Abstract
Poecilogony, or multiple developmental modes in a single species, is exceedingly rare. Several species described as poecilogenous were later demonstrated to be multiple (cryptic) species with a different developmental mode. The Southern Ocean is known to harbor a high proportion of brooders (Thorson’s Rule) but with an increasing number of counter examples over recent years. Here we evaluated poecilogony vs. crypticism in the brittle star Astrotoma agassizii across the Southern Ocean. This species was initially described from South America as a brooder before some pelagic stages were identified in Antarctica. Reproductive and mitochondrial data were combined to unravel geographic and genetic variation of developmental modes. Our results indicate that A. agassizii is composed of seven well-supported and deeply divergent clades (I: Antarctica and South Georgia; II: South Georgia and Sub-Antarctic locations including Kerguelen, Patagonian shelf, and New Zealand; III-VI-VII: Patagonian shelf, IV-V: South Georgia). Two of these clades demonstrated strong size dimorphism when in sympatry and can be linked to differing developmental modes (Clade V: dwarf brooder vs. Clade I: giant broadcaster). Based on their restricted geographic distributions and on previous studies, it is likely that Clades III-VI-VII are brooders. Clade II is composed of different morphological species, A. agassizii and A. drachi, the latter originally used as the outgroup. By integrating morphology, reproductive, and molecular data we conclude that the variation identified in A. agassizii is best described as crypticism rather than poecilogony.
Similar content being viewed by others
Introduction
Marine invertebrates display a wide diversity of developmental modes, with development occurring in both pelagic and non-pelagic habitats (Levin and Bridges 1995; Poulin and Féral 1996; Marshall et al. 2012). It is generally accepted that developmental mode has an influence on dispersal distance, and that brooding species have a shorter distance of dispersal than broadcasters (Kelly and Palumbi 2010; White et al. 2010; Selkoe and Toonen 2011; Moreau et al. 2017). Developmental mode strongly affects connectivity and demographic patterns within or among populations and, consequently, how independently these populations evolve (Kelly and Palumbi 2010; Allcock and Strugnell 2012; Riesgo et al. 2015).
Marine invertebrates have long been a model for studying life-history evolution because of the diversity of their developmental modes (Emlet et al. 1987; McEdward 1995), with phylogenetic analyses in some taxa showing rapid transition in developmental mode (Hart et al. 1997, 2003; Duda and Palumbi 1999; Collin et al. 2007). Poecilogony, defined as an “intraspecific variation in developmental mode” (Knott and McHugh 2012), is a rare life-history strategy that has been reported in a variety of marine invertebrates, primarily polychaetes and molluscs (e.g., see Blake and Arnofsky 1999; Krug 2007). Typically, this involves a species producing both planktotrophic and lecithotrophic larvae, but under Levin and Bridges’ (1995) classification scheme for invertebrate larval development, a shift in the site of development (e.g., pelagic to benthic lecithotrophy) would also be considered a variation in developmental mode. In many cases, species described as poecilogenous have later been shown with molecular analyses to be morphologically cryptic species (Byrne et al. 2003; Hart et al. 2003; Knott and McHugh 2012). Differentiating between cases of true poecilogony and the presence of cryptic species is critical to understanding evolutionary transitions in developmental mode and the implications this has for the dispersal potential of a single species (Knott and McHugh 2012; Moon et al. 2017).
A case in point is the brittle star Astrotoma agassizii Lyman, 1875 (Euryalida: Gorgonocephalidae), which has a southern circumpolar distribution (Antarctic and sub-Antarctic through to southern temperate regions) at depths ranging from 40 to 1700 m (Bernasconi 1965; Sands et al. 2013; Berecoechea et al. 2017) and has been recognized as a morphologically uniform species throughout its range (Hunter and Halanych 2008; Galaska et al. 2017). This species was first described as a brooder using a small number of South American specimens (three to four individuals/study; Bernasconi 1965; De La Serna De Estaban 1966). A more large-scale study from several Antarctic and sub-Antarctic locations (806 specimens from Antarctic Peninsula, Ross Sea, South Georgia, and Patagonia) revealed brooding specimens in each region, but with a very low frequency in Antarctica (2 individuals out of 325 investigated; Monteiro and Tommasi 1983). Molecular analyses—using mitochondrial genes (Hunter and Halanych 2008) and RAD-seq data (Galaska et al. 2017)—have revealed three clades in A. agassizii, one from the Antarctic continent (Antarctic Peninsula) and two from the Patagonian shelf of South America. Recent evidence using DNA barcoding of plankton samples has shown that A. agassizii from the Ross Sea region of Antarctica has a pelagic lecithotrophic larva (Heimeier et al. 2010) and all sequences were within the Antarctic clades presented by Hunter and Halanych (2008). This suggests that A. agassizii might be composed of cryptic species with distinct developmental modes: a broadcasting species in Antarctica and two brooding species on the Patagonian shelf. None of the previous studies focused on both genetic and reproductive data, so the potential for poecilogony at some locations cannot be excluded.
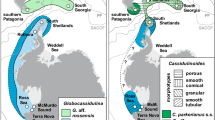
In the current work, we investigated the likelihood of poecilogony in A. agassizii by examining the relationship between genetic diversity and developmental mode in specimens collected across the Southern Ocean (Fig. 1). Genetic variation was evaluated using the mitochondrial gene Cytochrome c Oxidase subunit I, whereas developmental mode was investigated through direct (brooding in mature individuals) and indirect (egg size) evidence. This combination of distinct methods allowed us to address two evolutionary questions: (1) Can we identify different developmental modes in A. agassizii? (2) Can these modes be linked to genetic clades or is this a rare example of poecilogony? (3) When compared with the previous Antarctic Peninsula focus of Hunter and Halanych (2008), and Galaska et al. (2017), is there further genetic structure in A. agassizii when taking into account the wider distribution of the species? (4) When compared with the related species Astrotoma drachi (Guille 1979), is the variation among clades equivalent to that seen between the ingroup clades and A. drachi?
Material and methods
Sampling
Specimens were collected during several international expeditions (ACE, JR144, JR179, JR262, TAN0402, TAN0602, TAN0705, TAN0802, TRIP, POKER II, PS77, PS82, and REVOLTA I-II-III) from 2004 to 2017. Samples were immediately frozen or stored in 96% ethanol. In total, 298 specimens were sampled to obtain both reproductive and genetic data. Sampling locations (Fig. 1) include Antarctic locations (Adélie Land: 17, Amundsen Sea: 15, Balleny Islands: 5, Ross Sea: 26, Scott Island: 2, and Weddell Sea: 38), Patagonian shelf (33), South Georgia (78) and Shag Rocks (50), Kerguelen (26), Prince Edward (1), and Subantarctic New Zealand (7). All the samples were identified as A. agassizii by Sadie Mills, Rafael Martin-Ledo, and Chester Sands, except the New Zealand specimens that were identified as A. drachi.
Genetic data
Cytochrome oxidase 1 (CO1) sequences (603 base fragment) were obtained for 227 individuals. DNA extractions were performed on arm tip tissues and were based on the salting-out protocol from Sunnucks and Hales (1996). Amplification used the primers “F-LCO1490-OPHIUROID” (5′-TTTCAACNAAACAYAAGGAYATWGG-3′) and “R-HCO” (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′; Folmer et al. 1994). Each PCR mix (10 μl) included 8.5 μl of Mastermix (dNTP 25 mM, ammonium sulfate 0.1 M, Tris HCl 1 M, bovine serum albumin 25 mg/ml, β-mercaptoethanol 1 M, MgCl2 50 mM, H2O), 0.5 µl of Taq Polymerase (5 U/µl), 0.5 µl each primer (10 µM), and 0.5 µl of the DNA extract. PCR conditions consisted of 35 cycles for each of the three temperature steps [30 s at 94 °C (denaturation), 30 s at 48 °C (annealing), and 45 s at 72 °C (elongation)]. These cycles were preceded by a step of 2 min at 94 °C and were followed by a step of 10 min at 72 °C. PCR products were then sent to the MACROGEN sequencing service for purification and sequencing. Sequence editing and alignment were performed using BioEdit 7.0 (Hall 1999) and MEGA7 (Kumar et al. 2016). The absence of stop codon in the sequence was checked in MEGA7, whereas the absence of base compositional heterogeneity was examined in SeqVis v1.5 (Ho et al. 2006). A TCS (Templeton Crandall Singh) haplotype network was produced in PopART 1.7 (Leigh and Bryant 2015). Molecular diversity indeces were calculated using Arlequin 3.5 (Excoffier and Lischer 2010), including the number of haplotypes, the haplotype diversity, the number of polymorphic sites, the nucleotide diversity, and Tajima’s D and Fu’s Fs. Significance of D and F were evaluated through a permutation test (10,000 randomizations). Only clades with at least six individuals were considered for the calculations of these statistics, as it has been demonstrated that lower sampling sizes might lead to inaccurate estimates (Goodall-Copestake et al. 2012). JModeltestv2 (AIC - "Akaike information criterion", Darriba et al. 2012) was used to determine that GTR + G was the most suitable substitution model for our dataset. A Bayesian phylogeny was then produced in MrBayes 3.2 (Ronquist and Huelsenbeck 2003) using 100 million generations. Every 100,000 generations, the tree was sampled, thus obtaining a total of 1000 trees. Node support was assessed through posterior probability values. Initially, the closely related species A. drachi was used as the outgroup but our first results showed that it was in fact an ingroup. We therefore selected another outgroup, namely Ophiura lymani and other more closely related taxa (Asteronyx, Astrochlamys, Astrohamma, Gorgonocephalus). Mean pairwise distances between clades (p-distances, corrected, and uncorrected) were calculated between clades using the software Mega X (Kumar et al. 2018).
Two methods of “automatic” single locus species delimitation were used to assess how clade divergence matched with theoretical views on the speciation process. The multirate General Mixed Yule Coalescent (GMYC) was implemented using the Splits package (Ezard et al. 2009; Fujisawa, Barraclough 2013) with R (V 3.5.1, R Core Team 2016). First, an ultrametric phylogeny was produced in BEAST 2.4 (Bouckaert et al. 2014) using a GTR model, a relaxed uncorrelated molecular clock, and a yule process prior. Several runs were conducted and after each run prior value performance was assessed in TRACER V1.6; prior values were then “tweaked” until performance was deemed satisfactory. The final run was 108 generations with 25% burnin, sampling every 105 generations. Sampled trees were combined in LogCombiner V2.4 and summarized in TreeAnnotator V2.4, both part of the BEAST 2 package. Species delimitation was also explored using the Poisson Tree Process (PTP; Zhang et al. 2013). Although GMYC uses branch coalescing patterns to group putative species, PTP delimits species by judging relative branch lengths within and among putative groups. We used the more sophisticated multirate (mPTP) package of Kapli et al. (2017), using the maximum likelihood tree and a maximum likelihood heuristic search, taking into account minimum branch lengths.
Reproductive data
A total of 233 specimens were dissected and the disc diameter was measured using Vernier calipers. For each individual we described the presence of gonads, as well as any direct evidence of brooding, such as the presence of juveniles inside the bursae. In order to compare brooding and sexual maturity at different locations, we used the definition of Monteiro and Tommasi (1983) as sexually mature “individuals incubating juveniles or harboring ovaries with distinguishable eggs” and extracted some reproductive data from Monteiro and Tommasi (1983). In ten specimens from different locations/clades, we also measured the size of the largest eggs from the ovaries using a binocular microscope. Egg diameter was determined from digital photographs using the software ImageJ (Schneider et al. 2012). Statistical tests were performed using STATISTICA 7.0 (statsoft.com) or in R 3.3.2 (R Core Team 2016) using the Vegan package (Oksanen et al. 2010).
Results
Genetic data
We successfully obtained a 603 base alignment of 227 CO1 sequences resulting in 75 haplotypes distributed among 7 discrete genetic lineages (posterior probabilities = 1, Figs. 2 and 3). These seven lineages are based on the mPTP analysis, whereas the other species delimitation method (GMYC) provided a number of 35 hypothesized species. The largest clade (Clade I, 128 individuals) included samples from both the Antarctic continental shelf and from around the shelf of South Georgia and Shag Rocks. All other clades formed a polytomy and were sister to Clade I (Fig. 3). Clade II (divided into three geographically specific sub-clades) included samples from the Kerguelen Plateau and Prince Edward Island (sub-clade A), South Georgia, Shag Rocks and the Patagonian Shelf (sub-clade B), and the “outgroup” taxon A. drachi from New Zealand with one specimen from Scott Island (sub-clade C). Clade III was exclusively from the Patagonian Shelf. Clades IV and V share a common ancestor and are exclusively from the South Georgia/Shag Rocks shelf region, whereas Clade VI and VII, also sharing a common ancestor, are exclusively from Patagonian Shelf. The degree of divergence between Clade I and the other six clades is considerable (>10% average corrected pairwise distance, >6% uncorrected p-distance; Supplementary Material 1), and the corrected distances between the other clades ranged between 2.4% (Clades IV and V) and 8.3% (Clades II and VII), respectively (2.1% and 5.0% uncorrected p-distances, Supplementary Material 1). Within clade, distances ranged between 0.1% (Clade I) and 0.5% (Clade VII), demonstrating a clear diversity “gap” within and among clades.
Haplotype network based on 227 CO1 sequences (603 bp) of Astrotoma. Different colors denote different locations (see legend in Fig. 1). Circle sizes at the bottom right denote the number of samples exhibiting the haplotype (outer circle: 10 samples, inner circle: 1 sample). Clades I–VII are based on the mPTP analysis (species delimitation method)
Bayesian phylogeny of 227 Astrotoma CO1 sequences. Values at each node are the posterior probabilities. Nodes with support less than 0.75 were collapsed. Different colors denote different locations (see legend in Figure 1). For better visualization, some outgroups were removed and the branch closest to the basal node was shortened (a link to the complete and unmodified phylogeny can befound in Data availability section). Clades I to VII are based on the mPTP analysis (species delimitation method)
The relationship between haplotype diversity and nucleotide diversity in the combined dataset (all samples in one group) was high compared with that expected from a panmictic species (see Fig. 3 from Goodall-Copestake et al. 2012), indicating that it is likely there are cryptic species present. When analyzed on a clade-by-clade basis, haplotype diversity vs. nucleotide diversity fits within the expected variation of a panmictic species in each clade (Table 1). Nucleotide diversity of Clade I was, however, low compared with haplotype diversity (Table 1). As a result, both Tajima’s D and Fu’s S significantly deviated from neutral expectations, indicating either selection or population expansion (Table 1). When separated out into the two geographic components “Antarctic Clade I” and “South Georgia Clade I,” the result for each group was similar (Tajima’s D and Fu’s S significantly negative, data not shown). Clade V, also found on South Georgia, was significantly different from neutral expectations, suggesting a common mechanism or shared history with Clade I (Table 1).
Reproductive data
Among the 233 specimens dissected in our study, 173 were assigned to a genetic clade (116 individuals to Clade I, 19 to Clade II, 1 to Clade III, 2 to Clade IV, 19 to Clade V, and 4 to Clade VII). Thirty-eight percent of specimens were observed to be mature with a disc diameter (at maturity) ranging between 1.3 and 6.4 cm (Table 2). Mature specimens were present in Clade I in February (Antarctica), April (South Georgia, Antarctica), and November (South Georgia); Clade V had mature specimens in April and November (Supplementary Material 3). Only 2% of the total specimens were observed to carry young in the bursae, much fewer than expected (following Monteiro and Tommasi (1983), the expectation was closer to 11%) (Table 2). Brooding was only recorded from five specimens from South Georgia/Shag Rocks (4% of total individuals in this region, Table 2). Monteiro and Tommasi (1983) found evidence of brooding on the Patagonian shelf (22%), in South Georgia/Shag Rocks (14%) and in Antarctica (0.6 %) (Table 2).
We focused our quantitative comparisons using specimens from Clade I (Antarctica, South Georgia) and Clade V (South Georgia). From the Antarctic continental shelf region, the mean disc diameter of A. agassizii was 3.9 cm (range 2.1–5.1, SE ± 0.2), whereas from the shelf areas around South Georgia and Shag Rocks there was a larger range in disc diameter (mean: 3.5 cm, 1.3–6.4, SE ± 0.2). Disc diameter frequency showed a bimodal distribution around South Georgia, suggesting either separate cohorts or distinct size-specific populations on the South Georgia shelf (Fig. 4). As molecular data indicated that there were distinct clades on the South Georgia shelf, we investigated whether the size categories identified were related to the genetic lineages: this analysis showed a clear size bimodality within the South Georgia specimens: the smaller morphotype is associated with Clade V, whereas the larger morphotype is associated with the Antarctic Clade I (Mann–Whitney test, p < 0.001). Within Clade I, we also observed a size dimorphism between Antarctic and South Georgia specimens (Fig. 4, Mann–Whitney test: p < 0.001).
Monteiro and Tommasi (1983) also had a bimodal distribution in disc diameter of South Georgia specimens, although it was not so pronounced as in Fig. 4, because they had fewer large specimens. A Mann–Whitney test indicated that their South Georgia collections were not directly comparable to ours (disc diameter significantly different, p < 0.0001). However, their collection did not significantly differ in disc diameter from our Clade V specimens. Monteiro and Tommasi (1983) found that only individuals with small disc size (<3 cm) were brooders. All the brooding individuals from our collection had a considerably smaller disc diameter (mean 1.82 cm, range 1.6–2 cm) than expected from the average of our South Georgia collection (p < 0.05) but their disc size did not deviate significantly from the Monteiro and Tommasi (1983) collection.
Due to the condition of many of the ethanol samples, egg size measurements were only possible on very few individuals. Of these six were from Clade I (mean 452 µm, SE ± 20), and four were from Clade V (mean 749 µm, SE ± 41), indicating even with this small dataset that eggs from Clade V are considerably larger than those from Clade I (Mann–Whitney test, p < 0.015).
Discussion
Developmental modes and their genetic distinctness
Although historically identified as a brooding species (Bernasconi 1965), Heimeier et al. (2010) found strong evidence for a dispersing larva in Antarctic A. agassizii. The previous work of Monteiro and Tommasi (1983) was able to identify brooding individuals around South Georgia and the Patagonian Shelf with moderate frequency (14% and 22%, respectively) but found brooding to be exceedingly rare around the Antarctic continental shelf with only 2 specimens out of 325 investigated (0.6%). We were unable to find any evidence for brooding among the 79 specimens dissected from around the Antarctic Continental Shelf, supporting the idea that Antarctic Astrotoma tend toward a dispersive larval stage.
Further support for this hypothesis was provided by the egg size measurements obtained from the few well-preserved mature Astrotoma specimens. It is generally accepted that smaller eggs result in a feeding pelagic larval stage, whereas larger eggs produce non-feeding larvae or are brooded and develop directly into young adults (Stearns 1977), a pattern supported by empirical data on ophiuroid egg sizes (Sewell and Young 1997). The ovarian egg size that we measured from Clade I Antarctic individuals of ≈450 µm suggests a non-feeding larva and is in the size range of DNA-barcoded A. agassizii-fertilized eggs collected from the plankton (mean 463 µm, range 436–513 µm, Heimeier et al. 2010). Species with a planktonic larva, which usually indicates substantial dispersal ability, are expected to have few barriers to gene flow (although this varies depending on the species, see Shanks 2009; Kelly and Palumbi 2010; Selkoe and Toonen 2011; Faurby and Barber (2012)). Our molecular data (Antarctic shelf individuals all belonging to Clade I), as well as that of Galaska et al. (2017), show no population structure across the whole of the sampled range, again strongly supporting a clade with a planktonic larval stage.
Brooding was detected in our study in a few individuals from around South Georgia and Shag Rocks, but at a considerably lower prevalence (4%) than reported by Monteiro and Tommasi (1983) from South Georgia (14%). Our brooded specimens had much larger eggs (≈750 µm), and in both our dataset and Monteiro and Tommasi (1983), brooding was detected in the smaller mature individuals. This in accordance with the generally accepted hypothesis that correlates brooding with small adult size (e.g., Strathmann and Strathmann 1982).
Detailed analysis showed that our South Georgia specimens had a bimodal size-frequency distribution in disc diameters, with the brooding individuals always within the smaller size class, and from Clade V, whereas the larger mode represented Clade I, but with a considerably larger disc diameter on average than Clade 1 individuals from the Antarctic continental shelf (Fig. 4). We hypothesize that the size-frequency patterns in A. agassizii in South Georgia represent a case of character divergence, as described by Darwin (1859) or character displacement as refined by Brown and Wilson (1956); both refer to accentuated change in a character state (here the disc size) between two closely related species in a zone of sympatry that is not observed outside of the zone of overlap (Brown and Wilson 1956). Although earlier research focused on resource (ecological) or sexual selection (reinforcement) as driving character displacement (Brown and Wilson 1956), more recent studies have found subtle variations and combinations of these themes to play a part (see review in Table 1 of Stuart et al. 2017). Here, as in situ observations of A. agassizii are extremely difficult due to their depth distribution (200–1500 m), we will avoid speculation on the reasons for character displacement, but instead focus on the implications.
If we accept that character displacement occurs between closely related species (Brown and Wilson 1956), this suggests that Clade I and V of A. agassizii represent cryptic (unrecognized) sympatric species, although because of overlapping reproductive seasons there is, as yet, no obvious barrier to interbreeding and we are uncertain how species cohesion and morphological uniqueness are maintained. We find little support for poecilogony as the two developmental modes (pelagic lecithotrophy, brooding) occur in separate clades, rather than within clades (e.g., Ellingson and Krug 2006).
The North Atlantic ophiuroid Ophioderma longicauda tells a very similar story to A. agassizii (Stöhr et al. 2009; Boissin et al. 2011; Weber et al. 2014). Originally thought to have a dispersive pelagic larva, further research revealed brooding individuals in a monophyletic clade. In the area of sympatry between brooding and non-brooding forms, the mature brooding individuals (<13 mm) were smaller than the non-brooding individuals (20–26 mm). The authors concluded that the brooding form is a cryptic species in what is likely to be a species complex. Similarly, Ophiacantha wolfarntzi (Martín-Ledo et al. 2013), another ophiuroid found around the South Georgia/Shag Rocks continental shelf, is sympatric with a sister clade, which is yet to be formally described but appears to be identical to O. wolfarntzi (Martín-Ledo et al. 2013). It is likely that if the two clades do represent different diverging species, some morphological shift might become apparent once sufficient samples are examined.
Although the most parsimonious hypothesis for the presence of two developmental modes within A. agassizii is the presence of cryptic species, the report by Monteiro and Tommasi (1983) of two brooding specimens of A. agassizii in Antarctica (northern Ross Sea) is perplexing. These 2 specimens (disc diameter < 3 cm) were found to be brooding 182 and 923 juveniles in the bursae. Although rare (2 out of 235 Antarctic specimens), records of such large numbers of young within the bursae make it likely that this is a true record of A. agassizii brooding in the Ross Sea. How might this be explained? Three non-mutually exclusive answers are proposed as follows: (1) that there is more than one species of A. agassizii on the Antarctic continental shelf: one that broadcasts lecithotrophic larvae (Heimeier et al. 2010) and a rarer species that broods (this was, however, not revealed in our genetic data); (2) that, rarely, some individuals of Clade I are poecilogenous with a dual developmental mode (broadcasting-brooding) as reported or suggested for other echinoderms (McClary and Mladenov 1989; Mercier and Hamel 2008; O’Hara et al. 2014); or (3) that there is connectivity of adult specimens between a brooding A. agassizii species in South America and/or the Subantarctic with the Antarctic continent, through rafting. Connectivity by rafting has been previously proposed for A. agassizii within Antarctica by Hunter and Halanych (2008), and across the Antarctic Polar Front by Galaska et al. (2017), to explain a South American haplotype being found on the Antarctic Peninsula.
High levels of diversity across the sub-Antarctic
It is clear from the previous research of Hunter and Halanych (2008) and Galaska et al. (2017) that there is considerable genetic variation within A. agassizii between Antarctic and Patagonian shelf samples, as well as among Patagonian Shelf individuals. The divergence between the Antarctic and Patagonian shelf samples is, however, maintained, despite some indication of gene flow, and there were few putative hybrids between two identified Patagonian shelf populations, despite overlap of distributions (Hunter and Halanych 2008, Galaska et al. (2017).
Our sampling of A. agassizii extended more broadly around Antarctica and to sub-Antarctic regions, and unsurprisingly identified further deep genetic differentiation with what appears to be a rapid radiation of the sub-Antarctic clades. This pattern bears some resemblance to the polytomy identified by Sands et al. (2015) in the similarly distributed Southern Ocean ophiuroid O. lymani, where there was strong support for each geographically specific clade, but low support for timing of divergence order. In addition, both species show a low diversity Antarctic-specific clade, although in Astrotoma the Antarctic clade is shared with South Georgia, whereas the Antarctic clade in Ophiura is specific to the Antarctic continental shelf (Sands et al. 2015). A third similarity between studies is that in both cases the chosen “outgroup”, or assumed sister species, has ended up being an “ingroup”. In the case of Ophiura, the Antarctic clade is better known as Ophiura carinifera and is morphologically distinct from O. lymani, but apparently nested within the group’s genetic variation. In Astrotoma, A. drachi from southern New Zealand waters nests in Clade II.
A. drachi was initially described in the Philippines by Guille (1979) who noted the absence of multiple conical spines on the aboral areas (the opposite of Astrotoma manilense, also described from the Philippines, which harbors such spines). Despite numerous surveys, there is no recorded specimen between the Philippines and New Zealand (disjunct distribution), meaning that the New Zealand specimens are unlikely to be conspecific with any species from the Philippines (O’Hara and Harding 2014). After further morphological investigations of A. drachi from New Zealand waters and A. manilense from Tasmanian waters, O’Hara and Harding (2014) were hesitant to synonymize these two species and suggested the need for molecular data to investigate species boundaries inside the genus. Our molecular data from A. drachi (New Zealand) groups the species with an individual sampled from Scott Island (Northern Ross Sea, Antarctica) in what we have labeled Clade II sub-clade C. The three sub-clades of Clade II are also a polytomy with sub-clade A, restricted to islands of the Indian Ocean sector of the Southern Ocean (Kerguelen and Prince Edward Islands), sub-clade B from the Atlantic region of the Southern Ocean (Patagonian Shelf and South Georgia/Shag Rocks), and sub-clade C from the Pacific region of the Southern Ocean (New Zealand and Scott Island). Uncorrected among group genetic differences were <1% (Supplementary Material 1).
Of interest is the identification of four distinct genetic clades on the Patagonian Shelf (Clades III, VI, VII, and representatives of Clade II sub-clade B), where other studies have only identified two (Hunter and Halanych 2008; Galaska et al. 2017). A closer inspection a posteriori may nevertheless bring out the existence of the other two groups in the previous studies (as suggested by Figs. 3 and 4 from Galaska et al. 2017). The holotype of A. agassizii, described from the Strait of Magellan (Lyman 1875), is likely to belong to one of these four clades. We have found between group genetic-corrected distances between 5% and 8% for these Patagonian clades (Supplementary Material 2). The discrete genetic groupings in sympatry may indicate species boundaries, despite limited gene flow as identified in Hunter and Halanych (2008).
Galaska et al. (2017) used 2bRAD to generate genomic-wide single-nucleotide polymorphisms to characterize their populations. Although a powerful technique within and between closely related populations, the power of this technique rapidly diminishes as genetic divergences increase, because the necessary restriction sites become more and more prone to mutation over time. We suggest a more conservative approach to understanding the evolutionary and demographic history of Astrotoma, particularly the Patagonian Shelf populations, would be to use more conserved sequencing techniques such as the exon capture system recently developed by Hugall et al. (2015). Exon capture would still bring the power of many genomic markers, but have the advantage of these markers being homologous across multiple divergent populations and species.
Conclusions
Following the suggestion of “species as hypotheses” proposed by Pante et al. (2015), our work combined with the historic papers cited above lead us to propose that A. agassizii is a species complex, where Clade I to VII are hypothesized species that encompass both brooding and dispersing developmental modes. We can be confident that Clade I, distributed across the Antarctic continental shelf and in South Georgia, is a genetically discrete entity with a dispersing pelagic larva, whereas Clade V, endemic to South Georgia, is a smaller brooding species. It is clear from Monteiro and Tommasi (1983) that some or all of the other clades found on the Patagonian Shelf may also brood; however, sample preservation problems and sample size erosion as a consequence of many divergent lineages has made further meaningful comment on clade-specific developmental mode or poecilogony impossible.
We do acknowledge that our suggestion of crypsis is based on a single mitochondrial marker, and that caution is advised when using mitochondrial markers as a proxy for species evolution (Ballard and Whitlock 2004; Weber et al. 2019). However, such markers can be a good predictor of crypsis where multiple strongly supported clades are found in morphologically uniform species (Held 2003, 2005; Linse et al. 2007; Leese et al. 2008; Janosik and Halanych 2010; Hemery et al. 2012; Sands et al. 2015; Verheye et al. 2016; Brasier et al. 2017; Jossart et al. 2017). Between Clades I and V, the genetic differentiation is corroborated by a morphological differentiation that we hypothesize as a case of character displacement. Further research using nuclear markers will be needed to further clarify the species status of the other clades.
This research also has two broader implications. First, Clade II of A. agassizii includes the recognized outgroup A. drachi. If included as an ingroup, then we must reconsider A. agassizii as a complex in which A. drachi (A manilense) are elements in much the same way as Sands et al. (2015) found that O. carinifera is an element of the O. lymani complex (Sands et al. 2015). Another similarity with O. lymani is the large diversity of A. agassizii haplotypes and clades identified from Patagonian Shelf and South Georgia (the Atlantic sector of the Southern Ocean), which hints at a South Georgia/Patagonian shelf origin for the A. agassizii complex. It was proposed that O. lymani originated in the sub-Antarctic and expanded its range onto the Patagonian Shelf in more recent times, and it may be that A. agassizii, having shared geographic range and some congruence in mitochondrial phylogeny, had a similar (shared) evolutionary history (Sands et al. 2015).
Finally, the identification of multiple hypothetical species has consequences for conservation management, particularly in South Georgia where Clade V appears to be endemic, but one of three distinct clades present. If all three of these clades are later recognized as distinct species, then South Georgia now has an increased biodiversity (1 species to 3) but shifts in terms of management from a single robust faunal element (wide distribution, high connectivity, and large population size) to three separate evolutionary units, one with no connectivity and lower population size. Further detailed testing is thus required of the specific species hypotheses we propose here.
Data archiving
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.n7s0g10.
References
Allcock AL, Strugnell JM (2012) Southern Ocean diversity: new paradigms from molecular ecology. Trends Ecol Evol 27(9):520–528
Ballard JWO, Whitlock MC (2004) The incomplete natural history of mitochondria. Mol Ecol 13(4):729–744
Berecoechea JJ, Brogger MI, Penchaszadeh PE (2017) New evidence of brooding in the deep-sea brittle star Astrotoma agassizii Lyman, 1876 from aSouth Western Atlantic Canyon. Deep Sea Res I 127:105–110
Bernasconi I (1965) Astrotoma agassizii Lyman, especie vivípara del Atlántico sur (Ophiuroidea, Gorgonocephalidae). Contrib Científica 15:1–5
Blake JA, Arnofsky PL (1999) Reproduction and larval development of the spioniform Polychaeta with application to systematics and phylogeny. Hydrobiologia 402:57–106
Boissin E, Stöhr S, Chenuil A (2011) Did vicariance and adaptation drive cryptic speciation and evolution of brooding in Ophioderma longicauda (Echinodermata: Ophiuroidea), a common Atlanto‐Mediterranean ophiuroid? Mol Ecol 20(22):4737–4755
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10(4):e1003537
Brasier MJ, Harle J, Wiklund H, Jeffreys RM, Linse K, Ruhl HA, Glover AG (2017) Distributional patterns of polychaetes across the West Antarctic based on DNA barcoding and particle tracking analyses. Front Mar Sci 4:356
Brown WL, Wilson EO (1956) Character displacement. Syst Zool 5(2):49–64
Byrne M, Hart MW, Cerra A, Cisternas P (2003) Reproduction and larval morphology of broadcasting and viviparous species in the Cryptasterina species complex. Biol Bull 205(3):285–294
Collin R, Chaparro OR, Winkler F, Veliz D (2007) Molecular phylogenetic and embryological evidence that feeding larvae have been reacquired in a marine gastropod. Biol Bull 212(2):83–92
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9(8):772
Darwin C (1859) The origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. John Murray, London, p 1–502.
De La Serna De Estaban CJ (1966) Consideraciones preliminares sobre la reproduccion de Astrotoma agassizii Lyman, 1876 (Ophiuroidea, Euryalae). Physis 26(71):89–94
Duda TF, Palumbi SR (1999) Developmental shifts and species selection in gastropods. Proc Natl Acad Sci USA 96(18):10272–10277
Ellingson RA, Krug PJ (2006) Evolution of poecilogony from planktotrophy: cryptic speciation, phylogeography, and larval development in the gastropod genus. Alder Evol 60(11):2293–2310
Emlet RB, McEdward LR, Strathmann RR (1987) Echinoderm larval ecology viewed from the egg. In: Jangoux M, Lawrence JM (eds) Echinoderm studies 2. Balkema, Leiden, p 55–136
Excoffier L, Lischer HE (2010) Arlequin suite ver 35: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567
Ezard T, Fujisawa T, Barraclough TG (2009) Splits: species limits by threshold statistics (version1.0-19). Available from: http://R-Forge.R-project.org/projects/splits/
Faurby S, Barber PH (2012) Theoretical limits to the correlation between pelagic larval duration and population genetic structure. Mol Ecol 21(14):3419–3432
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase sub- unit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3(5):294–299
Fujisawa T, Barraclough TG (2013) Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol 62(5):707–724
Galaska MP, Sands CJ, Santos SR, Mahon AR, Halanych KM (2017) Crossing the divide: admixture across the Antarctic polar front revealed by the brittle star Astrotoma agassizii. Biol Bull 232(3):198–211
Goodall-Copestake WP, Tarling GA, Murphy EJ (2012) On the comparison of population-level estimates of haplotype and nucleotide diversity: a case study using the gene cox1 in animals. Heredity 109(1):50–56
Guille A (1979) Astrotoma drachi, nouvelle espèce bathyale d’ophiuride Gorgonocephalidae des iles Philippines. Vie Millieu 28:437–442
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hart MW, Byrne M, Johnson SL (2003) Patiriella pseudoexigua (Asteroidea: Asterinidae): a cryptic species complex revealed by molecular and embryological analyses. J Mar Biol Assoc UK 83(5):1109–1116
Hart MW, Byrne M, Smith MJ (1997) Molecular phylogenetic analysis of life‐history evolution in asterinid starfish. Evolution 51(6):1848–1861
Heimeier D, Lavery S, Sewell MA (2010) Molecular species identification of Astrotoma agassizii from planktonic embryos: further evidence for a cryptic species complex. J Hered 101(6):775–779
Held C (2003) Molecular evidence for cryptic speciation within the widespread Antarctic crustacean Ceratoserolis trilobitoides (Crustacea, Isopoda). In: Huiskes AHL, Gieskes WWC, Rozema J, Schorno RML, vanderVies SM, Wolff WJ (eds) Antarc Biol Glob Context, Backhuys, Leiden, p 135–139
Held C, Wägele JW (2005) Cryptic speciation in the giant Antarctic isopod Glyptonotus antarcticus (Isopoda: Valvifera: Chaetiliidae). Sci Mar 69:175–181
Hemery LG, Eléaume M, Roussel V, Améziane N, Gallut C, Steinke D, Wilson NG (2012) Comprehensive sampling reveals circumpolarity and sympatry in seven mitochondrial lineages of the Southern Ocean crinoid species Promachocrinus kerguelensis (Echinodermata). Mol Ecol 21(10):2502–2518
Ho JW, Adams CE, Lew JB, Matthews TJ, Ng CC, Shahabi-Sirjani A, Jermiin LS (2006) SeqVis: visualization of compositional heterogeneity in large alignments of nucleotides. Bioinformatics 22(17):2162–2163
Hugall AF, O’Hara TD, Hunjan S, Nilsen R, Moussalli A (2015) An exon-capture system for the entire class Ophiuroidea. Mol Biol Evol 33(1):281–294
Hunter RL, Halanych KM (2008) Evaluating connectivity in the brooding brittle star Astrotoma agassizii across the Drake Passage in the Southern Ocean. J Hered 99(2):137–148
Janosik AM, Halanych KM (2010) Unrecognized Antarctic biodiversity: a case study of the genus Odontaster (Odontasteridae; Asteroidea). Integr Comp Biol 50(6):981–992
Jossart Q, De Ridder C, Lessios HA, Bauwens M, Motreuil S, Rigaud T, David B (2017) Highly contrasted population genetic structures in a host–parasite pair in the Caribbean Sea. Ecol Evol 7(22):9267–9280
Kapli P, Lutteropp S, Zhang J, Kobert K, Pavlidis P, Stamatakis A, Flouri T (2017) Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 33(11):1630–1638
Kelly RP, Palumbi SR (2010) Genetic structure among 50 species of the northeastern Pacific rocky intertidal community. PLoS ONE 5(1):e8594
Knott KE, McHugh D (2012) Introduction to symposium: poecilogony—a window on larval evolutionary transitions in marine invertebrates. Int Comp Biol 52(1):120–127
Krug PJ (2007) Poecilogony and larval ecology in the gastropod genus Alderia. Am Malacol Bull 23(1):99–111
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 70 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Leese F, Kop A, Wägele JW, Held C (2008) Cryptic speciation in a benthic isopod from Patagonian and Falkland Island waters and the impact of glaciations on its population structure. Front Zool 5(1):19
Leigh JW, Bryant D (2015) popart: full‐feature software for haplotype network construction. Methods Ecol Evol 6(9):1110–1116
Levin LA, Bridges TS (1995) Pattern and diversity in reproduction and development. In: Mc Edward LR (ed.) Ecology of marine invertebrate larvae. CRC Press, Boca Raton, p 1–48
Linse K, Cope T, Lörz AN, Sands C (2007) Is the Scotia Sea a centre of Antarctic marine diversification? Some evidence of cryptic speciation in the circum-Antarctic bivalve Lissarca notorcadensis (Arcoidea: Philobryidae). Polar Biol 30(8):1059–1068
Lyman T (1875) Zoological results of the Hassler Expedition. 2 Ophiuridae Astro Illus Cat Mus Comp Zool Harv Coll 8(2):1–34
Marshall DJ, Krug PJ, Kupriyanova EK, Byrne M, Emlet RB (2012) The biogeography of marine invertebrate life histories. Annu Rev Ecol Evol Syst 43:97–114
Martín-Ledo R, Sands CJ, López-González PJ (2013) A new brooding species of brittle star (Echinodermata: Ophiuroidea) from Antarctic waters. Polar Biol 36(1):115–126
McClary DJ, Mladenov PV (1989) Reproductive pattern in the brooding and broadcasting sea star Pteraster militaris. Mar Biol 103:531–540
McEdward LR (1995) Ecology of marine invertebrate larvae. CRC Press, Boca Raton, p 480
Mercier A, Hamel JF (2008) Depth-related shift in life history strategies of a brooding and broadcasting deep-sea asteroid. Mar Biol 156(2):205–223
Monteiro AMG, Tommasi LR (1983) Ophiuroidea das regiões Antárctica e Subantarctica. 1. Sobre três espécies de Gorgonocephalidae e Ophiacanthidae. Bolm Inst Oceano 32(1):33–54
Moon KL, Chown SL, Fraser CI (2017) Reconsidering connectivity in the sub‐Antarctic. Biol Rev 92(4):2164–2181
Moreau C, Saucède T, Jossart Q, Agüera A, Brayard A, Danis B (2017) Reproductive strategy as a piece of the biogeographic puzzle: a case study using Antarctic sea stars (Echinodermata, Asteroidea). J Biogeogr 44(4):848–860
O’Hara TD, England PR, Gunasekera RM, Naughton KM (2014) Limited phylogeographic structure for five bathyal ophiuroids at continental scales. Deep Sea Res I 84:18–28
O’Hara TD, Harding C (2014) A new species of Sigsbeia and additional records of ophiuroids from the Great Australian Bight. Mem Mus Vic 72:131–140
Oksanen J, Blanchet FG, Kindt R, Legendre P, O’hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2010) vegan: Community Ecology Package. R package version 1.17-2. Available from: http://cran.r-project.org
Pante E, Puillandre N, Viricel A, Arnaud‐Haond S, Aurelle D, Castelin M, Viard F (2015) Species are hypotheses: avoid connectivity assessments based on pillars of sand. Mol Ecol 24(3):525–544
Poulin E, Féral JP (1996) Why are there so many species of brooding Antarctic echinoids? Evolution 50(2):820–830
R Core Team (2016) R: a language and environment for statistical computing. Available from: https://www.R-project.org/.
Riesgo A, Taboada S, Avila C (2015) Evolutionary patterns in Antarctic marine invertebrates: an update on molecular studies. Mar Genom 23:1–13
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574
Sands CJ, Griffiths HJ, Downey RV, Barnes DK, Linse K, Martín-Ledo R (2013) Observations of the ophiuroids from the West Antarctic sector of the Southern Ocean. Antar Sci 25(1):3–10
Sands CJ, O’Hara T, Barnes DK, Martín-Ledo R (2015) Against the flow: evidence of multiple recent invasions of warmer continental shelf waters by a Southern Ocean brittle star. Front Ecol Evol 3:1–13
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7):671
Selkoe KA, Toonen RJ (2011) Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Mar Ecol Prog Ser 436:291–305
Sewell MA, Young CM (1997) Are echinoderm egg size distributions bimodal? Biol Bull 193(3):297–305
Shanks AL (2009) Pelagic larval duration and dispersal distance revisited. Biol Bull 216(3):373–385
Stearns SC (1977) The evolution of life history traits: a critique of the theory and a review of the data. Annu Rev Ecol Syst 8(1):145–171
Stöhr S, Boissin E, Chenuil A (2009) Potential cryptic speciation in Mediterranean populations of Ophioderma (Echinodermata: Ophiuroidea). Zootaxa 2071:1–20
Strathmann RR, Strathmann MF (1982) The relationship between adult size and brooding in marine invertebrates. Am Nat 119(1):91–101
Stuart YE, Inkpen SA, Hopkins R, Bolnick DI (2017) Character displacement is a pattern: so, what causes it? Biol J Linn Soc 121(3):711–715
Sunnucks P, Hales DF (1996) Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol Biol Evol 13(3):510–524
Verheye ML, Backeljau T, d’Udekem d’Acoz C (2016) Looking beneath the tip of the iceberg: diversification of the genus Epimeria on the Antarctic shelf (Crustacea, Amphipoda). Polar Biol 39:925–945
Weber AAT, Stöhr S, Chenuil A (2014) Genetic data, reproduction season and reproductive strategy data support the existence of biological species in Ophioderma longicauda. Comptes Rendus Biol 337:553–560
Weber AAT, Stöhr S, Chenuil A (2019) Species delimitation in the presence of strong incomplete lineage sorting and hybridization: Lessons from Ophioderma (Ophiuroidea: Echinodermata). Mol Phylogenet Evol 131:138–148
White C, Selkoe KA, Watson J, Siegel DA, Zacherl DC, Toonen, RJ (2010) Ocean currents help explain population genetic structure. Proc R Soc Lond B Biol Sci 277:1685–1694
Zhang J, Kapli P, Pavlidis P, Stamatakis A (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876
Acknowledgements
This work was supported by a New Zealand Antarctic Research Institute (NZARI) postdoctoral fellowship (contract number: NZARI 2016-1-8, Type A—Research Projects of Excellence in Antarctic, Subantarctic, and Southern Ocean Research). We thank the following people for their help: S. Mills and M. Éléaume for the sharing of samples; C. Moreau, S. Lavery, W. Goodall-Copestake, D. Barnes, and G. Stowasser for helpful discussions; S. Mills and R. Martin-Ledo for their taxonomic expertise; S. Stöhr and M. Jangoux for their help in the identification of the reproductive structures; E. Frost, V. Thakur, D. Montenegro, M. Clark, and G. Caulier for their assistance in the laboratory; The BAS Research Development and Support team, with a special thanks to P. Geissler and G. Hillyard; and R. McLeod and G. Wilson for their friendly and professional following up of the NZARI project. We thank the NIWA Invertebrate Collection for providing the specimens and the following projects, which collected the material: voyage TAN0802, funded by the New Zealand Government under the NZ International Polar Year Census of Antarctic Marine Life Project (Phase 1: So001IPY; Phase 2; IPY2007–01). We gratefully acknowledge project governance provided by the Ministry of Fisheries (MFish) Science Team and the Ocean Survey 20/20 CAML Advisory Group (Land Information New Zealand (LINZ), MFish, Antarctica New Zealand, Ministry of Foreign Affairs and Trade, and National Institute of Water and Atmospheric Research (NIWA)); TAN0602, “Antarctic Geophysical & Scientific Studies” funded by MFish; TAN0402, A biodiversity survey of the western Ross Sea and Balleny Islands in 2004 undertaken by NIWA and financed by MFish; TAN0705, Ocean Survey 20/20 Chatham/Challenger Biodiversity and Seabed Habitat Project, jointly funded by MFish, LINZ, NIWA, and Department of Conservation; TRIP specimens collected under the Scientific Observer Program funded by the New Zealand Ministry for Primary Industries. We thank the master, the crew, and the scientific team of the FV “Austral” to have collected, sorted, and made available for studies the benthic samples of the POKER 2 (2010) cruise. We are grateful to the leader of the cruise, G. Duhamel (MNHN) and Echinodermata curator M. Eléaume to have granted access to POKER 2 collection. Work at sea was supported by the Terres Australes et Antarctiques Françaises (TAAF), the Syndicat des Armateurs Réunionnais de Palangriers Congélateurs (SARPC), the Direction des Pêches Maritimes et de l’Aquaculture, Ministère de l’Agriculture et de l’Alimentation (DPMA), the Réserve Nationale Naturelle of TAAF, and the Muséum national d’Histoire naturelle, Paris (MNHN). We thank the scientific team of IPEV (Institut Polaire Français Paul-Emile Victor) program 1124 REVOLTA (Radiation EVOLutives en Terre Adélie) cruises 2009–12 (chief scientists M. Eléaume, N. Améziane, and G. Lecointre). Financial and technical support was granted by IPEV, MNHN, CNRS, and Sorbonne Université. We thank the captains, crews, and science leaders who contributed to the material collected from the RRS James Clarke Ross (JR144, JR179, and JR262), RV Polarstern (PS77), and Akademik Treshnikov (Antarctic Circumnavigation Expedition, Swiss Polar Institute).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Jossart, Q., Sands, C.J. & Sewell, M.A. Dwarf brooder versus giant broadcaster: combining genetic and reproductive data to unravel cryptic diversity in an Antarctic brittle star. Heredity 123, 622–633 (2019). https://doi.org/10.1038/s41437-019-0228-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41437-019-0228-9
This article is cited by
-
A pioneer morphological and genetic study of the intertidal fauna of the Gerlache Strait (Antarctic Peninsula)
Environmental Monitoring and Assessment (2023)