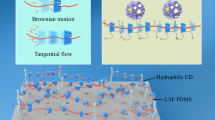
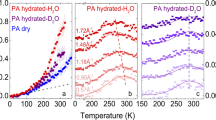
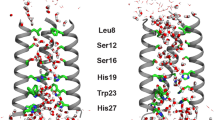
Abstract
The behavior of water molecules significantly influences the effectiveness of protein stabilizers and biomaterials. Although the polymerization of low-molecular-weight molecules enhances their functionality, the hydration states and water dynamics around polymers and small molecules are typically examined separately. Therefore, the effect of polymerization on water dynamics at the molecular level remains unclear. By density functional tight-binding molecular dynamics (DFTB-MD) simulations of five zwitterionic solute solutions, (trimethylamine N-oxide) (TMAO), the N-[3-(dimethylamino)propyl]acrylamide N-oxide (DMAO) monomer, poly(N-[3-(dimethylamino)propyl]acrylamide N-oxide) (PDMAO), the 2-methacryloyloxyethyl phosphorylcholine (MPC) monomer, and poly(2-methacryloyloxyethyl phosphorylcholine) (PMPC), the effects of polymerization on water dynamics were investigated. DMAO and MPC polymerization (to PDMAO and PMPC, respectively) promote the slow and rapid rotation of water molecules, respectively. In PDMAO, water molecules are trapped between side chains due to the formation of hydrogen bonds between water and PDMAO, resulting in slow water dynamics, whereas in PMPC, a reduction in the solvent-accessible surface area due to polymerization disrupts the hydrogen-bond network among the water molecules, resulting in acceleration of the rotational dynamics of water molecules. The hydration amount determined using differential scanning calorimetry (DSC) and terahertz time-domain spectroscopy (THz-TDS) is consistent with the MD simulation results, which provide molecular-level insights that advance the current understanding of water dynamics in small-molecule polymerization for potential functional enhancement.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Li Q, Wen C, Yang J, Zhou X, Zhu Y, Zheng J, et al. Zwitterionic Biomaterials. Chem Rev. 2022;122:17073–154.
Dargaville BL, Hutmacher DW. Water as the often neglected medium at the interface between materials and biology. Nat Commun. 2022;13:4222.
Tanaka M, Mochizuki A. Clarification of the blood compatibility mechanism by controlling the water structure at the blood-poly(meth)acrylate interface. J Biomater Sci Polym Ed. 2010;21:1849–63.
Tanaka M, Mochizuki A. Effect of water structure on blood compatibility— thermal analysis of water in poly(meth)acrylate. J Biomed Mater Res A. 2004;68A:684–95.
Hishida M, Anjum R, Anada T, Murakami D, Tanaka M. Effect of Osmolytes on Water Mobility Correlates with Their Stabilizing Effect on Proteins. J Phys Chem B. 2022;126:2466–75.
Anjum R, Nishimura SN, Kobayashi S, Nishida K, Anada T, Tanaka M. Protein Stabilization Effect of Zwitterionic Osmolyte-bearing Polymer. Chem Lett. 2021;50:1699–702.
Fedotova MV. Compatible osmolytes - bioprotectants: Is there a common link between their hydration and their protective action under abiotic stresses?. J Mol Liq. 2019;292:111339.
Hishida M, Kanno R, Terashima T. Hydration State on Poly(ethylene glycol)-Bearing Homopolymers and Random Copolymer Micelles: In Relation to the Thermoresponsive Property and Micellar Structure. Macromolecules. 2023;56:7587–96.
Israelachvili J, Wennerström H. Role of hydration and water structure in biological and colloidal interactions. Nature. 1996;379:219–25.
Hishida M. Correlation between Hydration States and Self-assembly Structures of Phospholipid and Surfactant Studied by Terahertz Spectroscopy. J Oleo Sci. 2024;73:419–27.
Bagchi B. Water Dynamics in the Hydration Layer around Proteins and Micelles. Chem Rev. 2005;105:3197–219.
Laage D, Elsaesser T, Hynes JT. Water Dynamics in the Hydration Shells of Biomolecules. Chem Rev. 2017;117:10694–725.
Leitenstorfer A, Moskalenko AS, Kampfrath T, Kono J, Castro-Camus E, Peng K, et al. The 2023 terahertz science and technology roadmap. J Phys D Appl Phys. 2023;56:223001.
Shiraga K, Adachi A, Nakamura M, Tajima T, Ajito K, Ogawa Y. Characterization of the hydrogen-bond network of water around sucrose and trehalose: Microwave and terahertz spectroscopic study. J Chem Phys. 2017;146:105102.
Rahman MK, Yamada T, Yamada NL, Higuchi Y, Seto H. Hydration Water Dynamics in Zwitterionic Phospholipid Membranes Mixed with Charged Phospholipids. J Phys Chem B. 2025;129:18.
Schirò G, Fichou Y, Gallat FX, Wood K, Gabel F, Moulin M, et al. Translational diffusion of hydration water correlates with functional motions in folded and intrinsically disordered proteins. Nat Commun. 2015;6:1–8.
Talon C, Smith LJ, Brady JW, Lewis BA, Copley JRD, Price DL, et al. Dynamics of Water Molecules in Glucose Solutions. J Phys Chem B. 2004;108:5120–6.
Li B, Jain P, Ma J, Smith JK, Yuan Z, Hung HC, et al. Trimethylamine N-oxide–derived zwitterionic polymers: A new class of ultralow fouling bioinspired materials. Sci Adv. 2019;5:9562–76.
Senaratne W, Andruzzi L, Ober CK. Self-assembled monolayers and polymer brushes in biotechnology: Current applications and future perspectives. Biomacromolecules. 2005;6:2427–48.
Raynor JE, Capadona JR, Collard DM, Petrie TA, García AJ. Polymer brushes and self-assembled monolayers: Versatile platforms to control cell adhesion to biomaterials (Review). Biointerphases. 2009;4:FA3–16.
Zhang Z, Zhang M, Chen S, Horbett TA, Ratner BD, Jiang S. Blood compatibility of surfaces with superlow protein adsorption. Biomaterials. 2008;29:4285–91.
Kawabe S, Seki M, Tabata H. Evaluation of hydration in a water-soluble polymer by terahertz spectroscopy. Appl Phys Lett. 2016;108:81103.
Tominaga T, Hishida M, Murakami D, Fujii Y, Tanaka M, Seto H. Experimental evidence of slow mode water in the vicinity of poly (ethylene oxide) at physiological temperature. J Phys Chem B. 2022;2022:1758–67.
Nakada M, Ishida H, Furushima Y. Structural and dynamical characterisation of intermediate water interacting polyvinyl pyrrolidone. Materialia (Oxf). 2020;12:100743.
Keefe AJ, Jiang S. Poly(zwitterionic)protein conjugates offer increased stability without sacrificing binding affinity or bioactivity. Nat Chem. 2011;4:59–63.
Baldwin RL. How Hofmeister ion interactions affect protein stability. Biophys J. 1996;71:2056–63.
Parsons DF, Boström M, Nostro P, Lo, Ninham BW. Hofmeister effects: interplay of hydration, nonelectrostatic potentials, and ion size. Phys Chem Chem Phys. 2011;13:12352–67.
Mazzini V, Craig VSJ. What is the fundamental ion-specific series for anions and cations? Ion specificity in standard partial molar volumes of electrolytes and electrostriction in water and non-aqueous solvents. Chem Sci. 2017;8:7052–65.
Ma G, Ji F, Lin W, Chen S. Determination of non-freezing water in different nonfouling materials by differential scanning calorimetry. J Biomater Sci Polym Ed. 2022;33:1012–24.
Leng C, Sun S, Zhang K, Jiang S, Chen Z. Molecular level studies on interfacial hydration of zwitterionic and other antifouling polymers in situ. Acta Biomater. 2016;40:6–15.
Wu J, Lin W, Wang Z, Chen S, Chang Y. Investigation of the hydration of nonfouling material poly(sulfobetaine methacrylate) by low-field nuclear magnetic resonance. Langmuir. 2012;28:7436–41.
Higuchi Y, Saleh MA, Anada T, Tanaka M, Hishida M. Rotational Dynamics of Water near Osmolytes by Molecular Dynamics Simulations. J Phys Chem B. 2024;128:5017.
Meng EC, Kollman PA. Molecular dynamics studies of the properties of water around simple organic solutes. J Phys Chem. 1996;100:11460–70.
Stirnemann G, Duboué-Dijon E, Laage D. Ab Initio Simulations of Water Dynamics in Aqueous TMAO Solutions: Temperature and Concentration Effects. J Phys Chem B. 2017;121:11189–97.
Saladino G, Marenchino M, Pieraccini S, Campos-Olivas R, Sironi M, Gervasio FL. A simple mechanism underlying the effect of protecting osmolytes on protein folding. J Chem Theory Comput. 2011;7:3846–52.
Usui K, Hunger J, Sulpizi M, Ohto T, Bonn M, Nagata Y. Ab Initio Liquid Water Dynamics in Aqueous TMAO Solution. J Phys Chem B. 2015;119:10597–606.
Hower JC, He Y, Bernards MT, Jiang S. Understanding the nonfouling mechanism of surfaces through molecular simulations of sugar-based self-assembled monolayers. J Chem Phys. 2006;125:214704.
Kuo AT, Urata S, Koguchi R, Yamamoto K, Tanaka M. Analyses of equilibrium water content and blood compatibility for Poly(2-methoxyethyl acrylate) by molecular dynamics simulation. Polymer (Guildf). 2019;170:76–84.
Kuo AT, Sonoda T, Urata S, Koguchi R, Kobayashi S, Tanaka M. Elucidating the Feature of Intermediate Water in Hydrated Poly(ω-methoxyalkyl acrylate)s by Molecular Dynamics Simulation and Differential Scanning Calorimetry Measurement. ACS Biomater Sci Eng. 2020;6:3915–24.
Shikata K, Kikutsuji T, Yasoshima N, Kim K, Matubayasi N. Revealing the hidden dynamics of confined water in acrylate polymers: Insights from hydrogen-bond lifetime analysis. J Chem Phys. 2023;158:174901.
Kuo AT, Urata S, Koguchi R, Sonoda T, Kobayashi S, Tanaka M. Molecular Dynamics Study on the Water Mobility and Side-Chain Flexibility of Hydrated Poly(ω-methoxyalkyl acrylate)s. ACS Biomater Sci Eng. 2020;6:6690–700.
Yang C, Lu D, Liu Z. How PEGylation enhances the stability and potency of insulin: A molecular dynamics simulation. Biochemistry. 2011;50:2585–93.
Martinez L, Andrade R, Birgin EG, Martínez JM. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J Comput Chem. 2009;30:2157–64.
Hourahine B, Aradi B, Blum V, Bonafé F, Buccheri A, Camacho C, et al. DFTB+, a software package for efficient approximate density functional theory based atomistic simulations. J Chem Phys. 2020;152:124101.
Gaus M, Goez A, Elstner M. Parametrization and benchmark of DFTB3 for organic molecules. J Chem Theory Comput. 2013;9:338–54.
Kubillus M, Kubař T, Gaus M, Řezáč J, Elstner M. Parameterization of the DFTB3 method for Br, Ca, Cl, F, I, K, and Na in organic and biological systems. J Chem Theory Comput. 2015;11:332–42.
Goyal P, Qian H-J, Irle S, Lu X, Roston D, Mori T, et al. Molecular simulation of water and hydration effects in different environments: Challenges and developments for DFTB based models. J Phys Chem B. 2014;118:11007–27.
Řezáč J. Empirical Self-Consistent Correction for the Description of Hydrogen Bonds in DFTB3. J Chem Theory Comput. 2017;13:4804–17.
Sakti AW, Nishimura Y, Nakai H. Divide-and-conquer-type density-functional tight-binding simulations of hydroxide ion diffusion in bulk water. J Phys Chem B. 2017;121:1362–71.
Choi TH, Liang R, Maupin CM, Voth GA. Application of the SCC-DFTB method to hydroxide water clusters and aqueous hydroxide solutions. J Phys Chem B. 2013;117:5165–79.
Higuchi Y, Asano Y, Kuwahara T, Hishida M. Rotational Dynamics of Water at the Phospholipid Bilayer Depending on the Head Groups Studied by Molecular Dynamics Simulations. Langmuir. 2021;37:5329–38.
Abascal JLF, Vega C. A general purpose model for the condensed phases of water: TIP4P/2005. J Chem Phys. 2005;123:234505.
Martyna GJ, Tuckerman ME, Tobias DJ, Klein ML. Explicit reversible integrators for extended systems dynamics. Mol Phys. 1996;87:1117–57.
Berendsen H, Postma J, Van Gunsteren W, DiNola A, Haak J. Molecular dynamics with coupling to an external bath. J Phys Chem. 1984;81:3684–90.
Martí J, Padro JA, Guàrdia E. Molecular dynamics simulation of liquid water along the coexistence curve: Hydrogen bonds and vibrational spectra. J Chem Phys. 1996;105:639–49.
Hatakeyama H, Hatakeyama T. Interaction between water and hydrophilic polymers. Thermochim Acta. 1998;308:3–22.
Tanaka M, Hayashi T, Morita S. The roles of water molecules at the biointerface of medical polymers. Polym J. 2013;45:701–10.
Tanaka M, Motomura T, Ishii N, Shimura K, Onishi M, Mochizuki A, et al. Cold crystallization of water in hydrated poly (2-methoxyethyl acrylate)(PMEA). Polym Int. 2000;49:1709–13.
Shiomoto S, Inoue K, Higuchi H, Nishimura SN, Takaba H, Tanaka M, et al. Characterization of Hydration Water Bound to Choline Phosphate-Containing Polymers. Biomacromolecules. 2022;23:2999–3008.
Hishida M, Tanaka K. Long-range hydration effect of lipid membrane studied by terahertz time-domain spectroscopy. Phys Rev Lett. 2011;106:158102.
Das N, Tarif E, Dutta A, Sen P. Associated Water Dynamics Might Be a Key Factor Affecting Protein Stability in the Crowded Milieu. J Phys Chem B. 2023;127:3151–63.
Negi KS, Das N, Khan T, Sen P. Osmolyte induced protein stabilization: modulation of associated water dynamics might be a key factor. Phys Chem Chem Phys. 2023;25:32602–12.
Hishida M, Kaneko A, Yamamura Y, Saito K. Contrasting Changes in Strongly and Weakly Bound Hydration Water of a Protein upon Denaturation. J Phys Chem B. 2023;127:6296–305.
Privalov PL, Makhatadze GI. Contribution of hydration to protein folding thermodynamics: II. The entropy and gibbs energy of hydration. J Mol Biol. 1993;232:660–79.
Acknowledgements
We are grateful to the Japanese government (MEXT) for providing a scholarship to pursue higher education and research at Kyushu University. This research was partially supported by JSPS KAKENHI (Grant Numbers JP19H05717, JP19H05718, JP19H05720, and JP22H00591). We thank the Supercomputer Center at the Institute for Solid–State Physics, University of Tokyo, for the use of its facilities. We also thank the Cooperative Research Program “Dynamic Alliance for Open Innovation Bridging Human, Environment, and Materials” for their support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saleh, M.A., Higuchi, Y., Shiomoto, S. et al. Effect of zwitterionic monomer polymerization on water dynamics: a molecular dynamics simulation study supported by differential scanning calorimetry and terahertz spectroscopy. Polym J 57, 1127–1139 (2025). https://doi.org/10.1038/s41428-025-01066-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41428-025-01066-0