Abstract
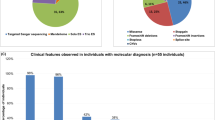
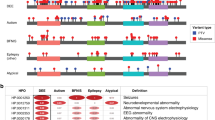
Zhu–Tokita–Takenouchi–Kim (ZTTK) syndrome, an intellectual disability syndrome first described in 2016, is caused by heterozygous loss-of-function variants in SON. Its encoded protein promotes pre-mRNA splicing of many genes essential for development. Whereas individual phenotypic traits have previously been linked to erroneous splicing of SON target genes, the phenotypic spectrum and the pathogenicity of missense variants have not been further evaluated. We present the phenotypic abnormalities in 52 individuals, including 17 individuals who have not been reported before. In total, loss-of-function variants were detected in 49 individuals (de novo in 47, inheritance unknown in 2), and in 3, a missense variant was observed (2 de novo, 1 inheritance unknown). Phenotypic abnormalities, systematically collected and analyzed in Human Phenotype Ontology, were found in all organ systems. Significant inter-individual phenotypic variability was observed, even in individuals with the same recurrent variant (n = 13). SON haploinsufficiency was previously shown to lead to downregulation of downstream genes, contributing to specific phenotypic features. Similar functional analysis for one missense variant, however, suggests a different mechanism than for heterozygous loss-of-function. Although small in numbers and while pathogenicity of these variants is not certain, these data allow for speculation whether de novo missense variants cause ZTTK syndrome via another mechanism, or a separate overlapping syndrome. In conclusion, heterozygous loss-of-function variants in SON define a recognizable syndrome, ZTTK, associated with a broad, severe phenotypic spectrum, characterized by a large inter-individual variability. These observations provide essential information for affected individuals, parents, and healthcare professionals to ensure appropriate clinical management.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the findings of this study are available in the Supplementary material of this article and online at https://databases.lovd.nl/shared/genes/SON and https://humandiseasegenes.nl/son/.
References
de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9.
Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–12.
Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18.
Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17:774–81.
Tokita MJ, Braxton AA, Shao Y, Lewis AM, Vincent M, Küry S, et al. De novo truncating variants in SON cause intellectual disability, congenital malformations, and failure to thrive. Am J Hum Genet. 2016;99:720–7.
Kim JH, Shinde DN, Reijnders MRF, Hauser NS, Belmonte RL, Wilson GR, et al. De novo mutations in SON disrupt RNA splicing of genes essential for brain development and metabolism, causing an intellectual-disability syndrome. Am J Hum Genet. 2016;99:711–9.
Takenouchi T, Miura K, Uehara T, Mizuno S, Kosaki K. Establishing SON in 21q22.11 as a cause a new syndromic form of intellectual disability: possible contribution to Braddock-Carey syndrome phenotype. Am J Med Genet A. 2016;170:2587–90.
Hickey CJ, Kim JH, Ahn EY. New discoveries of old SON: a link between RNA splicing and cancer. J Cell Biochem. 2014;115:224–31.
Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, et al. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol Cell. 2011;42:185–98.
Sharma A, Markey M, Torres-Muñoz K, Varia S, Kadakia M, Bubulya A, et al. Son maintains accurate splicing for a subset of human pre-mRNAs. J Cell Sci. 2011;124:4286–98.
Kim JH, Park EY, Chitayat D, Stachura DL, Schaper J, Lindstrom K, et al. SON haploinsufficiency causes impaired pre-mRNA splicing of CAKUT genes and heterogeneous renal phenotypes. Kidney Int. 2019;95:1494–504.
Lu X, Göke J, Sachs F, Jacques P, Liang H, Feng B, et al. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat Cell Biol. 2013;15:1141–52.
Ueda M, Matsuki T, Fukada M, Eda S, Toya A, Iio A, et al. Knockdown of Son, a mouse homologue of the ZTTK syndrome gene, causes neuronal migration defects and dendritic spine abnormalities. Mol Brain. 2020;13:80.
Kim JH, Baddoo MC, Park EY, Stone JK, Park H, Butler TW, et al. SON and its alternatively spliced isoforms control MLL complex-mediated H3K4me3 and transcription of leukemia-associated genes. Mol Cell. 2016;61:859–73.
Ahn EE, Higashi T, Yan M, Matsuura S, Hickey CJ, Lo MC, et al. SON protein regulates GATA-2 through transcriptional control of the microRNA 23a~27a~24-2 cluster. J Biol Chem. 2013;288:5381–8.
Quintana Castanedo L, Sánchez Orta A, Maseda Pedrero R, Santos Simarro F, Palomares Bralo M, Feito Rodríguez M, et al. Skin and nails abnormalities in a patient with ZTTK syndrome and a de novo mutation in SON. Pediatr Dermatol. 2020;37:517–9.
Tan Y, Duan L, Yang K, Liu Q, Wang J, Dong Z, et al. A novel frameshift variant in SON causes Zhu-Tokita-Takenouchi-Kim Syndrome. J Clin Lab Anal. 2020;34:e23326.
Yang Y, Xu L, Yu Z, Huang H, Yang L. Clinical and genetic analysis of ZTTK syndrome caused by SON heterozygous mutation c.394C>T. Mol Genet Genom Med. 2019;7:e953.
Chiu C, Loth S, Kuhlen M, Ginzel S, Schaper J, Rosenbaum T, et al. Mutated SON putatively causes a cancer syndrome comprising high-risk medulloblastoma combined with café-au-lait spots. Fam Cancer. 2019;18:353–8.
Slezak R, Smigiel R, Rydzanicz M, Pollak A, Kosinska J, Stawinski P, et al. Phenotypic expansion in Zhu-Tokita-Takenouchi-Kim syndrome caused by de novo variants in the SON gene. Mol Genet Genom Med. 2020;8:e1432.
Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet. 2008;83:610–5.
Dingemans AJM, Stremmelaar DE, Vissers L, Jansen S, Nabais Sá MJ, van Remortele A, et al. Human Disease Genes website series: an international, open and dynamic library for up-to-date clinical information. Am J Med Genet A. 2021;185:1039–46.
Portal SJCsRHPD. ProteinPaint. 2015.
van der Donk R, Jansen S, Schuurs-Hoeijmakers JHM, Koolen DA, Goltstein L, Hoischen A, et al. Next-generation phenotyping using computer vision algorithms in rare genomic neurodevelopmental disorders. Genet Med. 2019;21:1719–25.
Dingemans AJM, Stremmelaar DE, van der Donk R, Vissers L, Koolen DA, Rump P, et al. Quantitative facial phenotyping for Koolen-de Vries and 22q11.2 deletion syndrome. Eur J Hum Genet. 2021. [epub ahead of print]
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
Acknowledgements
First and foremost, we are grateful to all individuals and their parents for participation in this study. Next to that, we are grateful to the Dutch Organisation for Health Research and Development: ZON-MW grants 912-12-109 (to BBAdV and LELMV), Donders Junior researcher grant 2019 (to BBAdV and LELMV) and Aspasia grant 015.014.066 (to LELMV). This study was also supported by the US National Institute of Health grants R01 CA190688 and R01 CA236911 (to E-YEA). This work has been generated through a collaboration with the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability (ERN-ITHACA) [EU Framework Partnership Agreement ID: 3HP-HP-FPA ERN-01-2016/739516]. The aims of this study contribute to the Solve-RD project (to LELMV) that has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 779257.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.J.M.D, K.M.G.T, E.Y.E.A., B.B.A.d.V, L.E.L.M.V; Data collection: all authors; Data Analysis: A.J.M.D, K.M.G.T; Funding acquisition: L.E.L.M.V, B.B.A.d.V, E.Y.E.A.; Writing – original draft: A.J.M.D, K.M.G.T, E.Y.E.A., B.B.A.d.V, L.E.L.M.V; Writing – review and editing: all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Consent for participation in this study was obtained via the individuals’ legal guardians, and the study was approved by the institutional review board of the Radboud University Medical Center (#2020–6763). For individuals whose facial photos are depicted in Fig. 2, additional informed signed consent was obtained for the publication of photographs.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dingemans, A.J.M., Truijen, K.M.G., Kim, JH. et al. Establishing the phenotypic spectrum of ZTTK syndrome by analysis of 52 individuals with variants in SON. Eur J Hum Genet 30, 271–281 (2022). https://doi.org/10.1038/s41431-021-00960-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-021-00960-4
This article is cited by
-
Recurrent myocardial injury in a de novo SON mutation ZTTK syndrome patient: a case report
BMC Pediatrics (2024)
-
Istore: a project on innovative statistical methodologies to improve rare diseases clinical trials in limited populations
Orphanet Journal of Rare Diseases (2024)
-
Nuclear speckleopathies: developmental disorders caused by variants in genes encoding nuclear speckle proteins
Human Genetics (2024)
-
Good genotype-phenotype relationships in rare disease are hard to find
European Journal of Human Genetics (2022)
-
Commentary on: Establishing the phenotypic spectrum of ZTTK syndrome by analysis of 52 individuals with variants in SON
European Journal of Human Genetics (2022)