Abstract
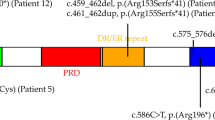
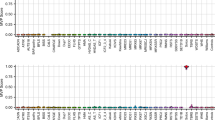
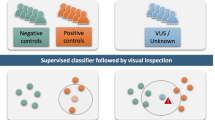
Smith-Magenis syndrome (SMS) and Potocki-Lupski syndrome (PTLS) are reciprocal genomic disorders caused by deletions and duplications of the 17p11.2 chromosomal region, respectively. This study aimed to identify and validate DNA methylation episignatures specific to SMS and PTLS, and to investigate their reciprocal relationship and shared molecular features with other neurodevelopmental disorders. Genome-wide DNA methylation was analyzed in individuals with an SMS (n = 26) or PTLS (n = 27) phenotype associated with copy number variation, and SMS patients with RAI1 sequence variants using the Infinium EPIC array. Differentially methylated CpG sites were identified and used to develop support vector machine (SVM)-based classifiers, which demonstrated high sensitivity and specificity for both syndromes. The analysis revealed a mirror-like episignature, with SMS showing predominant hypomethylation and PTLS displaying hypermethylation at shared loci. Functional correlation with other neurodevelopmental disorders highlighted significant overlap with known episignatures, including those associated with MEF2C-related disorders. Notably, individuals with RAI1 sequence variants did not exhibit the same DNA methylation patterns, suggesting that the epigenetic alterations are primarily driven by copy number changes. These findings establish SMS and PTLS as distinct yet interconnected epigenetic entities, offering valuable diagnostic biomarkers and insights into the molecular pathophysiology of 17p11.2-associated neurodevelopmental disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The deposition of individual epigenomic or any other personally identifiable data that has not previously been made publicly available for samples in the EpiSign Knowledge Database (EKD) is prohibited from deposition in publicly accessible databases due to institutional and ethical restrictions. Specifically, these include data and samples submitted from external institutions to London Health Sciences EKD that are subject to institutional material and data transfer agreements, data submitted to London Health Sciences for episignature assessment under Research Services Agreements, and research study cohorts under Institutional Research Ethics Approval (Western University REB 106302 and REB 116108). EpiSign is a commercial software and is not publicly available.
References
Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, Kimonis V, et al. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–7.
Moncla A, Piras L, Arbex OF, Muscatelli F, Mattei MG, Mattei JF, et al. Physical mapping of microdeletions of the chromosome 17 short arm associated with Smith-Magenis syndrome. Hum Genet. 1993;90:657–60.
Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet. 2003;33:466–8.
Lucas RE, Vlangos CN, Das P, Patel PI, Elsea SH. Genomic organisation of the approximately 1.5 Mb Smith-Magenis syndrome critical interval: transcription map, genomic contig, and candidate gene analysis. Eur J Hum Genet. 2001;9:892–902.
Girirajan S, Elsas LJ 2nd, Devriendt K, Elsea SH. RAI1 variations in Smith-Magenis syndrome patients without 17p11.2 deletions. J Med Genet. 2005;42:820–8.
Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, Friedman EM, et al. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am J Hum Genet. 2007;80:633–49.
Zhang F, Potocki L, Sampson JB, Liu P, Sanchez-Valle A, Robbins-Furman P, et al. Identification of uncommon recurrent Potocki-Lupski syndrome-associated duplications and the distribution of rearrangement types and mechanisms in PTLS. Am J Hum Genet. 2010;86:462–70.
Linders CC, van Eeghen AM, Zinkstok JR, van den Boogaard MJ, Boot E. Intellectual and behavioral phenotypes of Smith-Magenis syndrome: comparisons between individuals with a 17p11.2 deletion and pathogenic RAI1 variant. Genes. 2023;14:1514.
Poisson A, Nicolas A, Bousquet I, Raverot V, Gronfier C, Demily C. Smith-Magenis syndrome: molecular basis of a genetic-driven melatonin circadian secretion disorder. Int J Mol Sci. 2019;20:3533.
Srividhya D, Parambath SV, Sathyanarayanan R, Huligerepura Sosalegowda A, Korlimarla A, Niranjana Murthy AS, et al. Whole exome sequencing of a multiplex family of indian origin identifies variants in the RAI1 and FLII genes within the 17p11.2 region in siblings with autism and Smith Magenis syndrome. Mol Syndromol. 2024;15:537–44.
May PA, Blankenship J, Marais AS, Gossage JP, Kalberg WO, Barnard R, et al. Approaching the prevalence of the full spectrum of fetal alcohol spectrum disorders in a South African population-based study. Alcohol Clin Exp Res. 2013;37:818–30.
Sadikovic B, Aref-Eshghi E, Levy MA, Rodenhiser D. DNA methylation signatures in Mendelian developmental disorders as a diagnostic bridge between genotype and phenotype. Epigenomics. 2019;11:563–75.
Sadikovic B, Levy MA, Kerkhof J, Aref-Eshghi E, Schenkel L, Stuart A, et al. Clinical epigenomics: genome-wide DNA methylation analysis for the diagnosis of Mendelian disorders. Genet Med. 2021;23:1065–74.
Kerkhof J, Rastin C, Levy MA, Relator R, McConkey H, Demain L, et al. Diagnostic utility and reporting recommendations for clinical DNA methylation episignature testing in genetically undiagnosed rare diseases. Genet Med. 2024;26:101075.
Rooney K, Sadikovic B. DNA methylation episignatures in neurodevelopmental disorders associated with large structural copy number variants: clinical implications. Int J Mol Sci. 2022;23:7862.
van der Laan L, Rooney K, Trooster TM, Mannens MM, Sadikovic B, Henneman P. DNA methylation episignatures: insight into copy number variation. Epigenomics. 2022;14:1373–88.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22:245–57.
Zhou W, Triche TJ Jr., Laird PW, Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018;46:e123.
Aref-Eshghi E, Bend EG, Colaiacovo S, Caudle M, Chakrabarti R, Napier M, et al. Diagnostic utility of genome-wide DNA methylation testing in genetically unsolved individuals with suspected hereditary conditions. Am J Hum Genet. 2019;104:685–700.
Ho D, Imai K, King G, Stuart EA. MatchIt: nonparametric preprocessing for parametric causal inference. J Stat Softw. 2011;42:1–28.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, Lord R V, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6.
Levy MA, McConkey H, Kerkhof J, Barat-Houari M, Bargiacchi S, Biamino E, et al. Novel diagnostic DNA methylation episignatures expand and refine the epigenetic landscapes of Mendelian disorders. HGG Adv. 2022;3:100075.
Levy MA, Relator R, McConkey H, Pranckeviciene E, Kerkhof J, Barat-Houari M, et al. Functional correlation of genome-wide DNA methylation profiles in genetic neurodevelopmental disorders. Hum Mutat. 2022;43:1609–28.
Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2.
Cardoso MA, Rizzardi LEA, Kume LW, Groeneveld CS, Trefflich S, Morais DAA, et al. TreeAndLeaf: an R/Bioconductor package for graphs and trees with focus on the leaves. Bioinformatics. 2022;38:1463–4.
Cavalcante RG, Sartor MA. annotatr: genomic regions in context. Bioinformatics. 2017;33:2381–3.
Cal S, Quesada V, Llamazares M, Díaz-Perales A, Garabaya C, López-Otín C. Human polyserase-2, a novel enzyme with three tandem serine protease domains in a single polypeptide chain. J Biol Chem. 2005;280:1953–61.
Agrawal K, Chauhan S, Kumar D. Expression analysis and regulation of GLI and its correlation with stemness and metabolic alteration in human brain tumor. 3 Biotech. 2023;13:10.
Spineli-Silva S, de Leeuw N, Pontes LB, Leijsten N, Ruiterkamp-Versteeg M, Prota JRM, et al. A nonsense variant in the C-terminal transactivation domain of the EBF3 gene in an individual with intellectual disability and behavioural disorder: case report and literature review. Psychiatr Genet. 2025;35:75–78.
Aref-Eshghi E, Kerkhof J, Pedro VP, France GD, Barat-Houari M, Ruiz-Pallares N, et al. Evaluation of DNA methylation episignatures for diagnosis and phenotype correlations in 42 mendelian neurodevelopmental disorders. Am J Hum Genet. 2021;108:1161–3.
Acknowledgements
We wish to thank all the individuals, family members and staff from all the units that participated in the study.
Funding
Funding for this study is provided in part by the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-188) and Italian Ministry of Health (PNRR-MR1-2022-12376811 and RF-2021-12374963, to MT). Liselot van der Laan was awarded with the AR&D Travel grant from the Amsterdam UMC, which provided financial support for this work. This work was generated in part within the European Reference Network for Rare Malformation Syndromes, Intellectual and Other Neurodevelopmental Disorders (ERN ITHACA).
Author information
Authors and Affiliations
Contributions
Conceptualization; LVDL, KS, KK, and BS.; Data Curation; KR, LVDL, KK, MAL, and RR.; Formal analysis; KR, LVDL, KK, MAL, and RR.; Investigation; LVDL, KR, MA, AB, ALA, NBP, AMCG, BRD, GC, CD, DE, VG, BG, DG, and GBF. MK, MN, AN, VO, SO, WGP, AMP, TR, NRP, QS, ST, MT, MAT, AT, UK, IV, JMVH, AMVDKK, PH, MMAMM, and MMVH.; Methodology; LVDL, KR, KK, and BS.; Project administration; BS.; Supervision; BS and MMVH.; Validation; KR, LVDL, BS, and MMVH.; Visualization; LVDL, KK, and KR.; Writing-original draft; LVDL, KK, and KR.; Writing-review and editing; LVLD, KK, BS, and MMVH.
Corresponding authors
Ethics declarations
Competing interests
B.S. is a shareholder in EpiSign Inc.
ETHICAL approval
Written informed consent was obtained from all individuals or family members prior to inclusion in this study including for the use of DNA and clinical information. The study was conducted in accordance with the regulations of the Western University Research Ethics Board (REB116108, and REB106302).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
van der Laan, L., Karimi, K., Rooney, K. et al. DNA methylation episignature for Smith-Magenis and Potocki-Lupski syndromes: a mirror perspective. Eur J Hum Genet (2025). https://doi.org/10.1038/s41431-025-01956-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41431-025-01956-0