Abstract
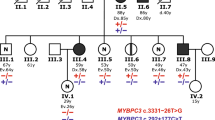
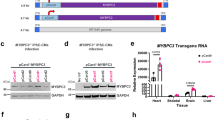
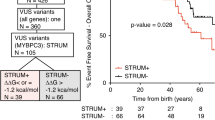
MYBPC3 pathogenic variants are the most common cause of hypertrophic cardiomyopathy (HCM) and are associated with significant phenotypic heterogeneity. Despite their pathogenic potential, MYBPC3 founder variants persist within specific populations. This study investigates the MYBPC3 c.2309-2 A > G splice variant hypothesizing its founder origin in central Italy. The aim was to confirm the presence of a common haplotype, assess its molecular and clinical impact, and compare the phenotype with that of other MYBPC3 founder variants. Among the 5251 HCM patients recruited at eight Italian referral centers, 1108 probands (21.1%) were identified as carriers of pathogenic or likely pathogenic MYBPC3 variants, and among these, 11.6% carried the c.2309-2 A > G variant. Haplotype reconstruction using short tandem repeats and tag-SNPs revealed a unique 5.2 Mb haplotype segregating with the c.2309-2 A > G variant in all carriers. Age estimation suggested that the variant originated approximately 481 years ago, likely in the Lazio region with clustering in Rome. Clinically, carriers exhibited variable expressivity with age-and sex-dependent penetrance. Males showed earlier onset, higher penetrance and greater disease severity compared to females. RNA analysis showed the retention of both introns 23 and 24, and significantly reduced MYBPC3 expression consistent with haploinsufficiency. Comparative analysis with other MYBPC3 founder variants highlighted differences in phenotypic expression, particularly in left ventricular wall thickness and clinical outcomes. This study establishes c.2309-2 A > G as an Italian MYBPC3 founder mutation, enhancing the understanding of HCM genetics and regional founder effects. These findings emphasize the importance of targeted genetic screening and personalized management for MYBPC3 c.2309-2 A > G carriers.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are included in the supplementary materials and are also available from the corresponding author upon request.
References
Carrier L. Targeting the population for gene therapy with MYBPC3. J Mol Cell Cardiol. 2021;150:101–8.
Cirino AL, Harris SL, Murad AM, Hansen B, Malinowski J, Natoli JL, et al. The uptake and utility of genetic testing and genetic counseling for hypertrophic cardiomyopathy—A systematic review and meta-analysis. J Genet Couns. 2022;31:1290–305.
Ariga R, Tunnicliffe EM, Manohar SG, Mahmod M, Raman B, Piechnik SK, et al. Identification of Myocardial Disarray in Patients With Hypertrophic Cardiomyopathy and Ventricular Arrhythmias. J Am Coll Cardiol. 2019;73:2493–502.
Marian AJ, Asatryan B, Wehrens XHT. Genetic basis and molecular biology of cardiac arrhythmias in cardiomyopathies. Cardiovasc Res. 2020;116:1600–19.
Tudurachi BS, Zăvoi A, Leonte A, Țăpoi L, Ureche C, Bîrgoan SG, et al. An Update on MYBPC3 Gene Mutation in Hypertrophic Cardiomyopathy. Int J Mol Sci. 2023;24:10510.
Schlossarek S, Mearini G, Carrier L. Cardiac myosin-binding protein C in hypertrophic cardiomyopathy: Mechanisms and therapeutic opportunities. J Mol Cell Cardiol. 2011;50:613–20.
Alders M. The 2373insG mutation in the MYBPC3 gene is a founder mutation, which accounts for nearly one-fourth of the HCM cases in the Netherlands. Eur Heart J. 2003;24:1848–53.
Helms AS, Thompson AD, Glazier AA, Hafeez N, Kabani S, Rodriguez J, et al. Spatial and Functional Distribution of MYBPC3 Pathogenic Variants and Clinical Outcomes in Patients With Hypertrophic Cardiomyopathy. Circ Genom Precis Med. 2020;13:396–405.
Marian AJ, Braunwald E. Hypertrophic Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy. Circ Res. 2017;121:749–70.
Ribeiro M, Furtado M, Martins S, Carvalho T, Carmo-Fonseca M. RNA Splicing Defects in Hypertrophic Cardiomyopathy: Implications for Diagnosis and Therapy. Int J Mol Sci. 2020;21:1329.
Ito K, Patel PN, Gorham JM, McDonough B, DePalma SR, Adler EE, et al. Identification of pathogenic gene mutations in LMNA and MYBPC3 that alter RNA splicing. Proc Natl Acad Sci. 2017;114:7689–94.
Frank-Hansen R, Page SP, Syrris P, McKenna WJ, Christiansen M, Andersen PS. Micro-exons of the cardiac myosin binding protein C gene: flanking introns contain a disproportionately large number of hypertrophic cardiomyopathy mutations. Eur J Hum Genet. 2008;16:1062–9.
Torrado M, Maneiro E, Lamounier Junior A, Fernández-Burriel M, Sánchez Giralt S, Martínez-Carapeto A, et al. Identification of an elusive spliceogenic MYBPC3 variant in an otherwise genotype-negative hypertrophic cardiomyopathy pedigree. Sci Rep. 2022;12:7284.
Caroselli S, Fabiani M, Micolonghi C, Savio C, Tini G, Musumeci B, et al. Reanalysis of Next-generation Sequencing Data in Patients With Hypertrophic Cardiomyopathy: Contribution of Spliceogenic MYBPC3 Variants in an Italian Cohort. Ann Lab Med. 2025;45:96–100.
Girolami F, Olivotto I, Passerini I, Zachara E, Nistri S, Re F, et al. A molecular screening strategy based on β-myosin heavy chain, cardiac myosin binding protein C and troponin T genes in Italian patients with hypertrophic cardiomyopathy. J Cardiovasc Med. 2006;7:601–7.
Calore C, De Bortoli M, Romualdi C, Lorenzon A, Angelini A, Basso C, et al. A founder MYBPC3 mutation results in HCM with a high risk of sudden death after the fourth decade of life. J Med Genet. 2015;52:338–47.
Vodnjov N, Maver A, Teran N, Peterlin B, Toplišek J, Writzl K. Clinical Outcome of Hypertrophic Cardiomyopathy in Probands with the Founder Variant c.913_914del in MYBPC3: A Slovenian Cohort Study. J Cardiovasc Transl Res [Internet]. 2024 Aug [cited 2024 Dec 12]; Available from: https://link.springer.com/10.1007/s12265-024-10551-5.
Adalsteinsdottir B, Burke M, Maron BJ, Danielsen R, Lopez B, Diez J, et al. Hypertrophic cardiomyopathy in myosin-binding protein C (MYBPC3) Icelandic founder mutation carriers. Open Heart. 2020;7:e001220.
Redin C, Pavlidou DC, Bhuiyan Z, Porretta AP, Monney P, Bedoni N, et al. The «Amish» NM_000256.3:c.3330+2T>G splice variant in MYBPC3 associated with hypertrophic cardiomyopathy is an ancient Swiss mutation. Eur J Med Genet. 2022;65:1204627.
Boen HM, Alaerts M, Van Laer L, Saenen JB, Goovaerts I, Bastianen J, et al. Phenotypic spectrum of the first Belgian MYBPC3 founder: a large multi-exon deletion with a varying phenotype. Front Genet. 2024;15:1392527.
Coppini R, Ho CY, Ashley E, Day S, Ferrantini C, Girolami F, et al. Clinical Phenotype and Outcome of Hypertrophic Cardiomyopathy Associated With Thin-Filament Gene Mutations. J Am Coll Cardiol. 2014;64:2589–600.
Roncarati R, Latronico MVG, Musumeci B, Aurino S, Torella A, Bang M, et al. Unexpectedly low mutation rates in beta-myosin heavy chain and cardiac myosin binding protein genes in italian patients with hypertrophic cardiomyopathy. J Cell Physiol. 2011;226:2894–900.
Cecconi M, Parodi MI, Formisano F, Spirito P, Autore C, Musumeci MB, et al. Targeted next-generation sequencing helps to decipher the genetic and phenotypic heterogeneity of hypertrophic cardiomyopathy. Int J Mol Med. 2016;38:1111–24.
Rubattu S, Bozzao C, Pennacchini E, Pagannone E, Musumeci B, Piane M, et al. A Next-Generation Sequencing Approach to Identify Gene Mutations in Early- and Late-Onset Hypertrophic Cardiomyopathy Patients of an Italian Cohort. Int J Mol Sci. 2016;17:1239.
Sabater-Molina M, Saura D, García-Molina Sáez E, González-Carrillo J, Polo L, Pérez-Sánchez I, et al. A Novel Founder Mutation in MYBPC3: Phenotypic Comparison With the Most Prevalent MYBPC3 Mutation in Spain. Rev Esp Cardiol Engl Ed. 2017;70:105–14.
Arbelo E, Protonotarios A, Gimeno JR, Arbustini E, Barriales-Villa R, Basso C, et al. 2023 ESC Guidelines for the management of cardiomyopathies. Eur Heart J. 2023;44:3503–626.
Micolonghi C, Perrone F, Fabiani M, Caroselli S, Savio C, Pizzuti A, et al. Unveiling the Spectrum of Minor Genes in Cardiomyopathies: A Narrative Review. Int J Mol Sci. 2024;25:9787.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Barrett JC. Haploview: Visualization and Analysis of SNP Genotype Data. Cold Spring Harb Protoc. 2009;2009:pdb.ip71.
Duan S, Zhang W, Cox NJ, Dolan ME. FstSNP-HapMap3: a database of SNPs with high population differentiation for HapMap3. Bioinformation. 2008;3:139–41.
Little S. Amplification-Refractory Mutation System (ARMS) Analysis of Point Mutations. Curr Protoc Hum Genet [Internet]. 1995 Nov [cited 2024 Dec 12];7. Available from: https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/0471142905.hg0908s07.
Mertes C, Scheller IF, Yépez VA, Çelik MH, Liang Y, Kremer LS, et al. Detection of aberrant splicing events in RNA-seq data using FRASER. Nat Commun. 2021;12:529.
Holliday M, Singer ES, Ross SB, Lim S, Lal S, Ingles J, et al. Transcriptome Sequencing of Patients With Hypertrophic Cardiomyopathy Reveals Novel Splice-Altering Variants in MYBPC3. Circ Genom Precis Med. 2021;14:e003202.
Slatkin M, Rannala B. Estimating the age of alleles by use of intraallelic variability. Am J Hum Genet. 1997;60:447–58.
Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet. 2004;41:445–9.
Adalsteinsdottir B, Teekakirikul P, Maron BJ, Burke MA, Gudbjartsson DF, Holm H, et al. Nationwide Study on Hypertrophic Cardiomyopathy in Iceland: Evidence of a MYBPC3 Founder Mutation. Circulation. 2014;130:1158–67.
Christiaans I, Nannenberg EA, Dooijes D, Jongbloed RJE, Michels M, Postema PG, et al. Founder mutations in hypertrophic cardiomyopathy patients in the Netherlands. Neth Heart J. 2010;18:248–54.
Méndez I, Fernández AI, Espinosa MÁ, Cuenca S, Lorca R, Rodríguez JF, et al. Founder mutation in myosin-binding protein C with an early onset and a high penetrance in males. Open Heart. 2021;8:e001789.
Olivotto I, Maron MS, Adabag AS, Casey SA, Vargiu D, Link MS, et al. Gender-Related Differences in the Clinical Presentation and Outcome of Hypertrophic Cardiomyopathy. J Am Coll Cardiol. 2005;46:480–7.
Pioner JM, Vitale G, Steczina S, Langione M, Margara F, Santini L, et al. Slower Calcium Handling Balances Faster Cross-Bridge Cycling in Human MYBPC3 HCM. Circ Res. 2023;132:628–44.
Jansen M, Schmidt AF, Jans JJM, Christiaans I, Van Der Crabben SN, Hoedemaekers YM, et al. Circulating Acylcarnitines Associated with Hypertrophic Cardiomyopathy Severity: an Exploratory Cross-Sectional Study in MYBPC3 Founder Variant Carriers. J Cardiovasc Transl Res. 2023;16:1267–75.
Lorca R, Gómez J, Martín M, Cabanillas R, Calvo J, León V, et al. Insights Into Hypertrophic Cardiomyopathy Evaluation Through Follow-up of a Founder Pathogenic Variant. Rev Esp Cardiol Engl Ed. 2019;72:138–44.
Funding
This work was partially funded by Progetto PRIN 2022 (from the Italian Ministry of Instruction, University and Research, no. 2022E75TWB) to S.R. The research leading to these results has also received funding from the European Union-NextGenerationEU through the Italian Ministry of University and Research under PNRR–M4C2-I1.3 Project PE_00000019 “HEAL ITALIA”, CUP B53C22004000006 to S.R. The views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them.
Author information
Authors and Affiliations
Contributions
MF and MP conceptualized and designed the study and wrote the original draft. MF, CM, CS, LV and MP performed haplotype analyses. VV, AP, and MP coordinated the study, with MP and SR providing writing review and editing. SC and CS provided methodology and software. CS, IB, MPC, and VO performed genetic analyses. AL, EM, and RM performed RT-PCR analyses, with EM and RM also providing samples and clinical data. CL, AS, LG, and FC performed RNAseq analyses and provided samples and clinical data, while MI coordinated participant recruitment and also coordinated RNAseq analyses. AGE carried out data curation and statistical analyses, and LV provided genomics expertise. CM, SP, LP, AB, AGO, GTI, BM, EP, LDF, and CPC provided samples collected clinical data. DD, FGU, GTO, MM, AN, CR, PG, FS, FGI, IO, and CA collected clinical data and coordinated the recruitment of study participants. European Union-NextGenerationEU.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of A.O.U. Sant’Andrea (Ref 7310 Protocol number 0955/2023 29/11/2023) for studies involving humans. Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fabiani, M., Micolonghi, C., Caroselli, S. et al. MYBPC3 c.2309-2A>G: exploring a founder variant in Italian hypertrophic cardiomyopathy patients. Eur J Hum Genet (2025). https://doi.org/10.1038/s41431-025-01873-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41431-025-01873-2