Abstract
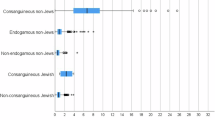
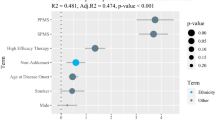
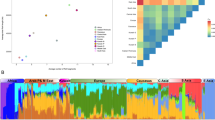
Recent studies have used genome-wide single-nucleotide polymorphisms (SNPs) to investigate relationships among various Jewish populations and their non-Jewish historical neighbors, often focusing on small subsets of populations from a limited geographic range or relatively small samples within populations. Here, building on the significant progress that has emerged from genomic SNP studies in the placement of Jewish populations in relation to non-Jewish populations, we focus on population structure among Jewish populations. In particular, we examine Jewish population-genetic structure in samples that span much of the historical range of Jewish populations in Europe, the Middle East, North Africa, and South Asia. Combining 429 newly genotyped samples from 29 Jewish and 3 non-Jewish populations with previously reported genotypes on Jewish and non-Jewish populations, we investigate variation in 2789 individuals from 114 populations at 486,592 genome-wide autosomal SNPs. Using multidimensional scaling analysis, unsupervised model-based clustering, and population trees, we find that, genetically, most Jewish samples fall into four major clusters that largely represent four culturally defined groupings, namely the Ashkenazi, Mizrahi, North African, and Sephardi subdivisions of the Jewish population. We detect high-resolution population structure, including separation of the Ashkenazi and Sephardi groups and distinctions among populations within the Mizrahi and North African groups. Our results refine knowledge of Jewish population-genetic structure and contribute to a growing understanding of the distinctive genetic ancestry evident in closely related but historically separate Jewish communities.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Mourant AE, Kopec AC, Dmaniewska-Sobczak K. The Genetics of the Jews. Oxford: Clarendon Press; 1978.
Carmelli D, Cavalli-Sforza LL. The genetic origin of the Jews: a multivariate approach. Hum Biol. 1979;51:41–61.
Goodman RM. Genetic disorders among the Jewish people. Baltimore: Johns Hopkins University Press; 1979.
Karlin S, Kenett R, Bonné-Tamir B. Analysis of biochemical genetic data on Jewish populations: II. Results and interpretations of heterogeneity indices and genetic distance measures with respect to standards. Am J Hum Genet. 1979;31:341–65.
Ostrer H. A genetic profile of contemporary Jewish populations. Nat Rev Genet. 2001;2:891–8.
Klitz W, Gragert L, Maiers M, Fernandez-Vina M, Ben-Naeh Y, Benedek G, et al. Genetic differentiation of Jewish populations. Tissue Antigens. 2010;76:442–58.
Ostrer H, Skorecki K. The population genetics of the Jewish people. Hum Genet. 2013;132:119–27.
Rosenberg NA, Weitzman SP. From generation to generation: the genetics of Jewish populations. Hum Biol. 2013;85:817–23.
Atzmon G, Hao L, Pe’er I, Velez C, Pearlman A, Palamara PF, et al. Abraham’s children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern ancestry. Am J Hum Genet. 2010;86:850–9.
Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–42.
Seldin MF, Shigeta R, Villoslada P, Selmi C, Tuomilehto J, Silva G, et al. European population substructure: clustering of northern and southern populations. PLoS Genet. 2006;2:e143.
Bauchet M, McEvoy B, Pearson LN, Quillen EE, Sarkisian T, Hovhannesyan K, et al. Measuring European population stratification with microarray genotype data. Am J Hum Genet. 2007;80:948–56.
Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236.
Tian C, Plenge RM, Ransom M, Lee A, Villoslada P, Selmi C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4.
Kopelman NM, Stone L, Wang C, Gefel D, Feldman MW, Hillel J, et al. Genomic microsatellites identify shared Jewish ancestry intermediate between Middle Eastern and European populations. BMC Genet. 2009;10:80.
Need AC, Kasperaviciute D, Cirulli ET, Goldstein DB. A genome-wide genetic signature of Jewish ancestry perfectly separates individuals with and without full Jewish ancestry in a large random sample of European Americans. Genome Biol. 2009;10:R7.
Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S, Warren ST. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc Natl Acad Sci USA. 2010;107:16222–7.
Listman JB, Hasin D, Kranzler HR, Malison RT, Mutirangura A, Sughondhabirom A, et al. Identification of population substruture among Jews using STR markers and dependence on reference populations studied. BMC Genet. 2010;11:48.
Campbell CL, Palamara PF, Dubrovsky M, Botigué LR, Fellous M, Atzmon G, et al. North African Jewish and non-Jewish populations form distinctive, orthogonal clusters. Proc Natl Acad Sci USA. 2012;109:13865–70.
Behar DM, Metspalu M, Baran Y, Kopelman NM, Yunusbayev B, Gladstein A, et al. No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Hum Biol. 2013;85:859–900.
Waldman YY, Biddanda A, Davidson NR, Billing-Ross P, Dubrovsky M, Campbell CL, et al. The genetics of Bene Israel from India reveals both substantial Jewish and Indian ancestry. PLoS ONE. 2016;11:e0152056.
Waldman YY, Biddanda A, Dubrovsky M, Campbell CL, Oddoux C, Friedman E, et al. The genetic history of Cochin Jews from India. Hum Genet. 2016;135:1127–43.
Rosenberg NA, Woolf E, Pritchard JK, Schaap T, Gefel D, Shpirer I, et al. Distinctive genetic signatures in the Libyan Jews. Proc Natl Acad Sci USA. 2001;98:858–63.
Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4.
International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58.
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, et al. Genetic structure of human populations. Science. 2002;298:2381–5.
Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung H-C, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003.
Mountain JL, Cavalli-Sforza LL. Multilocus genotypes, a tree of individuals, and human evolutionary history. Am J Hum Genet. 1997;61:705–18.
Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Curr Biol. 2005;15:R159–R160.
Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–7.
Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–87.
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 2015;15:1179–91.
Chouraqui AN. Between east and west: a history of the Jews of North Africa. Philadelphia: Jewish Publication Society of America; 1968.
Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987.
Timm NH. Applied multivariate analysis. New York: Springer; 2002.
Le Roux B, Rouanet H. Geometric data analysis: from correspondence analysis to structured data analysis. Dordrecht: Kluwer; 2004.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Seattle, WA: Department of Genome Sciences, University of Washington; 2005.
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–7.
Minch E, Ruiz Linares A, Goldstein DB, Feldman MW, Cavalli-Sforza LL. MICROSAT version 1.5d: a program for calculating statistics on microsatellite data. Stanford, CA: Department of Genetics, Stanford University; 1998.
Acknowledgements
We acknowledge support from the CNRS-PICS program on genetic diversity in Central Asian populations and the University of Michigan Life Sciences Institute/Israeli Universities Partnership.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kopelman, N.M., Stone, L., Hernandez, D.G. et al. High-resolution inference of genetic relationships among Jewish populations. Eur J Hum Genet 28, 804–814 (2020). https://doi.org/10.1038/s41431-019-0542-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-019-0542-y