the PIPn signaling cycle has since been recognized as a pivotal pathway in biology despite PIs representing a minor component of cellular membranes [3]. PIPns are present in the plasma membrane, Golgi apparatus, endoplasmic reticulum, endosomes, lysosomes, and the nuclear envelope [4]. Early studies focused on the biochemical and functional roles of membrane-associated PIPns, but research in the 1960s and 1970s revealed phospholipids within the nuclear matrix and chromatin [5, 6]. In 1983, Smith and Wells detected PI kinase and PIP kinase activity in the nucleus [7], leading to Cocco et al.‘s discovery in 1987 of nuclear PIPn generation [8]. These discoveries expanded the scope of membrane-bound PIPn signaling into non-membranous regions within the nucleus. Over the past 30 years, extensive evidence has demonstrated that PIPns also localize in distinct non-membranous subcellular compartments, including the nucleoplasm, nuclear matrix, nucleolus, nuclear speckles, nuclear PtdIns(4,5)P2 islets, chromatin, ribosomes, centrosomes, cytoskeleton, and inflammasomes, with specific PI transfer proteins (PITPs), lipid kinases and phosphatases regulating these regional PIPn pools [2, 9,10,11,12,13,14,15,16,17]. The cytoskeleton is one such non-membranous compartment where PIPn signaling not only regulates cytoskeletal dynamics but also provides a link with cellular membranes [18].
In the context of cell dynamics, PIPn signaling interfaces with the cytoskeleton to orchestrate the formation and retraction of cellular protrusions, such as lamellipodia and filopodia, which are primarily driven by the polymerization and depolymerization of actin microfilaments [18,19,20]. The coordinated action of PIPn signaling and the cytoskeleton is essential for cell polarization, directional movement, and the establishment of stable adhesion sites with the extracellular matrix or neighboring cells. This review aims to dissect the intricate relationship between PIPn signaling and the cytoskeleton, focusing on the role of PIPns in regulating the dynamics and function of microfilaments, IFs, and microtubules in cell motility and adhesion. We will explore the molecular mechanisms by which PIPn signaling influences cytoskeletal remodeling and the consequences of these interactions for cellular operations.
PIPn signaling at microfilaments
Microfilaments, also known as actin filaments, are essential cytoskeleton components with a diameter of ~7 nm [21]. These thin, flexible filaments comprise actin monomers (G-actin) that polymerize into filamentous actin (F-actin) in a highly regulated process [22, 23]. Actin filaments are inherently polarized, with a fast-growing barbed (+) end and a slower-growing pointed (−) end [22, 23]. The polarity of microfilaments is central to their dynamic behavior; ATP-bound G-actin monomers are preferentially added to the barbed end, promoting filament elongation, while ADP-bound G-actin dissociates from the pointed end [23, 24]. This dynamic process, known as treadmilling, allows for the continuous turnover of actin filaments with an evident influence on processes like cell motility, shape changes, and intracellular transport [25].
Functionally, microfilaments are crucial for maintaining cell shape and providing mechanical support through the actin cortex beneath the plasma membrane [21, 23]. They facilitate cell motility by driving the extension of lamellipodia and filopodia, which are essential for migration [1, 26, 27]. During cytokinesis, microfilaments form a contractile ring that constricts the cell membrane; however, the process also involves complex membrane trafficking from endosomes and a tightly regulated abscission phase to complete cell division. They also serve as tracks for myosin motor proteins to transport vesicles and organelles [24, 28, 29]. Also, microfilaments are critical in signal transduction, linking extracellular signals to intracellular responses by interacting with membrane receptors and influencing cellular responses [24, 30]. Furthermore, microfilaments are crucial in cell adhesion, connecting cells to the extracellular matrix and each other via structures like focal adhesions (FAs). These functions underscore the importance of microfilaments in inter- and intra-cellular dynamics, in which PIPn signaling pathways play a significant role.
The rapid polymerization and depolymerization of microfilaments enable cells to respond efficiently to environmental cues, especially during cell migration, where actin filaments form protrusions such as lamellipodia and filopodia at the leading edge [31,32,33]. Additionally, actin filaments provide mechanical support to the cell cortex, maintaining cell integrity and facilitating processes like cytokinesis [33, 34]. The interaction of actin with various actin-binding proteins, including profilin, cofilin, and actin-related proteins-2/3 (ARP2/3), further regulates filament dynamics and architecture, allowing cells to execute complex mechanical tasks, such as membrane deformation, vesicle transport, and contractile ring formation during cell division [31, 33, 35, 36].
The actin cytoskeleton is sculpted by many actin-binding proteins (ABPs) that bind, twist, sever, branch, or cap actin filaments (e.g., cofilin, gelsolin, profilin, etc.) [35, 36]. The activities of ABPs are regulated by PIPns, which in turn influence cellular activities [22, 23, 37, 38]. PIPn signaling is integral to regulating actin dynamics, mainly through the actions of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in conjunction with ABPs [36, 39].
PtdIns(4,5)P2 is a critical PIPn located at the plasma membrane that modulates actin dynamics by interacting with a range of ABPs [20, 39,40,41,42]. PtdIns(4,5)P2 generally inhibits ABPs that promote actin filament disassembly and activates those that enhance actin polymerization. Consequently, PtdIns(4,5)P2 facilitates the formation of actin structures, particularly under the plasma membrane and around organelles enriched with PIPns (Fig. 2).
This figure illustrates the critical role of PtdIns(4,5)P2 in regulating actin filament assembly and disassembly through its interaction with various ABPs. At the plasma membrane, PtdIns(4,5)P2 interacts with ABPs such as cofilin and twinfilin, which promote filament depolymerization, while binding with profilin enhances actin polymerization. Cofilin and twinfilin are sequestered by PtdIns(4,5)P2, preventing filament disassembly, while profilin-G-actin complexes are regulated by PtdIns(4,5)P2 to control actin monomer availability. N-WASP contains a WASP Homology 1 domain (WH1) that binds WIP-family proteins, a basic sequence for PtdIns(4,5)P2 binding (B), a GTPase-binding domain (GBD) that interacts with CDC42-GTP, a proline-rich region (Pro), and a VCA domain, which consists of a V (verprolin homology) region for actin binding, a C (central) domain, and an A (acidic) motif that binds the ARP2/3 complex to promote actin nucleation. The interaction of N-WASP with PtdIns(4,5)P2 and CDC42 leads to the activation of ARP2/3 and subsequent actin polymerization. Additionally, PtdIns(4,5)P2 modulates the activity of gelsolin, inhibiting its filament-severing functions, thereby maintaining actin filament stability under dynamic conditions of cellular movement and membrane deformation. This interplay is critical for processes such as cell motility and structural organization. This diagram is generated using BioRender.
For instance, cofilin, a protein responsible for actin filament depolymerization, binds to ADP-actin filaments to promote their disassembly [1]. Competitively, PtdIns(4,5)P2 sequesters cofilin, preventing its interaction with actin, thus stabilizing the filaments [42,43,44]. Upon a decrease in PtdIns(4,5)P2 levels, cofilin is released, leading to heightened filament turnover and disassembly. Profilin, another actin-binding protein, facilitates actin polymerization by binding G-actin and promoting the exchange of ADP for ATP [22, 45, 46]. PtdIns(4,5)P2 interacts with profilin and regulates its availability, thereby controlling the pool of actin monomers available for filament formation [47, 48]. PtdIns(4,5)P2 suffices to disrupt the actin-profilin complex and inhibit profilin’s actin monomer sequestration activity. Through this mechanism, PtdIns(4,5)P2 helps regulate the rate of actin polymerization. Like cofilin, changes in the concentration of PtdIns(4,5)P2 on the membrane may cause profilin to switch between membrane-bound and actin-bound forms, making it an efficient mediator of external signals to microfilaments [49]. Twinfilin is a ubiquitous actin monomer-binding protein. Studies have found that in yeast and murine cells, twinfilin interacts with PtdIns(4,5)P2, and its actin monomer-sequestering activity is inhibited by PtdIns(4,5)P2 [50, 51]. Gelsolin, a severing and capping protein, inhibits further actin polymerization by capping the barbed ends of filaments [22, 52]. Interaction with PtdIns(4,5)P2 inhibits gelsolin’s severing activity, stabilizing the actin network [53,54,55]. Upon hydrolysis of PtdIns(4,5)P2, gelsolin becomes active, leading to filament severing and turnover.
PtdIns(4,5)P2 can also enhance de novo actin polymerization by modulating neural Wiskott–Aldrich Syndrome protein (N-WASP) activity, an essential regulator of actin dynamics. N-WASP is autoinhibited due to intramolecular interactions between its N-terminal and C-terminal regions, which mask the VCA (verprolin, central, acidic) domain—responsible for actin nucleation [56]. This autoinhibition is released when PtdIns(4,5)P2 and the small GTPase cell division control protein 42 (CDC42) simultaneously bind to N-WASP, leading to a conformational change that unmasks the VCA region [57, 58]. Once exposed, the VCA domain of N-WASP can bind to and activate the ARP2/3 complex, which promotes actin nucleation and polymerization [59]. PtdIns(4,5)P2 binding facilitates N-WASP activation and helps position N-WASP at membrane sites rich in actin nucleation activity, allowing for rapid and localized polymerization of actin filaments [59]. This coordinated regulation by PtdIns(4,5)P2 and CDC42 ensures precise spatiotemporal control of actin dynamics, particularly during cell movement, vesicle trafficking, and membrane protrusion formation [27, 60]. Through these interactions, PtdIns(4,5)P2 plays a pivotal role in reorganizing the actin cytoskeleton in response to extracellular signals.
PtdIns(3,4,5)P3, generated via class I phosphatidylinositol-3-kinase (PI3K)-mediated phosphorylation of PtdIns(4,5)P2, is a pivotal regulator of actin cytoskeletal dynamics, particularly in the processes of cell migration, signal transduction, and polarization [20, 61]. PtdIns(3,4,5)P3, accumulated at the membrane, creates a gradient that promotes the recruitment and activation of small GTPases such as Rac and CDC42, essential for initiating actin-rich structures like lamellipodia and filopodia, which drive directional movement [62,63,64]. Additionally, evidence suggests that PtdIns(3,4,5)P3 interacts with the WASP family verprolin homologous protein 2 (WAVE2) through its basic domain. WAVE2 is known to promote actin nucleation and branching, which are essential for the formation of dynamic actin networks [65,66,67]. The amino-terminal region of WAVE contains the PtdIns(3,4,5)P3-binding sequence, allowing PtdIns(3,4,5)P3 generated by PI3K at the cell membrane to recruit WAVE to polarized membrane regions directly [68,69,70]. This recruitment is critical for forming lamellipodia at the leading edge, an essential step in directed cell migration.
Like PtdIns(4,5)P2, PtdIns(3,4,5)P3 modulates ABPs, including profilin, by altering its interactions with actin monomers [48, 71]. The affinity of profilin for PtdIns(3,4,5)P3 is slightly higher than for PtdIns(4,5)P2, further influencing actin filament dynamics by shifting profilin from its monomer-sequestering function to actin polymerization [48]. Thus, PtdIns(3,4,5)P3 is a critical mediator linking external signals to cytoskeletal reorganization, supporting cell migration, polarization, and membrane trafficking.
PtdIns(3,4)P2 plays a critical role in regulating actin dynamics and adhesion processes, particularly in FA assembly [72]. Locally produced by enzymes such as class II PI3K and SHIP, PtdIns(3,4)P2 accumulates at specific subcellular sites to coordinate the recruitment of actin-binding and scaffolding proteins. In FAs, PtdIns(3,4)P2 generated by class II PI3K-C2β can promote FA disassembly [73]. PtdIns(3,4)P2 promotes RhoA-dependent stress fiber turnover by recruiting the PtdIns(3,4)P2-dependent RhoA-GTPase activating protein ARAP3. Furthermore, Studies have enriched the details of the molecular interactions of PtdIns(3,4)P2 during FA transformation [72, 74]. The PtdIns(3,4)P2, present in minimal amounts, mediates the recruitment of the Tks5-Grb2 scaffold, followed by N-WASP accumulation and podosome formation near newly formed FAs, affecting cell motility.
PtdIns(3,4)P2 regulates actin remodeling during phagocytosis by recruiting Lamellipodin (Lpd) to nascent phagosomes [75]. Lpd, in turn, organizes actin filaments via vasodilator-stimulated phosphoprotein (VASP). Depleting PtdIns(3,4)P2 disrupts pseudopod extension and blocks particle engulfment, underscoring its essential role in actin dynamics during phagosome formation [75].
By regulating these actin-associated proteins, PIPn signaling maintains a critical balance between actin filament assembly and disassembly, governing essential cellular operations, including motility, morphological changes, and membrane trafficking. This regulatory mechanism is vital for the cell’s ability to respond to extracellular signals and maintain structural integrity. Overall, PIPn signaling orchestrates the dynamic remodeling of the actin cytoskeleton, enabling fundamental activities like migration, division, and trafficking.
PIPn signaling at intermediate filaments (IFs)
IFs are a diverse assembly of protein fibers that form the cytoskeleton with microtubules and microfilaments. These filaments, composed of various proteins made up of amino acid chains, are characterized by a diameter of ~10 nm [76] and provide mechanical strength to cells and tissues, playing a crucial role in maintaining cellular integrity [77]. While microfilaments and microtubules are polymers of single types of proteins—actin and tubulin, respectively—IFs consist of a diverse array of proteins expressed across various cell types. Over 50 different IF proteins have been identified, and they are classified into six groups based on similarities in their amino acid sequences. IFs create an extensive network within the cytoplasm, extending from a ring that encircles the nucleus to the plasma membrane [78]. The maturation of IFs begins with the formation of parallel dimers, which associate in an anti-parallel, staggered manner to form tetramers, ultimately culminating in mature filaments consisting of about eight tetramers [79]. Known for their remarkable stability, IFs are vital for tensile strength, particularly in robust structures such as hair, scales, and fingernails [80]. PIPn signaling is deeply involved in regulating cell growth, differentiation, survival, and migration [81]. These pathways are intricately linked to IFs, which, beyond being critical components of the cytoskeleton, also contribute to the modulation of cellular responses [82] (Fig. 3).
This figure illustrates the pivotal role of PIPn signaling in regulating cell proliferation and migration through its interaction with IFs. Activation of cell-surface receptors triggers the production of PIPns (e.g., PtdIns4P, PtdIns(4,5)P2, PtdIns(3,4,5)P3), leading to AKT activation. This activation prompts AKT to phosphorylate PKP1, causing it to disengage from desmosomal IFs and diminishing cell adhesion. The phosphorylated PKP1 then interacts with 14-3-3γ, which fosters cell proliferation and migration. The stimulation of S6K and inhibition of 4E-BP by mTORC1-mediated phosphorylation downstream of the PI3K/AKT signaling pathway promote protein synthesis by influencing the eIF4 complex. The phosphorylation of PKP1 enhances eIF4 activity, which relaxes the complex structures in the 5’-UTR of mRNA, leading to increased protein production and cell growth. Within the nucleus, Lamin A/C interacts with PtdIns(4,5)P2, likely generated through the sequential actions of nuclear class I PITPα/β, PI4KIIα, and PIPKIα. The binding of PtdIns(4,5)P2 promotes the phosphorylation of Lamin A/C, leading to the formation of a complex with NM1. This complex activates RNA polymerase II (Pol II) transcription, thereby driving cell proliferation. This diagram is generated using BioRender.
Keratins are a diverse family of IF proteins primarily found in epithelial cells [83]. They provide essential structural support, maintaining the mechanical integrity of cells and tissues. Keratin 8 (K8) and Keratin 18 (K18), often co-expressed, are crucial for the function of simple epithelial cells, including normal hepatocytes in the liver [83]. Beyond their structural role, these keratins are also involved in cellular signaling pathways. Although K8/18 expression persists during tumorigenesis, their loss during epithelial-mesenchymal transition (EMT) is linked to metastasis and chemoresistance [84]. K8/18 depletion leads to hyperactivation of the PI3K/AKT/ nuclear factor kappa B (NF-κB) pathway and increased expression of matrix metalloproteinase 2 (MMP2) and matrix metalloproteinase 9 (MMP9), which prompts the tight junction protein claudin-1 to assume an active role in cancer progression [84]. In addition, K8/18 directly interacts with AKT, facilitating reciprocal AKT hyperglycosylation and hypophosphorylation, reducing AKT kinase activity [85]. Keratin 10 (K10) has also been reported to interact directly with AKT [86]. While K10 is not a substrate of AKT, it binds to AKT through its N-terminal domain and inhibits cell proliferation and tumorigenesis by impeding AKT activity [86, 87]. Although it remains unclear whether Keratin 17 (K17) directly interacts with AKT, K17 does affect AKT activity [88, 89]. Reduced K17 expression results in decreased AKT and mechanistic target of rapamycin (mTOR) kinase activity, leading to a lower proliferation rate than normal K17 expression levels. The complex interplay between keratins and the PI3K/AKT pathway underscores the importance of these proteins in cellular regulation. Understanding how keratins modulate the PIPn signaling could reveal new therapeutic targets for diseases characterized by aberrant keratin expression or AKT pathway activation, such as cancer.
Vimentin, a type III IF protein, maintains cellular structure and function. It self-assembles into a resilient cytoskeletal network composed of long, flexible filaments that preserve cell shape and resist mechanical stress, particularly in mesenchymal cells undergoing significant morphological changes [90, 91]. Vimentin also contributes to forming nuclear microdomains that facilitate gene expression and DNA repair [92]. Vimentin’s interaction with the PI3K/AKT pathway is significant in cancer progression and EMT [93]. Activation of growth factor receptors initiates a cascade that leads to vimentin phosphorylation at serine 39 by AKT. This promotes cell motility and invasion while protecting vimentin from caspase-mediated degradation, thereby enhancing cell survival [94, 95]. The phosphorylated vimentin also strengthens its binding to 14-3-3 proteins, inhibiting pro-apoptotic signaling pathways and contributing to chemoresistance in many cancer cells [96, 97]. During EMT, the hyperactivation of the PI3K/AKT pathway increases vimentin levels. It decreases the expression of epithelial markers like E-cadherin, fostering a mesenchymal phenotype characterized by increased motility and invasiveness. Vimentin’s interaction with mitogen-activated protein kinase kinase 4 (MAP2K4) propagates the effect of MAP2K4 on promoting breast cancer cell proliferation, migration, and invasion by activating the PI3K/AKT pathway, the downstream proteins such as c-JUN, the G1/S cell cycle, and the EMT [98]. Besides, patients with high expression levels of both MAP2K4 and vimentin exhibit poorer overall survival rates compared to those with low expression levels of these proteins [98].
Moreover, vimentin engages with the mTOR pathway, particularly mTORC1. Vimentin facilitates the activation of mTORC1 by modulating Rag GTPase activity, thereby enhancing ribosome biogenesis and protein synthesis, which is critical for supporting cell growth and survival [99]. This highlights vimentin’s potential as a therapeutic target for inhibiting cancer spread and improving patient outcomes. Further research is needed to elucidate how vimentin influences cancer biology and develop targeted therapies against it.
Lamin A/C belongs to the family of IF proteins known as nuclear lamins, located in the fibrous protein layer or network beneath the inner nuclear membrane of the cell nucleus [100]. They have a close structural relationship with the nuclear envelope, chromatin, and nuclear pore complexes. Lamin A and Lamin C are two different proteins produced by the same gene LMNA through alternative splicing [100, 101], and they play a crucial role in maintaining the structural stability of the cell nucleus, cell motility, mechanosensing, chromosome organization, gene regulation, cell differentiation, DNA damage repair, and telomere protection [102]. It functionally interacts with PtdIns(4,5)P2, a significant phospholipid involved in various cellular signaling processes, which is predominantly enriched in the nucleoplasm due to the orchestrated actions of class I PITPα/β, phosphatidylinositol-4-kinase II alpha (PI4KIIα), and phosphatidylinositol-4-phosphate 5-kinase type I alpha (PIPKIα) [9,10,11, 103].
Research has shown that Lamin A/C forms a complex with PtdIns(4,5)P2 and nuclear myosin I (NM1), with the assembly of this complex being influenced by Lamin A/C’s phosphorylation status; the formation of the complex facilitates the transcription of RNA polymerase II (Pol II), thereby promoting cell proliferation [103]. Besides PtdIns(4,5)P2, Lamin A/C is further linked to PIPn signaling as the mechanistic study indicates that Lamin A/C-related malignant behavior is regulated by modulation of the PI3K/AKT/phosphatase and tensin homolog (PTEN) signaling pathway [104]. Lamin A/C knockdown or overexpression decreased or increased the protein levels, respectively, of the PI3K subunits p110 and p85 in prostate tumor cell lines, including lymph node carcinoma of the prostate (LNCaP), DU145, and prostate cancer cell line-3 (PC3). Furthermore, small hairpin RNA-mediated knockdown or overexpression of Lamin A/C resulted in the inhibition or stimulation of cell growth, colony formation, migration, and invasion, suggesting that Lamin A/C influences cell motility via PIPn signaling [104].
In addition, the binding of Lamin A/C to PtdIns(4,5)P2 may help organize functional nuclear microdomains essential for gene expression and DNA repair [102, 103]. Furthermore, PI3K-C2β co-localizes with Lamin A/C in the nuclear matrix and is activated in the nuclear matrix [105, 106], suggesting the potential involvement of PtdIns3P and PtdIns(3,4)P2 in the functional role of nuclear lamina. These findings demonstrate a significant molecular link between PIPns and nuclear IFs. It accentuates the essential role of Lamin A/C, transcending its traditional function of providing structural integrity.
Desmosomes are specialized cell adhesion junctions crucial for maintaining tissue integrity and withstanding mechanical stress, particularly in epithelial tissues [107,108,109]. They consist of transmembrane glycoproteins known as desmosomal cadherins, including desmogleins and desmocollins, which are connected to the IF system through a complex of proteins in the cytoplasmic plaque, such as desmoplakin, plakoglobin, and plakophilin. These proteins interact with the carboxyl-terminal domain of desmosomal cadherins, linking them to IFs like keratins [109]. Beyond their structural function, desmosomes are dynamic entities capable of influencing cell signaling pathways, including the modulation of proteins such as p38 mitogen-activated protein kinase (MAPK), which plays an integral role in migration and differentiation, among other processes [110, 111]. Their interactions with signaling molecules suggest that desmosomes contribute to cell behavior, vital in biological projects such as wound healing and tissue regeneration.
Upon stimulation by agonizts such as insulin, cytosolic PIPns responsive to the cell surface receptors are produced, activating the downstream PI3K/AKT/mTOR/S6K signaling pathway, promoting cell proliferation, cell cycle progression [111]. Specifically, AKT2 phosphorylates desmosomal protein plakophilin 1 (PKP1), prompting its translocation from the cell membrane to the cytoplasm [112]. In the cytoplasm, phosphorylated PKP1 is stabilized and protected from degradation through binding with 14-3-3γ, leading to reduced intercellular adhesion but increased proliferation, migration, and anchorage-independent growth [113]. This phosphorylation event weakens PKP1 interactions with desmosomal proteins, such as desmoplakin (DSP) and desmoglein 1 (DSG1), thereby impairing intercellular adhesion. Further downstream, mTORC1 phosphorylates S6K and eukaryotic initiation factor 4E (eIF4E)-binding protein (4E-BP), resulting in the stimulation of S6K and inhibition of 4E-BP [111, 114]. This promotes the assembly of the eukaryotic initiation factor 4F (eIF4F) complex. Together, mTOR and S6K control the association of translation initiation factors with the eIF4 complex on mRNAs, thereby enhancing protein synthesis [111, 115]. Phosphorylated PKP1 interacts with this complex, stimulating eIF4 activity and facilitating the unwinding of secondary structures in the 5’untranslated region (5’-UTR), which correlates with increased protein biosynthesis, proliferation, and cell growth [115]. The connection between desmosomal proteins and the PI3K/AKT pathway suggests that desmosomes and their associated IFs may facilitate the transmission of signals from the cell membrane to the cytosol, influencing cellular activities such as proliferation and differentiation.
The intricate interplay between IFs, PIPn signaling, and cellular adhesion structures such as desmosomes is essential for maintaining cellular and tissue integrity. IFs, known for their remarkable mechanical strength, are the backbone for cellular resilience, while PIPn signaling pathways function as a regulatory hub, influencing growth, survival, and cellular behavior. Moreover, the dynamic nature of desmosomes not only ensures the structural stability of tissues but plays a crucial role in modulating cellular responses to external stimuli. Collectively, these components form a complex network fundamental to the health and functionality of cells and tissues, highlighting their importance in understanding and treating diseases where these systems are disrupted.
PIPn signaling at microtubules
Microtubules are cylindrical polymers composed of alpha- and beta-tubulin dimers that assemble into hollow tubes with a diameter of ~25 nm [116,117,118]. These highly dynamic structures exhibit intrinsic polarity, characterized by a fast-growing plus (+) end, where tubulin subunits are preferentially added, and a slower-growing minus (−) end, which is typically anchored to microtubule-organizing centers (MTOCs), such as centrosomes [116,117,118]. This polarity underpins many of the functions of microtubules, enabling directional transport and structural organization within the cell.
Microtubules are vital in resisting compressive forces within the cell, providing structural support, and maintaining cell shape, especially in cells with complex morphologies like neurons [118, 119]. In addition, they are essential in cellular processes such as intracellular transport and mitosis. During mitosis, microtubules form the mitotic spindle, a structure critical for segregating chromosomes into daughter cells, ensuring faithful cell division [116, 120]. Given their importance in maintaining cellular architecture, defects in microtubule function or regulation are associated with various diseases, including neurodegenerative disorders and cancers, highlighting the necessity of precise control over microtubule dynamics [121,122,123].
Microtubule-associated proteins (MAPs) are essential regulators of microtubule dynamics. MAPs stabilize microtubules by binding along their sides, reducing the depolymerization rate, or promoting polymerization at the plus end [124,125,126,127]. Other MAPs, such as kinesin and dynein, utilize microtubules as tracks for intracellular transport that direct cargoes like vesicles and organelles to specific locations within the cell [128,129,130]. For example, MAPs like tau stabilize long microtubule tracks in neurons, which are crucial for axonal transport. In contrast, in mitotic cells, MAPs, like the family of kinesin-related motor proteins, assist in forming the mitotic spindle, ensuring accurate chromosome segregation [131,132,133,134].
PIPn signaling pathways are crucial for regulating microtubules, specifically in their stabilization and modulation through MAPs [14, 55, 124, 135, 136]. Specifically, PtdIns(4,5)P2 has been shown to inhibit microtubule assembly by directly and selectively binding to tubulin, a process that is not observed with other phospholipids like PtdIns(3,4,5)P3, PtdIns3P, Phosphatidylcholine (PC), Phosphatidylethanolamine (PE), or Inositol (1,4,5)-trisphosphate (Ins(1,4,5)P3) [135]. The reduced PtdIns(4,5)P2 levels can lead to the disorganization of microtubule networks, as seen in Drosophila male germ cells, demonstrating its regulatory importance [137].
Various PIPns are modified by kinases and phosphatases, with several kinases implicated in microtubule homeostasis (Fig. 4). PI3K, for example, is associated with α/β-tubulin, and this interaction intensifies following insulin stimulation. Given that PtdIns(4,5)P2 is a primary substrate of PI3K, it likely plays a critical role in modulating microtubule responses to insulin signaling [136].
The schematic illustrates the interplay between activated receptors at the cell membrane, endocytosis, PIPn signaling, and microtubule dynamics. Receptor activation generates various PIPns (e.g., PtdIns3P, PtdIns(4,5)P2, PtdIns(3,4,5)P3) at endosomes, driving the recruitment and activation of AKT, which promotes cellular proliferation, migration, and invasion. Within this process, endosomal PtdIns3P recruits class I PI3K components (p110α/p85) by binding to the C2 domain of p110α. This complex is further linked to microtubules through interactions between MAP4 and PI3K. In cancer cells, the switch of the PI3K regulatory subunit from p85α to p85β enhances binding to PtdIns3P, amplifying complex recruitment and promoting constitutive PI3K/AKT signaling. At the microtubule-organizing centers (centrosomes), an accumulation of PtdIns4P occurs, where it interacts with TTBK2 and the accessory factor CEP164, preventing their interaction and inhibiting the removal of microtubule capping protein CP110 and microtubule extension. PIPKIγ reduces PtdIns4P levels by phosphorylating it to PtdIns(4,5)P2, which promotes TTBK2 recruitment and microtubule polymerization. In contrast, dephosphorylation of PtdIns(4,5)P2 back to PtdIns4P by INPP5E reverses this effect. This diagram is generated using BioRender.
Phosphatidylinositol-5-phosphate 4-kinase type II gamma (PIP4KIIγ), an enzyme that synthesizes PtdIns(4,5)P2, plays a role in regulating microtubule dynamics. During mitosis, the loss of PIP4KIIγ increases the accumulation of mitotic centromere-associated kinesin (MCAK), a microtubule-depolymerizing kinesin, at spindle poles, leading to destabilization of spindle pole-associated microtubules [16]. Additionally, PtdIns(4,5)P2 binds to polo-like kinase 1 (PLK1), reducing PLK1-mediated MCAK phosphorylation—a modification that activates MCAK in depolymerizing spindle microtubules. These findings suggest that PIP4KIIγ and PtdIns(4,5)P2 contribute to microtubule stability by preventing depolymerization [16]. In contrast, another kinase that generates PtdIns(4,5)P2, PIPKIα [138], directly interacts with kinesin superfamily protein 2A (KIF2A), a member of the kinesin-13 family known for its microtubule-depolymerizing activity [139, 140]. This interaction enhances the microtubule-depolymerizing activity of KIF2A [140]. In neurons, PIPKIα suppresses the elongation of axon branches in a KIF2A-dependent manner [140], suggesting a unique PIPKIα-mediated mechanism that controls microtubule dynamics during neuronal development.
PIPns engage with several MAPs to regulate microtubule dynamics, stability, and function. Studies from 1992 have demonstrated that PI can bind to MAP2 and inhibit microtubule assembly [124]. Our recent study has further revealed that PI3K binds to microtubules in a MAP4-dependent manner [14]. MAP4, a microtubule-associated protein, interacts with the C2 domain of the p110α catalytic subunit of PI3K through its microtubule-binding domain (MTBD). This interaction is essential for adequately functioning the PI3K pathway, which is often dysregulated in diseases such as cancer. The C2 domain of p110α is also known to bind with PtdIns3P enriched at the endosome surface [141], which is vital for the docking of class I PI3K to endosomes [142]. During oncogenesis, the regulatory subunit switch from p85α to p85β occurs in PI3K, ensuring induced activity [142]. These factors collectively contribute to the localization of class I PI3K to endosomal vesicles on microtubules, facilitating its association with receptor tyrosine kinases (RTKs). Activated RTKs are translocated to endosomes via endocytosis, where PI3K generates PtdIns(3,4,5)P3, activating the AKT signaling pathway and supporting microtubule homeostasis. As endosomes actively transport materials within the cell and facilitate their transfer to various compartments—such as the endoplasmic reticulum, mitochondria, the Golgi complex, and the nucleus—PIPn signaling is vital for their movement along microtubules [143,144,145,146,147]. This process impacts cargo delivery to subcellular organelles and plays a significant role in cell motility and adhesion.
Tau, a MAP responsible for microtubule stabilization, can be phosphorylated by Tau Tubulin kinase 2 (TTBK2), a serine/threonine protein kinase of the Casein kinase 1 (CK1) superfamily [148, 149]. TTBK2 is vital for removing the centriolar coiled-coil protein of 110 kDa (CP110) and caps the distal end of the centriole, thereby facilitating microtubule elongation and ciliogenesis [148, 150,151,152]. A study revealed that PtdIns4P accumulates at the centromeres of non-ciliated cells, where it binds to TTBK2 and the distal accessory protein CEP164 (Centrosomal protein of 164 kDa) [153]. This interaction inhibits the recruitment of TTBK2, thus preventing CP110 removal. PIPKIγ reduces PtdIns4P levels, promoting TTBK2 recruitment and the initiation of ciliogenesis. Additionally, inositol polyphosphate-5-phosphatase E (INPP5E) inhibits the recruitment of TTBK2 and ciliogenesis by dephosphorylating PtdIns(4,5)P2 to PtdIns4P.
Additionally, PtdIns(3,4)P2 biosynthesis regulates integrin endocytosis and thus affects microtubule dynamics [154]. PtdIns(3,4)P2-rich microdomains at invadopodia guide integrin β3 endocytosis through SNX9-dependent membrane invagination and dynein-mediated retrograde transport, facilitating adhesion disassembly and recycling. Furthermore, PtdIns(3,4)P2, produced by SHIP1 and class II PI3K, recruits SNX9 and then facilitates vesicle trafficking during apical surface remodeling [155]. Perturbing PtdIns(3,4)P2 disrupts polarization by causing subcortical vesicle retention, implicating its role in coordinating microtubule-based transport of apical vesicles.
These regulatory mechanisms underscore the critical role of PIPn signaling in maintaining microtubule integrity and ensuring proper cellular function across processes such as mitosis, intracellular trafficking, and cell migration. PIPn signaling is crucial in preserving cellular architecture and function through microtubule dynamics and associated pathways.
Discussion
PIPn signaling elucidates critical roles in regulating the dynamics of various cytoskeletal components, including microfilaments, microtubules, and IFs (Fig. 5). The findings indicate that PIPn signaling, mainly through PtdIns(4,5)P2 and PtdIns(3,4,5)P3, is essential for maintaining cytoskeletal integrity, facilitating intracellular transport, and enabling cell motility [156,157,158]. Specifically, PIPn signaling interacts with a range of ABPs and MAPs, highlighting its role as a central regulatory mechanism in coordinating cytoskeletal dynamics [14, 16, 22, 23, 37, 38, 55, 124, 135].
This figure provides an integrated overview of the regulatory mechanisms by which PIPns influence the cytoskeleton, including the dynamics of microfilaments, IFs, and microtubules. It highlights the complex interactions between PIPns, various effector proteins, and signaling pathways essential for cytoskeletal regulation, supporting critical cellular structures and processes such as FA, cell junction, proliferation, motility, migration, and invasion. This diagram is generated using BioRender.
PIPn microdomains form at specific subcellular sites through localized activities of kinases and phosphatases. Class I and class II PI3Ks, alongside PIPn phosphatases like SHIP1, regulate the accumulation of PIPns at distinct cortical regions [14, 105, 106, 155]. These microdomains compartmentalize signaling, directing processes like actin remodeling and cellular polarization. The localization of PIPn-generating kinases is regulated by interactions with scaffold proteins, cytoskeletal elements, and pre-existing lipids. For instance, class II PI3K-C2β co-localizes with Lamin A/C in the nuclear matrix, while class I PI3K associates with microtubules through MAP4 [105, 106, 142]. These mechanisms ensure spatiotemporal control of cytoskeletal polymerization and dynamics.
Different PIPns exhibit distinct roles in cytoskeletal regulation. Specificity in PIPn signaling is achieved through subcellular localization, differential binding affinities, and pathway interactions. PtdIns(4,5)P2 interacts with ABPs like profilin and cofilin to regulate actin turnover, while PtdIns(3,4,5)P3 influences microtubule dynamics via AKT signaling [22, 23, 37, 38, 142]. PtdIns(3,4)P2 specializes in integrin trafficking and cellular polarization by recruiting proteins like SNX9, linking membrane dynamics to cytoskeletal organization [154, 155]. PtdIns(4,5)P2 may regulate vimentin via AKT, whereas PtdIns(3,4,5)P3 stabilizes microtubules through distinct gradients [93, 142]. These mechanisms fine-tune cytoskeletal dynamics in processes like cell migration, division, and intracellular transport.
PIPn signaling not only influences cytoskeletal function but is also regulated by cytoskeletal components. IFs, such as keratins, vimentin, and lamin A/C, engage in reciprocal interactions with PIPn signaling, particularly impacting pathways like PI3K/AKT. For instance, keratins regulate AKT activity: K8 or K18 depletion hyperactivates the PI3K/AKT pathway, driving metastasis, while K10 inhibits AKT activity, suppressing tumorigenesis [84, 86, 87]. Vimentin phosphorylation by AKT enhances motility, invasion, and chemoresistance, especially during EMT [94, 95], while its interaction with MAP2K4 further propagates PI3K/AKT signaling [98]. Lamin A/C links to PIPn signaling in the nucleus, modulating PI3K/AKT activity and influencing proliferation and DNA repair [103, 104]. This bidirectional regulation underscores a feedback mechanism essential for maintaining cellular homeostasis.
The implications of these findings extend to various pathological conditions. Dysregulation of PIPn signaling pathways can contribute to diseases such as cancer, neurodegenerative disorders, and autoimmune conditions, highlighting the importance of understanding these signaling networks in cellular behavior and disease pathology [159,160,161,162,163]. The interdependence of microfilaments, IFs, and microtubules, regulated by PIPn signaling, is crucial for cellular functions and responses to environmental stimuli, including migration, division, and intracellular transport.
Despite advances in actin filament research, the current understanding of PIPn signaling in cytoskeletal dynamics highlights significant gaps regarding direct interactions with IFs and microtubules. Existing studies largely focus on indirect regulation via pathways like PI3K/AKT, leaving the mechanisms of direct PIPn-IF/microtubule modulation unclear. Future research should explore the interactions between PIPn signaling and IFs/microtubules, particularly their contributions to cytoskeletal stability and dynamics in various cellular contexts. Developing therapeutic strategies targeting PIPn signaling pathways also presents a promising avenue for restoring cytoskeletal integrity and function in diseases characterized by dysregulation. For instance, microtubule-targeting agents like paclitaxel (Taxol) stabilize microtubules and treat various cancers. Alternatively, agents like vincristine disrupt microtubule dynamics yet still show therapeutic value in the context of some cancers [164,165,166]. Moreover, drugs that enhance or mimic PIPn signaling, such as the PtdIns(3,4,5)P3 analogs, may offer novel therapeutic options in contexts where PIPn signaling is compromised [167]. In neurodegenerative diseases, drugs such as latrunculin, which sequesters actin monomers and disrupts actin polymerization, may offer insights into treatments that target actin dynamics [168,169,170,171]. Additionally, the role of PtdIns(3,4)P2 in coordinating microtubule and actin dynamics warrants further investigation to better understand its contributions to intracellular transport and membrane remodeling.
In conclusion, PIPn signaling is a fundamental regulator of cytoskeletal dynamics with significant implications for understanding cell biology and developing therapeutic interventions. Further investigation into these pathways and the clinical application of drugs targeting cytoskeletal stability will enhance our knowledge of cellular processes and inform strategies for addressing cytoskeletal-related diseases.
References
Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65.
Chen M, Wen T, Horn HT, Chandrahas VK, Thapa N, Choi S, et al. The nuclear phosphoinositide response to stress. Cell Cycle. 2020;19:268–89.
Hokin MR, Hokin LE. Enzyme secretion and the incorporation of P32 into phospholipides of pancreas slices. J Biol Chem. 1953;203:967–77.
Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–7.
Rose HG, Frenster JH. Composition and metabolism of lipids within repressed and active chromatin of interphase lymphocytes. Biochim Biophys Acta. 1965;106:577–91.
Manzoli FA, Maraldi NM, Cocco L, Capitani S, Facchini A. Chromatin phospholipids in normal and chronic lymphocytic leukemia lymphocytes. Cancer Res. 1977;37:843–9.
Smith CD, Wells WW. Phosphorylation of rat liver nuclear envelopes. II. Characterization of in vitro lipid phosphorylation. J Biol Chem. 1983;258:9368–73.
Cocco L, Gilmour RS, Ognibene A, Letcher AJ, Manzoli FA, Irvine RF. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem J. 1987;248:765–70.
Carrillo ND, Chen M, Wen T, Awasthi P, Wolfe TJ, Cryns VL, et al. Lipid transfer proteins and a PI 4-kinase initiate nuclear phosphoinositide signaling. bioRxiv. 2024.
Chen M, Choi S, Wen T, Chen C, Thapa N, Lee JH, et al. A p53-phosphoinositide signalosome regulates nuclear AKT activation. Nat Cell Biol. 2022;24:1099–113.
Choi S, Chen M, Cryns VL, Anderson RA. A nuclear phosphoinositide kinase complex regulates p53. Nat Cell Biol. 2019;21:462–75.
Chen M, Horn HT, Wen T, Cryns VL, Anderson RA. Assessing in situ phosphoinositide-protein interactions through fluorescence proximity ligation assay in cultured cells. Methods Mol Biol. 2021;2251:133–42.
Sobol M, Krausova A, Yildirim S, Kalasova I, Faberova V, Vrkoslav V, et al. Nuclear phosphatidylinositol 4,5-bisphosphate islets contribute to efficient RNA polymerase II-dependent transcription. J Cell Sci. 2018;131:jcs211094.
Thapa N, Chen M, Horn HT, Choi S, Wen T, Anderson RA. Phosphatidylinositol-3-OH kinase signalling is spatially organized at endosomal compartments by microtubule-associated protein 4. Nat Cell Biol. 2020;22:1357–70.
Chen KW, Boucher D, Broz P. Divide to conquer: NLRP3 is activated on dispersed trans-Golgi network. Cell Res. 2019;29:181–2.
Lin TC, Kuo HH, Wu YC, Pan TS, Yih LH. Phosphatidylinositol-5-phosphate 4-kinase gamma accumulates at the spindle pole and prevents microtubule depolymerization. Cell Div. 2019;14:9.
Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, et al. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–7.
Thapa N, Wen T, Cryns VL, Anderson RA. Regulation of cell adhesion and migration via microtubule cytoskeleton organization, cell polarity, and phosphoinositide signaling. biomolecules. 2023;13:1430.
Singh R. Central role of PI3K-SYK interaction in fibrinogen-induced lamellipodia and filopodia formation in platelets. FEBS Open Bio. 2016;6:1285–96.
Hilpelä P, Vartiainen M, Lappalainen P. Regulation of the actin cytoskeleton by PI (4, 5) P 2 and PI (3, 4, 5) P 3. Phosphoinositides in subcellular targeting and enzyme activation. 2004;282:117–63.
Cooper GM, Ganem D. The cell: a molecular approach. Nat Med. 1997;3:1042.
dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, et al. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev. 2003;83:433–73.
Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90:259–89.
Lappalainen P, Kotila T, Jegou A, Romet-Lemonne G. Biochemical and mechanical regulation of actin dynamics. Nat Rev Mol Cell Biol. 2022;23:836–52.
Neuhaus JM, Wanger M, Keiser T, Wegner A. Treadmilling of actin. J Muscle Res Cell Motil. 1983;4:507–27.
Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–21.
Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–22.
Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–60.
Naumanen P, Lappalainen P, Hotulainen P. Mechanisms of actin stress fibre assembly. J Microsc. 2008;231:446–54.
Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–45.
Svitkina T. The actin cytoskeleton and actin-based motility. Cold Spring Harb Perspect Biol. 2018;10:a018267.
Seetharaman S, Etienne-Manneville S. Cytoskeletal crosstalk in cell migration. Trends Cell Biol. 2020;30:720–35.
Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–63.
Kelkar M, Bohec P, Charras G. Mechanics of the cellular actin cortex: from signalling to shape change. Current Opin Cell Biol. 2020;66:69–78.
Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008;9:344–56.
Senju Y, Kalimeri M, Koskela EV, Somerharju P, Zhao H, Vattulainen I, et al. Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc Natl Acad Sci USA. 2017;114:E8977–86.
Vasilev F, Ezhova Y, Chun JT. Signaling enzymes and ion channels being modulated by the actin cytoskeleton at the plasma membrane. Int J Mol Sci. 2021;22:10366.
Pollard TD. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol. 2016;8:a018226.
Divecha N. Lipid kinases: charging PtdIns(4,5)P2 synthesis. Current Biol. 2010;20:R154–R7.
Ferrell JE Jr., Huestis WH. Phosphoinositide metabolism and the morphology of human erythrocytes. J Cell Biol. 1984;98:1992–8.
Sechi AS, Wehland J. The actin cytoskeleton and plasma membrane connection: PtdIns (4, 5) P 2 influences cytoskeletal protein activity at the plasma membrane. J Cell Sci. 2000;113:3679–80.
Yamaguchi H, Shiraishi M, Fukami K, Tanabe A, Ikeda-Matsuo Y, Naito Y, et al. MARCKS regulates lamellipodia formation induced by IGF-I via association with PIP2 and β-actin at membrane microdomains. J Cell Physiol. 2009;220:748–55.
Gorbatyuk VY, Nosworthy NJ, Robson SA, Bains NP, Maciejewski MW, Dos Remedios CG, et al. Mapping the phosphoinositide-binding site on chick cofilin explains how PIP2 regulates the cofilin-actin interaction. Mol Cell. 2006;24:511–22.
Zhao H, Hakala M, Lappalainen P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP2-density sensor. Biophys J. 2010;98:2327–36.
Pantaloni D, Carlier M-F. How profilin promotes actin filament assembly in the presence of thymosin β4. Cell. 1993;75:1007–14.
Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–9.
Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4, 5-bisphosphate and profilactin. Nature. 1985;314:472–4.
Lu P-J, Shieh W-R, Rhee SG, Yin HL, Chen C-S. Lipid products of phosphoinositide 3-kinase bind human profilin with high affinity. Biochemistry. 1996;35:14027–34.
van Rheenen J, Song X, van Roosmalen W, Cammer M, Chen X, Desmarais V, et al. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–59.
Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol. 2001;155:251–60.
Vartiainen MK, Sarkkinen EM, Matilainen T, Salminen M, Lappalainen P. Mammals have two twinfilin isoforms whose subcellular localizations and tissue distributions are differentially regulated. J Biol Chem. 2003;278:34347–55.
Kwiatkowski DJ. Functions of gelsolin: motility, signaling, apoptosis, cancer. Curr Opin Cell Biol. 1999;11:103–8.
Janmey PA, Stossel TP. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–4.
Xian W, Janmey PA. Dissecting the gelsolin-polyphosphoinositide interaction and engineering of a polyphosphoinositide-sensitive gelsolin C-terminal half protein. J Mol Biol. 2002;322:755–71.
Flanagan LA, Cunningham CC, Chen J, Prestwich GD, Kosik KS, Janmey PA. The structure of divalent cation-induced aggregates of PIP2 and their alteration by gelsolin and tau. Biophys J. 1997;73:1440–7.
Takenawa T, Itoh T. Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim Biophys Acta. 2001;1533:190–206.
Rohatgi R, Ho H-YH, Kirschner MW. Mechanism of N-Wasp activation by Cdc42 and phosphatidylinositol 4,5-bisphosphate. J Cell Biol. 2000;150:1299–310.
Higgs HN, Pollard TD. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol. 2000;150:1311–20.
Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–31.
Shewan A, Eastburn DJ, Mostov K. Phosphoinositides in cell architecture. Cold Spring Harb Perspect Biol. 2011;3:a004796.
Hirsch E, Braccini L, Ciraolo E, Morello F, Perino A. Twice upon a time: PI3K’s secret double life exposed. Trends Biochem Sci. 2009;34:244–8.
Srinivasan S, Wang F, Glavas S, Ott A, Hofmann F, Aktories K, et al. Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J Cell Biol. 2003;160:375–85.
Charest PG, Firtel RA. Big roles for small GTPases in the control of directed cell movement. Biochem J. 2007;401:377–90.
Tanna CE, Goss LB, Ludwig CG, Chen PW. Arf GAPs as regulators of the actin cytoskeleton-An update. Int J Mol Sci. 2019;20:442.
Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S, Fujiwara T, et al. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–6.
Yan C, Martinez-Quiles N, Eden S, Shibata T, Takeshima F, Shinkura R, et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–12.
Han KA, Ko J. Orchestration of synaptic functions by WAVE regulatory complex-mediated actin reorganization. Exp Mol Med. 2023;55:1065–75.
Malin J, Rosa-Birriel C, Hatini V. Pten, PI3K, and PtdIns(3,4,5)P3 dynamics control pulsatile actin branching in <em>Drosophila</em> retina morphogenesis. Dev Cell. 2024;59:1593–608.e6.
Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, et al. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat Cell Biol. 2004;6:420–6.
Desale SE, Chinnathambi S. Phosphoinositides signaling modulates microglial actin remodeling and phagocytosis in Alzheimer’s disease. Cell Commun Signal. 2021;19:28.
Moens PD, Bagatolli LA. Profilin binding to sub-micellar concentrations of phosphatidylinositol (4,5) bisphosphate and phosphatidylinositol (3,4,5) trisphosphate. Biochim Biophys Acta. 2007;1768:439–49.
Oikawa T, Takenawa T. PtdIns(3,4)P2 instigates focal adhesions to generate podosomes. Cell Adh Migr. 2009;3:195–7.
Posor Y, Kampyli C, Bilanges B, Ganguli S, Koch PA, Wallroth A, et al. Local synthesis of the phosphatidylinositol-3,4-bisphosphate lipid drives focal adhesion turnover. Dev Cell. 2022;57:1694–711.e7.
Yamaguchi H, Yoshida S, Muroi E, Yoshida N, Kawamura M, Kouchi Z, et al. Phosphoinositide 3-kinase signaling pathway mediated by p110α regulates invadopodia formation. J Cell Biol. 2011;193:1275–88.
Montaño-Rendón F, Walpole GFW, Krause M, Hammond GRV, Grinstein S, Fairn GD. PtdIns(3,4)P2, Lamellipodin, and VASP coordinate actin dynamics during phagocytosis in macrophages. J Cell Biol. 2022;221:e202207042.
Dutour-Provenzano G, Etienne-Manneville S. Intermediate filaments. Current Biol. 2021;31:R522–R9.
Kechagia Z, Eibauer M, Medalia O. Structural determinants of intermediate filament mechanics. Current Opin Cell Biol. 2024;89:102375.
Chang L, Goldman RD. Intermediate filaments mediate cytoskeletal crosstalk. Nat Rev Mol Cell Biol. 2004;5:601–13.
Lowery J, Kuczmarski ER, Herrmann H, Goldman RD. Intermediate filaments play a pivotal role in regulating cell architecture and function. J Biol Chem. 2015;290:17145–53.
Lazarus BS, Chadha C, Velasco-Hogan A, Barbosa JDV, Jasiuk I, Meyers MA. Engineering with keratin: a functional material and a source of bioinspiration. iScience. 2021;24:102798.
Lolicato F, Nickel W, Haucke V, Ebner M. Phosphoinositide switches in cell physiology - From molecular mechanisms to disease. J Biol Chem. 2024;300:105757.
Moujaber O, Stochaj U. The Cytoskeleton as Regulator of Cell Signaling Pathways. Trends Biochem Sci. 2020;45:96–107.
Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129:705–33.
Fortier AM, Asselin E, Cadrin M. Keratin 8 and 18 loss in epithelial cancer cells increases collective cell migration and cisplatin sensitivity through claudin1 up-regulation. J Biol Chem. 2013;288:11555–71.
Ku NO, Toivola DM, Strnad P, Omary MB. Cytoskeletal keratin glycosylation protects epithelial tissue from injury. Nat Cell Biol. 2010;12:876–85.
Paramio JM, Segrelles C, Ruiz S, Jorcano JL. Inhibition of protein kinase B (PKB) and PKCzeta mediates keratin K10-induced cell cycle arrest. Mol Cell Biol. 2001;21:7449–59.
Santos M, Paramio JM, Bravo A, Ramirez A, Jorcano JL. The expression of keratin k10 in the basal layer of the epidermis inhibits cell proliferation and prevents skin tumorigenesis. J Biol Chem. 2002;277:19122–30.
Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441:362–5.
Chivu-Economescu M, Dragu DL, Necula LG, Matei L, Enciu AM, Bleotu C, et al. Knockdown of KRT17 by siRNA induces antitumoral effects on gastric cancer cells. Gastric Cancer. 2017;20:948–59.
Sivagurunathan S, Vahabikashi A, Yang H, Zhang J, Vazquez K, Rajasundaram D, et al. Expression of vimentin alters cell mechanics, cell-cell adhesion, and gene expression profiles suggesting the induction of a hybrid EMT in human mammary epithelial cells. Front Cell Dev Biol. 2022;10:929495.
Alisafaei F, Mandal K, Saldanha R, Swoger M, Yang H, Shi X, et al. Vimentin is a key regulator of cell mechanosensing through opposite actions on actomyosin and microtubule networks. Commun Biol. 2024;7:658.
Patteson AE, Vahabikashi A, Pogoda K, Adam SA, Mandal K, Kittisopikul M, et al. Vimentin protects cells against nuclear rupture and DNA damage during migration. J Cell Biol. 2019;218:4079–92.
Ostrowska-Podhorodecka Z, Ding I, Norouzi M, McCulloch CA. Impact of vimentin on regulation of cell signaling and matrix remodeling. Front Cell Dev Biol. 2022;10:869069.
Bargagna-Mohan P, Hamza A, Kim YE, Khuan Abby Ho Y, Mor-Vaknin N, Wendschlag N, et al. The tumor inhibitor and antiangiogenic agent withaferin A targets the intermediate filament protein vimentin. Chem Biol. 2007;14:623–34.
Zhu QS, Rosenblatt K, Huang KL, Lahat G, Brobey R, Bolshakov S, et al. Vimentin is a novel AKT1 target mediating motility and invasion. Oncogene. 2011;30:457–70.
Pérez-Sala D, Oeste CL, Martínez AE, Carrasco MJ, Garzón B, Cañada FJ. Vimentin filament organization and stress sensing depend on its single cysteine residue and zinc binding. Nat Commun. 2015;6:7287.
Usman S, Waseem NH, Nguyen TKN, Mohsin S, Jamal A, Teh MT, et al. Vimentin is at the heart of epithelial mesenchymal transition (EMT) mediated metastasis. Cancers. 2021;13:4985.
Liu S, Huang J, Zhang Y, Liu Y, Zuo S, Li R. MAP2K4 interacts with Vimentin to activate the PI3K/AKT pathway and promotes breast cancer pathogenesis. Aging. 2019;11:10697–710.
Mohanasundaram P, Coelho-Rato LS, Modi MK, Urbanska M, Lautenschläger F, Cheng F, et al. Cytoskeletal vimentin regulates cell size and autophagy through mTORC1 signaling. PLoS Biol. 2022;20:e3001737.
Malashicheva A, Perepelina K. Diversity of nuclear lamin A/C action as a key to tissue-specific regulation of cellular identity in health and disease. Front Cell Dev Biol. 2021;9:761469.
Lin F, Worman HJ. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 1995;27:230–6.
Alcorta-Sevillano N, Macías I, Rodríguez CI, Infante A. Crucial role of lamin A/C in the migration and differentiation of MSCs in bone. Cells. 2020;9:1330.
Escudeiro-Lopes S, Filimonenko VV, Jarolimova L, Hozak P. Lamin A/C and PI(4,5)P2-A novel complex in the cell nucleus. Cells. 2024;13:399.
Kong L, Schafer G, Bu H, Zhang Y, Zhang Y, Klocker H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis. 2012;33:751–9.
Banfic H, Visnjic D, Mise N, Balakrishnan S, Deplano S, Korchev YE, et al. Epidermal growth factor stimulates translocation of the class II phosphoinositide 3-kinase PI3K-C2beta to the nucleus. Biochem J. 2009;422:53–60.
Sindic A, Aleksandrova A, Fields AP, Volinia S, Banfic H. Presence and activation of nuclear phosphoinositide 3-kinase C2beta during compensatory liver growth. J Biol Chem. 2001;276:17754–61.
Green KJ, Simpson CL. Desmosomes: new perspectives on a classic. J Investig Dermatol. 2007;127:2499–515.
Franz H, Rathod M, Zimmermann A, Stüdle C, Beyersdorfer V, Leal-Fischer K, et al. Unbiased screening identifies regulators of cell-cell adhesion and treatment options in pemphigus. Nat Commun. 2024;15:8044.
Johnson JL, Najor NA, Green KJ. Desmosomes: regulators of cellular signaling and adhesion in epidermal health and disease. Cold Spring Harb Perspect Med. 2014;4:a015297.
Bendrick JL, Eldredge LA, Williams EI, Haight NB, Dubash AD. Desmoplakin harnesses rho GTPase and p38 mitogen-activated protein kinase signaling to coordinate cellular migration. J Investig Dermatol. 2019;139:1227–36.
Müller L, Hatzfeld M, Keil R. Desmosomes as signaling hubs in the regulation of cell behavior. Front Cell Dev Biol. 2021;9:745670.
Wolf A, Rietscher K, Glass M, Huttelmaier S, Schutkowski M, Ihling C, et al. Insulin signaling via Akt2 switches plakophilin 1 function from stabilizing cell adhesion to promoting cell proliferation. J Cell Sci. 2013;126:1832–44.
Rietscher K, Keil R, Jordan A, Hatzfeld M. 14-3-3 proteins regulate desmosomal adhesion via plakophilins. J Cell Sci. 2018;131:jcs212191.
D’Abronzo LS, Ghosh PM. eIF4E Phosphorylation in prostate cancer. Neoplasia. 2018;20:563–73.
Wolf A, Krause-Gruszczynska M, Birkenmeier O, Ostareck-Lederer A, Huttelmaier S, Hatzfeld M. Plakophilin 1 stimulates translation by promoting eIF4A1 activity. J Cell Biol. 2010;188:463–71.
Dustin P. Microtubules (Springer Science & Business Media, 2012).
Stephens R, Edds K. Microtubules: structure, chemistry, and function. Physiol Rev. 1976;56:709–77.
Hawkins T, Mirigian M, Yasar MS, Ross JL. Mechanics of microtubules. J Biomech. 2010;43:23–30.
Baas PW, Rao AN, Matamoros AJ, Leo L. Stability properties of neuronal microtubules. Cytoskeleton. 2016;73:442–60.
Etienne-Manneville S. Microtubules in cell migration. Ann Rev Cell Dev Biol. 2013;29:471–99.
Sferra A, Nicita F, Bertini E. Microtubule dysfunction: a common feature of neurodegenerative diseases. Int J Mol Sci. 2020;21:7354.
Baird FJ, Bennett CL. Microtubule defects & Neurodegeneration. J Genet Syndr Gene Ther. 2013;4:203.
Marchisella F, Coffey ET, Hollos P. Microtubule and microtubule associated protein anomalies in psychiatric disease. Cytoskeleton. 2016;73:596–611.
Surridge CD, Burns RG. Phosphatidylinositol inhibits microtubule assembly by binding to microtubule-associated protein 2 at a single, specific, high-affinity site. Biochemistry. 1992;31:6140–4.
Maccioni RB, Cambiazo V. Role of microtubule-associated proteins in the control of microtubule assembly. Physiol Rev. 1995;75:835–64.
Goodson HV, Jonasson EM. Microtubules and microtubule-associated proteins. Cold Spring Harb Perspect Biol. 2018;10:a022608.
Bodakuntla S, Jijumon A, Villablanca C, Gonzalez-Billault C, Janke C. Microtubule-associated proteins: structuring the cytoskeleton. Trends Cell Biol. 2019;29:804–19.
Barlan K, Gelfand VI. Microtubule-based transport and the distribution, tethering, and organization of organelles. Cold Spring Harb Perspect Biol. 2017;9:a025817.
Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–80.
Ali I, Yang W-C. The functions of kinesin and kinesin-related proteins in eukaryotes. Cell Adhes Migr. 2020;14:139–52.
Scholz T, Mandelkow E. Transport and diffusion of Tau protein in neurons. Cell Mol Life Sci. 2014;71:3139–50.
Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2004;6:204.
Wordeman L. How kinesin motor proteins drive mitotic spindle function: lessons from molecular assays. Semin Cell Dev Biol. 2010;21:260–8.
Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–16.
Popova JS, Greene AK, Wang J, Rasenick MM. Phosphatidylinositol 4,5-bisphosphate modifies tubulin participation in phospholipase Cbeta1 signaling. J Neurosci. 2002;22:1668–78.
Kapeller R, Toker A, Cantley LC, Carpenter CL. Phosphoinositide 3-kinase binds constitutively to α/β-tubulin and binds to γ-tubulin in response to insulin. J Biol Chem. 1995;270:25985–91.
Wei HC, Rollins J, Fabian L, Hayes M, Polevoy G, Bazinet C, et al. Depletion of plasma membrane PtdIns(4,5)P2 reveals essential roles for phosphoinositides in flagellar biogenesis. J Cell Sci. 2008;121:1076–84.
Loijens JC, Anderson RA. Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J Biol Chem. 1996;271:32937–43.
Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089–118.
Noda Y, Niwa S, Homma N, Fukuda H, Imajo-Ohmi S, Hirokawa N. Phosphatidylinositol 4-phosphate 5-kinase alpha (PIPKα) regulates neuronal microtubule depolymerase kinesin, KIF2A and suppresses elongation of axon branches. Proc Natl Acad Sci USA. 2012;109:1725–30.
Singla A, Fedoseienko A, Giridharan SSP, Overlee BL, Lopez A, Jia D, et al. Endosomal PI(3)P regulation by the COMMD/CCDC22/CCDC93 (CCC) complex controls membrane protein recycling. Nat Commun. 2019;10:4271.
Thapa N, Chen M, Cryns VL, Anderson R. A p85 isoform switch enhances PI3K activation on endosomes by a MAP4- and PI3P-dependent mechanism. Cell Rep. 2024;43:114119.
Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–72.
Chen M, Zhang Y, Yu VC, Chong YS, Yoshioka T, Ge R. Isthmin targets cell-surface GRP78 and triggers apoptosis via induction of mitochondrial dysfunction. Cell Death Differ. 2014;21:797–810.
Chen M, Qiu T, Wu J, Yang Y, Wright GD, Wu M, et al. Extracellular anti-angiogenic proteins augment an endosomal protein trafficking pathway to reach mitochondria and execute apoptosis in HUVECs. Cell Death Differ. 2018;25:1905–20.
Burd CG. Physiology and pathology of endosome-to-Golgi retrograde sorting. Traffic. 2011;12:948–55.
Chaumet A, Wright GD, Seet SH, Tham KM, Gounko NV, Bard F. Nuclear envelope-associated endosomes deliver surface proteins to the nucleus. Nat Commun. 2015;6:8218.
Liao J-C, Yang TT, Weng RR, Kuo C-T, Chang C-W. TTBK2: a tau protein kinase beyond tau phosphorylation. BioMed Res Int. 2015;2015:575170.
Avila J, Lucas JJ, Perez M, Hernandez F. Role of tau protein in both physiological and pathological conditions. Physiol Rev. 2004;84:361–84.
Goetz SC, Liem KF, Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 2012;151:847–58.
Houlden H, Johnson J, Gardner-Thorpe C, Lashley T, Hernandez D, Worth P, et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat Genet. 2007;39:1434–6.
Taylor LM, McMillan PJ, Liachko NF, Strovas TJ, Ghetti B, Bird TD, et al. Pathological phosphorylation of tau and TDP-43 by TTBK1 and TTBK2 drives neurodegeneration. Mol Neurodegener. 2018;13:1–14.
Xu Q, Zhang Y, Wei Q, Huang Y, Hu J, Ling K. Phosphatidylinositol phosphate kinase PIPKIgamma and phosphatase INPP5E coordinate initiation of ciliogenesis. Nat Commun. 2016;7:10777.
Feng Z, Yu CH. PI(3,4)P(2)-mediated membrane tubulation promotes integrin trafficking and invasive cell migration. Proc Natl Acad Sci USA. 2021;118:e2017645118.
Román-Fernández Á, Roignot J, Sandilands E, Nacke M, Mansour MA, McGarry L, et al. The phospholipid PI(3,4)P2 is an apical identity determinant. Nat Commun. 2018;9:5041.
Prever L, Squillero G, Hirsch E, Gulluni F. Linking phosphoinositide function to mitosis. Cell Rep. 2024;43:114273.
Kölsch V, Charest PG, Firtel RA. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci. 2008;121:551–9.
Deng S, Leong HC, Datta A, Gopal V, Kumar AP, Yap CT. PI3K/AKT signaling tips the balance of cytoskeletal forces for cancer progression. Cancers. 2022;14:1652.
Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017;170:605–35.
Chu E, Mychasiuk R, Hibbs ML, Semple BD. Dysregulated phosphoinositide 3-kinase signaling in microglia: shaping chronic neuroinflammation. J Neuroinflam. 2021;18:276.
Glaviano A, Foo ASC, Lam HY, Yap KCH, Jacot W, Jones RH, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. 2023;22:138.
Ghavami S, Shojaei S, Yeganeh B, Ande SR, Jangamreddy JR, Mehrpour M, et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Progress Neurobiol. 2014;112:24–49.
Ren C, Carrillo ND, Cryns VL, Anderson RA, Chen M. Environmental pollutants and phosphoinositide signaling in autoimmunity. J Hazard Mater. 2024;465:133080.
Mukhtar E, Adhami VM, Mukhtar H. Targeting microtubules by natural agents for cancer therapy. Mol Cancer Ther. 2014;13:275–84.
Shelby RD, Hahn KM, Sullivan KF. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J Cell Biol. 1996;135:545–57.
Perez EA. Microtubule inhibitors: differentiating tubulin-inhibiting agents based on mechanisms of action, clinical activity, and resistance. Mol Cancer Ther. 2009;8:2086–95.
Zhang H, He J, Kutateladze TG, Sakai T, Sasaki T, Markadieu N, et al. 5-Stabilized phosphatidylinositol 3,4,5-trisphosphate analogues bind Grp1 PH, inhibit phosphoinositide phosphatases, and block neutrophil migration. Chembiochem. 2010;11:388–95.
Peng GE, Wilson SR, Weiner OD. A pharmacological cocktail for arresting actin dynamics in living cells. Mol Biol Cell. 2011;22:3986–94.
Bernstein BW, Bamburg JR. Actin-ATP hydrolysis is a major energy drain for neurons. J Neurosci. 2003;23:1–6.
Martín-Cámara O, Cores Á, López-Alvarado P, Menéndez JC. Emerging targets in drug discovery against neurodegenerative diseases: control of synapsis disfunction by the RhoA/ROCK pathway. Eur J Med Chem. 2021;225:113742.
Parisiadou L, Cai H. LRRK2 function on actin and microtubule dynamics in Parkinson’s disease. Commun Int Biol. 2010;3:396–400.
Ungewickell A, Hugge C, Kisseleva M, Chang SC, Zou J, Feng Y, et al. The identification and characterization of two phosphatidylinositol-4,5-bisphosphate 4-phosphatases. Proc Natl Acad Sci USA. 2005;102:18854–9.
Willett R, Martina JA, Zewe JP, Wills R, Hammond GRV, Puertollano R. TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat Commun. 2017;8:1580.
Acknowledgements
M.C. is supported by grant D2301007 from the Shenzhen Medical Research Fund, grant JCYJ20240813094605008 from Shenzhen Natural Science Foundation, grant 2023A1515110237 from the Guangdong Province Basic and Applied Basic Research Foundation, and grant 32400577 from the National Natural Science Foundation of China. S.W. is supported by grants 82030031, 92149301, and 82401082 from the National Natural Science Foundation of China, grant 2019–12M-5-031 from Chinese Academy of Medical Sciences Research Unit, grants PXM2021_014226_000041 and PXM2021_014226_000020 from Beijing Municipality Government, grant CXTD202201 from Innovation Research Team Project of Beijing Stomatological Hospital, Capital Medical University. J.S. is supported by grant 82071193 from the National Natural Science Foundation of China, grant 0920220233 from the Guangdong Zhujiang Program, grants JCYJ20220818102611025 and RCYX20210706092040044 from the Science and Technology Foundation of Shenzhen. This work was supported in part by a National Institutes of Health grant R35GM134955 (R.A.A.) and R01CA286492 (R.A.A. & V.L.C), Department of Defense Breast Cancer Research Program grants W81XWH-17-1-0258 (R.A.A.), W81XWH-17-1-0259 (V.L.C.), W81XWH-21-1-0129 (V.L.C.), HT9425-23-1-0553 (V.L.C.), and HT9425-23-1- 0554 (R.A.A.), and a grant from the Breast Cancer Research Foundation (V.L.C.).
Author information
Authors and Affiliations
Contributions
X.H. and Y.C.: drafting, visualization, and editing; N.D.C.: visualization and editing; R.A.A., V.L.C., and J.S.: editing and funding acquisition; S.W. and M.C.: supervision, drafting, visualization, editing, and funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Anastasis Stephanou
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hou, X., Chen, Y., Carrillo, N.D. et al. Phosphoinositide signaling at the cytoskeleton in the regulation of cell dynamics. Cell Death Dis 16, 296 (2025). https://doi.org/10.1038/s41419-025-07616-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-025-07616-x