Abstract
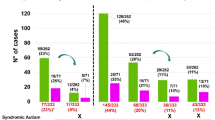
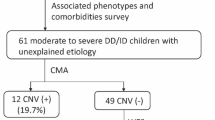
In children undergoing genetic testing for physical health concerns, we examined how often the results also revealed information about their risk for neurodevelopmental disorders. The study sample consisted of 3056 genetic tests (1686 chromosomal microarrays––CMAs, and 1378 next-generation sequencing––NGS panels) ordered at a tertiary pediatric hospital because of a physical/congenital health problem. Tests ordered to investigate developmental concerns were excluded. Pathogenic, or likely pathogenic variants were manually reviewed for diagnostic likelihood, and for evidence of an association with a neurodevelopmental disorder (e.g., autism or intellectual disability). A total of 169 CMAs (10%) and 232 NGS panels (17%) had likely diagnostic results. More than half (52%) of all diagnostic results had established evidence of a neurodevelopmental disorder association. In summary, there is a high prevalence of neurodevelopmental implications from genetic tests ordered for physical/congenital indications. This broad clinical utility suggests a growing need for genetics-first developmental care pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
Data generated and analysed during this study are included in this published article in the supplementary information files; some restrictions have been applied to de-identify variants and protect patient anonymity.
References
Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012;489:391–9.
Negi SK, Guda C. Global gene expression profiling of healthy human brain and its application in studying neurological disorders. Sci Rep. 2017;7:897.
Sigmon ER, Kelleman M, Susi A, Nylund CM, Oster ME. Congenital heart disease and autism: a case-control study. Pediatrics 2019;144:e20184114.
Timonen-Soivio L, Vanhala R, Malm H, Leivonen S, Jokiranta E, Hinkka-Yli-Salomäki S, et al. The association between congenital anomalies and autism spectrum disorders in a Finnish national birth cohort. Dev Med Child Neurol. 2015;57:75–80.
Tillman KK, Hakelius M, Höijer J, Ramklint M, Ekselius L, Nowinski D, et al. Increased risk for neurodevelopmental disorders in children with orofacial clefts. J Am Acad Child Adolesc Psychiatry. 2018;57:876–83.
Berg E, Haaland OA, Feragen KB, Filip C, Vindenes HA, Moster D, et al. Health status among adults born with an oral cleft in Norway. JAMA pediatrics. 2016;170:1063–70.
McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, et al. 22q11.2 deletion syndrome. Nat Rev Dis Prim. 2015;1:15071.
Butler MG, Miller JL, Forster JL. Prader-Willi syndrome-clinical genetics, diagnosis and treatment approaches: an update. Curr Pediatr Rev. 2019;15:207–44.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.
Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–5.
Manickam K, McClain MR, Demmer LA, Biswas S, Kearney HM, Malinowski J, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet in Med. 2021;2029–37.
Dimmock DP, Clark MM, Gaughran M, Cakici JA, Caylor SA, Clarke C, et al. An RCT of rapid genomic sequencing among seriously ill infants results in high clinical utility, changes in management, and low perceived harm. Am J Hum Genet. 2020;107:942–52.
Petrikin JE, Cakici JA, Clark MM, Willig LK, Sweeney NM, Farrow EG, et al. The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genom Med. 2018;3:6.
Holm IA, Agrawal PB, Ceyhan-Birsoy O, Christensen KD, Fayer S, Frankel LA, et al. The BabySeq project: implementing genomic sequencing in newborns. BMC Pediatr. 2018;18:225.
Biesecker LG, Green ED, Manolio T, Solomon BD, Curtis D. Should all babies have their genome sequenced at birth? Bmj. 2021;375:n2679.
Bennette CS, Gallego CJ, Burke W, Jarvik GP, Veenstra DL. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med. 2015;17:587–95.
Fuller EA, Kaiser AP. The effects of early intervention on social communication outcomes for children with autism spectrum disorder: a meta-analysis. J autism developmental Disord. 2020;50:1683–700.
McManus BM, Richardson Z, Schenkman M, Murphy N, Morrato EH. Timing and intensity of early intervention service use and outcomes among a safety-net population of children. JAMA Netw Open 2019;2:e187529–e.
Soleimani F, Azari N, Ghiasvand H, Shahrokhi A, Rahmani N, Fatollahierad S. Do NICU developmental care improve cognitive and motor outcomes for preterm infants? A systematic review and meta-analysis. BMC Pediatrics. 2020;20:67.
Lee JZ, Iverson JM, Roemer EJ, Plate S, Schneider JL. “I’m Worried About My Child”: a longitudinal investigation of parental concerns and repeat screening in toddlers with familial risk of autism spectrum disorder. Pediatrics 2021;147:73–4. 3 MeetingAbstract
Acknowledgements
DB acknowledges the Canadian Institutes of Health Research graduate scholarship and the O’Brien scholars program for supporting her fellowship training. SWS holds the Northbridge Chair in Paediatric Research at the Hospital for Sick Children. JV acknowledges the support from the Canadian Institutes of Health Research (PJT-162323).
Author information
Authors and Affiliations
Contributions
DAB and JV and NH conceptualized and designed the study, drafted the initial protocol and manuscript, and revised the manuscript. DJS coordinated and supervised data collection and extraction, supervised the genetic analyses/ variant calls, and critically reviewed the manuscript. GC, DAB and NH conducted the genetic analyses/variant calls and critically reviewed the manuscript. DAB, NH and TS extracted and analyzed the data. SWS, DAB, NH, and TS critically reviewed the manuscript for important intellectual content and scientific rigour. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
JV serves as a consultant for NoBias Therapeutics Inc. SWS is on the scientific advisory committee of Population Bio and is an academic consultant of the King Abdullaziz University. The other authors have no relevant conflicts of interest.
Ethical approval
The study was approved by the institution Research Ethics Boards.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Baribeau, D.A., Hoang, N., Selvanayagam, T. et al. Developmental implications of genetic testing for physical indications. Eur J Hum Genet 30, 1297–1300 (2022). https://doi.org/10.1038/s41431-022-01181-z
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41431-022-01181-z
This article is cited by
-
Integrative genetic analysis: cornerstone of precision psychiatry
Molecular Psychiatry (2025)
-
An integrated clinical approach to children at genetic risk for neurodevelopmental and psychiatric conditions: interdisciplinary collaboration and research infrastructure
Journal of Neurodevelopmental Disorders (2024)
-
Consensus reporting guidelines to address gaps in descriptions of ultra-rare genetic conditions
npj Genomic Medicine (2024)
-
Genome sequencing—do you know what you are getting into?
European Journal of Human Genetics (2022)