Abstract
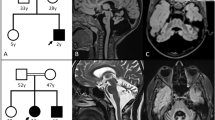
Pathogenic variants in the SRCAP (SNF2-related CREBBP activator protein) gene, which encodes a chromatin-remodeling ATPase, cause neurodevelopmental disorders including Floating Harbor syndrome (FLHS). Here, we report the discovery of a de novo transposon insertion in SRCAP exon 13 from trio genome sequencing in a 28-year-old female with failure to thrive, developmental delay, mood disorder and seizure disorder. The insertion was a full-length (~2.8 kb), antisense-oriented SVA insertion relative to the SRCAP transcript, bearing a 5’ transduction and hallmarks of target-primed reverse transcription. The 20-bp 5’ transduction allowed us to trace the source SVA element to an intron of a long non-coding RNA on chromosome 12, which is highly expressed in testis. RNA sequencing and qRT-PCR confirmed significant depletion of SRCAP expression and low-level exon skipping in the proband. This case highlights a novel disease-causing structural variant and the importance of transposon analysis in a clinical diagnostic setting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Sequencing data supporting the findings of this study are available upon reasonable request.
References
Hood RL, Lines MA, Nikkel SM, Schwartzentruber J, Beaulieu C, Nowaczyk MJM, et al. Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am J Hum Genet. 2012;90:308–13. https://doi.org/10.1016/j.ajhg.2011.12.001
Kehrer M, Beckmann A, Wyduba J, Finckh U, Dufke A, Gaiser U, et al. Floating-Harbor syndrome: SRCAP mutations are not restricted to exon 34. Clin Genet. 2014;85:498–9. https://doi.org/10.1111/cge.12199
Seifert W, Meinecke P, Krüger G, Rossier E, Heinritz W, Wüsthof A, et al. Expanded spectrum of exon 33 and 34 mutations in SRCAP and follow-up in patients with Floating-Harbor syndrome. BMC Med Genet. 2014;15:127. https://doi.org/10.1186/s12881-014-0127-0
Goff CL, Mahaut C, Bottani A, Doray B, Goldenberg A, Moncla A, et al. Not all floating-harbor syndrome cases are due to mutations in exon 34 of SRCAP. Hum Mutat2013;34:88–92. https://doi.org/10.1002/humu.22216
Nikkel SM, Dauber A, de Munnik S, Connolly M, Hood RL, Caluseriu O, et al. The phenotype of Floating-Harbor syndrome: clinical characterization of 52 individuals with mutations in exon 34 of SRCAP. Orphanet J Rare Dis. 2013;8:63. https://doi.org/10.1186/1750-1172-8-63
Robinson PL, Shohat M, Winter RM, Conte WJ, Gordon-Nesbitt D, Feingold M, et al. A unique association of short stature, dysmorphic features, and speech impairment (Floating-harbor syndrome). J Pediatrics. 1988;113:703–6. https://doi.org/10.1016/s0022-3476(88)80384-6
Rots D, Chater-Diehl E, Dingemans AJM, Goodman SJ, Siu MT, Cytrynbaum C, et al. Truncating SRCAP variants outside the Floating-Harbor syndrome locus cause a distinct neurodevelopmental disorder with a specific DNA methylation signature. Am J Hum Genet. 2021;108:1053–68. https://doi.org/10.1016/j.ajhg.2021.04.008
Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, et al. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–7. https://doi.org/10.1021/bi060043d
Wong MM, Cox LK, Chrivia JC. The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J Biol Chem 2007;282:26132–9. https://doi.org/10.1074/jbc.M703418200
Collins RL, Brand H, Karczewski KJ, Zhao X, Alfoldi J, Francioli LC, et al. A structural variation reference for medical and population genetics. Nature. 2020;581:444–51. https://doi.org/10.1038/s41586-020-2287-8
Borges-Monroy R, Chu C, Dias C, Choi J, Lee S, Gao Y, et al. Whole-genome analysis reveals the contribution of non-coding de novo transposon insertions to autism spectrum disorder. Mob DNA. 2021;12:28. https://doi.org/10.1186/s13100-021-00256-w
Feusier J, Watkins WS, Thomas J, Farrell A, Witherspoon DJ, Baird L, et al. Pedigree-based estimation of human mobile element retrotransposition rates. Genome Res. 2019;29:1567–77. https://doi.org/10.1101/gr.247965.118
Hancks DC, Kazazian HH Jr. Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9.https://doi.org/10.1186/s13100-016-0065-9
Aneichyk T, Hendriks WT, Yadav R, Shin D, Gao D, Vaine CA, et al. Dissecting the causal mechanism of X-Linked Dystonia-Parkinsonism by integrating genome and transcriptome assembly. Cell. 2018;172:897–909.e821. https://doi.org/10.1016/j.cell.2018.02.011
Kim J, Hu C, Moufawad El Achkar C, Black LE, Douville J, Larson A, et al. Patient-customized oligonucleotide therapy for a rare genetic disease. N Engl J Med. 2019. https://doi.org/10.1056/NEJMoa1813279
Juhl Corydon M, Vockley J, Rinaldo P, James Rhead W, Kjeldsen M, Winter V, et al. Role of common gene variations in the molecular pathogenesis of short-chain acyl-CoA dehydrogenase deficiency. Pediatr Res. 2001;49:18–23. https://doi.org/10.1203/00006450-200101000-00008
Nykamp K, Anderson M, Powers M, Garcia J, Herrera B, Ho YY, et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med2017;19:1105–17. https://doi.org/10.1038/gim.2017.37
Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–910.
Hancks DC, Kazazian HH Jr. SVA retrotransposons: evolution and genetic instability. Semin Cancer Biol. 2010;20:234–45. https://doi.org/10.1016/j.semcancer.2010.04.001
Yamamoto G, Miyabe I, Tanaka K, Kakuta M, Watanabe M, Kawakami S, et al. SVA retrotransposon insertion in exon of MMR genes results in aberrant RNA splicing and causes Lynch syndrome. Eur J Hum Genet. 2020. https://doi.org/10.1038/s41431-020-00779-5
Damert A, Raiz J, Horn AV, Lower J, Wang H, Xing J, et al. 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19:1992–2008. https://doi.org/10.1101/gr.093435.109
Ebert P, et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 2021. https://doi.org/10.1126/science.abf7117
Acknowledgements
We thank the patient and family for their participation and support. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal.
Funding
EAL was supported by the NIH (DP2 AG0724), Suh Kyungbae Foundation, Charles H. Hood foundation, and the Allen Discovery Center program of the Paul G. Allen Family Foundation. BZ was supported by the Manton Center Pilot Project Award and Rare Disease Research Fellowship. MHW is supported by an Early Career Award from the Thrasher Research Fund. PBA’s research work is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, R01AR068429-01, National Human Genome Research Institute, 1R01HG011798-01A1 and “Because of Bella” foundation. Sequencing and analysis were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1 HG008900 and in part by National Human Genome Research Institute grant R01 HG009141, as well as the National Institutes of Health grants MH115957 and HD081256. Sanger sequencing was performed by the Boston Children’s Hospital IDDRC Molecular Genetics Core Facility supported by NIH award U54HD090255 from the National Institute of Child Health and Human Development.
Author information
Authors and Affiliations
Contributions
BZ characterized the insertion variant, designed and analyzed experimental validation assays, performed RNA-seq analysis, qRT-PCR, and splicing analysis for pathogenicity confirmation, made all figures and supplementary materials, and wrote the manuscript. JAM coordinated samples, clinical record, and sequencing data collections, made Table 1 for clinical presentation, and wrote the manuscript. JL performed all experimental validation assays, and edited the manuscript. GTB enrolled the patient. MHW helped analyze genome sequencing results that identified the potential variant, and edited the manuscript. XZ, HB, and MT performed SV data processing, identified the potential variant, and edited the manuscript. EAL and PBA directed the overall research and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
PBA is on the Scientific Advisory Board of Illumina, Inc. and GeneDx, Inc. Other authors declare no competing interests.
Ethics approval and consent to participate
The proband and her family were enrolled into the Gene Discovery Core at The Manton Center for Orphan Disease Research approved by the IRB (10-02-0053) at Boston Children’s Hospital. Written informed consents were obtained from the patient and family members.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, B., Madden, J.A., Lin, J. et al. A neurodevelopmental disorder caused by a novel de novo SVA insertion in exon 13 of the SRCAP gene. Eur J Hum Genet 30, 1083–1087 (2022). https://doi.org/10.1038/s41431-022-01137-3
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-022-01137-3
This article is cited by
-
Pediatric floating-harbor syndrome: clinical features and treatment outcomes in a cohort of Chinese children
European Journal of Pediatrics (2026)
-
NERD-seq: a novel approach of Nanopore direct RNA sequencing that expands representation of non-coding RNAs
Genome Biology (2024)
-
Combined exome and whole transcriptome sequencing identifies a de novo intronic SRCAP variant causing DEHMBA syndrome with severe sleep disorder
Journal of Human Genetics (2024)
-
Investigating mobile element variations by statistical genetics
Human Genome Variation (2024)
-
PhenoSV: interpretable phenotype-aware model for the prioritization of genes affected by structural variants
Nature Communications (2023)