Abstract
Understanding the mating system of Eucalyptus species is necessary to accurately estimate genetic parameters and improve breeding programs. Eucalyptus species often exhibit mixed mating systems, leading to complex relationships among progenies. Traditional tree breeding programs that assume a half-sibling relationship for open-pollinated (OP) trials may overestimate genetic gains by neglecting the effects of inbreeding and dominance. This study focuses on Eucalyptus pellita, a species with a mixed mating system, to quantify the impact of selfing and dominance on breeding strategies. We simulated OP trial growth data for 100 randomly selected families in a randomized complete block design, using published estimated parameters for diameter at breast height (DBH). Our analysis indicated that marker-based models, particularly those incorporating dominance effects, provide more accurate genetic parameter estimates and larger predicted genetic gains than pedigree-based models. These results reveal pronounced genetic and genotypic gain gaps when traditional models are employed, underscoring the imperative for integrating dominance-informed genomic selection strategies. Thus, our study provides essential guidance for optimizing breeding programs to sustainably enhance productivity and genetic quality in Eucalyptus plantations.
Similar content being viewed by others
Introduction
Eucalyptus is among the most widely planted genera in tropical and subtropical forestry, with multiple species cultivated for timber, pulp, and other wood products (Grattapaglia and Kirst 2008; Varghese et al. 2024). Eucalyptus pellita F. Muell. stands out as an important species in tropical silviculture due its high productivity, economic value, and adaptability. E. pellita is native to northern Australia and parts of Papua New Guinea, and it has been extensively planted in Southeast Asia, particularly in Indonesia and Malaysia (Lukmandaru et al. 2016). The species is prized for its fast growth, broad environmental adaptability, and natural resistance to pests and diseases (Fadwati et al. 2023).
These traits, coupled with wood properties suitable for pulp, paper, veneers, and sawn timber (Thavamanikumar et al. 2020), have led to E. pellita being widely adopted in industrial plantations. Notably, this species has been used to replace Acacia mangium in over half a million hectares of plantations in Indonesia and neighboring countries (Thavamanikumar et al. 2020) that were devastated by disease outbreaks (Eyles et al. 2008; Mendham et al. 2015; Nambiar et al. 2018; Tarigan et al. 2011), underscoring its commercial importance.
Besides its silvicultural importance, E. pellita was also chosen as the model species for this study because it exemplifies the mixed mating system and genetic characteristics typical of the genus Eucalyptus. Like most eucalypts, E. pellita is predominantly allogamous (outcrossing) but maintains a considerable rate of self-fertilization in natural populations (Potts and Savva 1988). Open-pollinated seed from eucalypt trees often includes a considerable proportion of selfed progeny, typically ranging from 5% to over 50%, depending on the species and population conditions (Eldridge et al. 1993; Griffin et al. 2019; House and Bell 1996; Quezada et al. 2022), resulting in inbreeding and complex pedigree relationships among seedlings within family. Furthermore, E. pellita exhibts quantitative genetic parameters in line with other commercial eucalypt species, such as moderate heritability for growth and wood quality traits (Brawner et al. 2010; Hung et al. 2015; Thavamanikumar et al. 2020) and pronounced inbreeding depression in fitness-related traits. These commonalities support the use of E. pellita as a representative model for the broader Eucalyptus genus in simulations of breeding strategies.
Historically, tree breeders have not accounted for all of the relevant sources of genetic variance and/or have ignored the assumptions that underpin the models used in forest tree improvement programs (Lebedev et al. 2020; Tambarussi et al. 2018; Tambarussi et al. 2022). As a result, inaccurate estimates may have limited the realized genetic gains in key tree species, especially when non-additive genetic effects and mating system complexity are not properly accounted for (Araújo et al. 2012; Bouvet et al. 2015; Costa e Silva et al. 2004). A clear understanding of a species’ mating system is essential to accurately estimate genetic parameters and their components and to quantify the expected gains from selection in a breeding program (Bush et al. 2015; Tambarussi et al. 2018; Tambarussi et al. 2022). The magnitude of genetic gain is partially dependent on the species mating system. Therefore, it should be considered when estimating genetic parameters, offering a clear understanding of the transmission of genetic information between generations (Bush et al. 2015; Namkoong 1966; Wright 1921).
Information on mating systems should be directly applied to breeding strategies, genetic conservation practices, and the production of improved seeds (Gusson et al. 2006). Mating systems can be allogamous, autogamous, or mixed (Goodwillie et al. 2005). Species of the genus Eucalyptus L’Hér. have a mixed mating system (Griffin et al. 2019; Hardner and Potts 1997; Kennington and James 1997; Sampson and Byrne 2008; Yeh et al. 1983). Under favorable conditions, i.e., flowering synchronicity and sufficient pollinators, most offspring are the result of panmictic biparental mating, where crosses between unrelated individuals predominate and there is a small proportion of offspring resulting from self-fertilization and mating between related parents.
This complex mating system influences the evolutionary patterns of species (Griffin et al. 2019; Hardner and Tibbits 1998; Sampson et al. 1995) and breeding strategies (Gonzaga et al. 2016; Griffin and Cotterill 1988). It also leads to different levels of relatedness between progenies within families, creating varying patterns of growth and survival (Burgess et al. 1996). Thus, the amount of additive variance is directly affected by natural patterns of inbreeding and biparental mating. As a result, phenotypic variation in Eucalyptus is not only influenced by additive effects, but also dominance and epistatic effects (Araújo et al. 2012; Costa e Silva et al., 2004; Tan et al. 2018). Failure to account for non-additive variance can inflate heritability, resulting in incorrect estimates of genetic gain (Wright Stephen et al., 2008).
A decline in the phenotypic performance of fitness-related quantitative traits is known as inbreeding depression (ID) (Byers and Waller 1999; Costa e Silva et al., 2010; Costa e Silva et al., 2011; Falconer and Mackay 1996; Lohr and Haag 2015). Inbreeding results in decreased genetic variability within progeny, whereas biparental inbreeding increases kinship between parents and progeny (Griffin and Eckert 2003; Ronfort and Couvet 1995). The spatial structure of natural populations also influences the likelihood of inbreeding, as individuals are more likely to mate with nearby genotypes than individuals in more distant populations (Epperson 1992; Tambarussi et al. 2017). This is typical for eucalypts, as seeds are dispersed over short distances and neighborhood relatedness and inbreeding are common (Jones et al. 2006; McDonald et al. 2003; Pupin et al. 2019). In nature, the intrapopulation spatial genetic structure resulting from restricted gene flow may explain the existence and maintenance of the mixed mating system (Tambarussi et al. 2017). Consequently, there is a decrease in cross-fertilization, and the genes for self-fertilization are transmitted without losing the adaptive value. Thus, Wright’s equilibrium may explain the maintenance of genetic variability in these populations (Coelho and Vencovsky 2003). In mixed systems, individuals with vigorous growth are present in some populations, even if they are inbred, and are similar to those found in panmictic populations in Hardy-Weinberg equilibrium (Coelho and Vencovsky 2003; Vencovsky and Crossa 1999).
Inbreeding patterns create different degrees of kinship between individuals, which may be higher than expected for half-siblings, leading to biased genetic estimates due to the species’ mating system (Ismail and Kokko 2019; Tambarussi et al. 2017; Vencovsky et al. 2001). Most tree breeding programs still use the pedigree-based matrix which assumes half-sib relationships between trees from open-pollinated trials. This assumption has an impact on the true value of genetic estimates (Tambarussi et al. 2022; Tambarussi et al. 2018). As the quantification of inbreeding and dominance effects continues to be neglected by breeders, it is necessary to take a closer look at this problem to improve the accuracy of genetic parameters (Tambarussi et al. 2022). The main objective of the present study is to provide a better understanding of the effects of selfing and dominance for Eucalyptus pellita F.Muell., a species with a mixed mating system (Eldridge et al. 1993). We performed simulations considering different levels of dominance and inbreeding to explore their impacts on breeding programs under different selection intensities. Subsequently, we sought to quantify the effect of dominance under different inbreeding scenarios.
Material and methods
Simulations
We simulated growth data for open-pollinated (OP) breeding populations for a randomized trial design with 20 complete-block replications of 100 families in single-tree plots (STP). Simulated parameters included trial mean (μ), and additive, dominance, plot, and phenotypic variances (\({\sigma }_{a}^{2}\), \({\sigma }_{d}^{2}\), \({\sigma }_{c}^{2}\), and \({\sigma }_{p}^{2}\), respectively). Simulations were based on several published diameter at breast height (DBH) datasets for Eucalyptus pellita evaluated at approximately four years after planting (Brawner et al. 2010; Hardiyanto 2003; Harwood et al. 1997; Leksono et al. 2006; Leksono et al. 2008; Leksono et al. 2009; Luo et al. 2006; Nieto et al. 2016; Thavamanikumar et al. 2020). For all simulations, we set all parameters (variances and overall mean) to the median of the analyzed studies for E. pellita four years after planting.
Simulations were carried out for the breeding population and next generation breeding population (progeny test) using the package AlphaSimR (Gaynor et al. 2021), in the R software environment, as outlined below.
Genetic structure of the breeding population
The first step in the simulation was to generate unrelated founder haplotypes for the breeding population (Bp). The method used was Markovian Coalescent Simulation (MaCS), which effectively simulates haplotypes under any population history model (Chen et al. 2008). Simulations of the OP population started with 1000 trees as founders in the breeding population and the quantitative trait under selection was DBH. We assumed 1000 segregating sites (loci) per chromosome, of which 600 were related to the simulated DBH trait. These loci were also assumed to be evenly distributed across 11 chromosomes (as found for all Eucalyptus species) (Myburg et al. 2014). Therefore, the whole-genome sequences for 11 chromosome pairs were constructed by randomly extracting 1000 biallelic single-nucleotide polymorphisms (SNP) as markers per chromosome and randomly assigning 600 SNP as QTL per chromosome. The genotypes were coded as 0 for reference (ancestral) homozygote, 1 for heterozygote, and 2 for alternative (derived) homozygote.
The breeding population for the OP population was assumed to be a breeding seed orchard consisting of 1000 superior trees selected for random mating, with the poorest performing trees removed. The strategy was chosen because it is widely used in eucalypt breeding programs.
Global model parameters for the simulations were set assuming that genetic control of DBH (g), can be decomposed into additive (a) and dominance (d) effects. To simplify the simulation and analysis, epistasis (e) was not simulated and interactions with the environment (s) (i.e., a x s, d x s, e x s) were assumed as zero. In summary, the parameter estimates used in the simulations (based on the literature, as described above) were as follows: μ = 10.5 0 ± 2 cm; \({\sigma }_{p}^{2}\) = 12 ± 2; \({\sigma }_{a}^{2}\) = 3 ± 0.01; \({\sigma }_{d}^{2}\) = from 1.2 to 2.4 ± 0.01. From these parameters, the narrow-sense heritability and the coefficient of determination for dominance (\({h}_{d}^{2}\)) were: \({h}_{a}^{2}\) = 0.25 ± 0.02 and \({h}_{d}^{2}\) = from 0.1 to 0.2 ± 0.01.
Breeding values (a), dominance effects (d), genotypic values (g), and phenotypic values (p)
The simulation of the additive (a) and dominance (d) effects follows the parametrization of classical quantitative trait models (Falconer and Mackay 1996). The a and d effects are defined as genotype dosage scaling with an individual’s raw genotype dosage as the number of copies of the ‘1’ allele at a locus (Gaynor et al. 2021).
The additive effect is sampled in two stages: (1) sampling initial values from a standard normal distribution (stochastic simulation); (2) scaling the magnitude of the initial values to achieve a desired genetic variance. This process began by first calculating the variance in the founder population (breeding population in this study) that accounted for both a and d effects, using the initial sampled effects. Then, a scaling constant was calculated and applied to all effects to achieve the target variance in the breeding population.
The individual breeding values (\({BV}(a)\)) were calculated as follows:
Where, \({a}_{q}\) is the additive effect of the q-th QTL and \({x}_{A}\) is the scaled additive genotype dosage (which scales the relative dosage to set the values for opposite homozygotes to −1 and 1), calculated as:
The individual dominance effects (\(D\left(d\right)\)) were calculated as follows:
Where, \({d}_{q}\) is the dominance effect of the q-th QTL and \({x}_{D}\) is the scaled dominance dosage (which scales the relative dosage to set the values for opposite homozygotes to 0 and the middlemost heterozygote to 1), calculated as:
The dominance effects were calculated as partial dominance. The dominance degree (δ) was assumed to be 0.2 (i.e., partial dominance) and d was calculated as:
In summary, the simulations of d were performed by scaling the magnitude of the sampled initial values for the supplied parameters. These supplied parameters are the mean and variance for a normal distribution used to sample the degrees of dominance and are then scaled as described for the additive effects to calculate the scaling constant. The scaling constant is then applied to d. Although this scaling changes the value of the dominance effect, the dominance degree remains unchanged. This makes the specification of the dominance degree distribution independent of the desired genetic variance.
Finally, the individual genotypic value (\({GV}(x)\)) for the simulated DBH trait was:
For more details of the a and d simulation, see Traits in AlphaSimR (r-project.org).
To simulate the phenotypic values (P), we simulated the environmental effects (E), calculated based on the broad-sense heritability (\({h}_{g}^{2}\)). The \({h}_{g}^{2}\) was calculated as the ratio between genotypic variance (\({\sigma }_{g}^{2}={\sigma }_{a}^{2}+{\sigma }_{d}^{2}\,\)) and phenotypic variance (\({\sigma }_{p}^{2}={\sigma }_{g}^{2}+{{\rm{environmental\; variance}}\,(\sigma }_{e}^{2})\); under the assumption that there is no interaction between genotype and environment, and that there are no other sources of variation in the phenotype), as follows:
The individual E was calculated as \(\sim N[\mu \left({given\; above}\right);{\sigma }_{e}^{2}]\). Therefore, P was calculated as:
Crossing simulations for the progeny test
The progeny test was simulated using the “gene drop” method, which is used to create new haplotypes from simulated haplotypes of the original founder population (breeding population in this study). Based on the whole-genome (distributions of QTL effects, and with sequence and SNP phased alleles and genotypes) and the pedigree of the breeding population generated by the MaCS method (described above), the new haplotypes were created by modeling genetic recombination during meiosis (Hickey and Gorjanc 2012).
A sample of haplotypes with sequence information for each chromosome according to the specified breeding population was then dropped through a pedigree with genetic recombination according to the gamma model (McPeek and Speed 1995). In AlphaSimR, a genetic map is constructed and used to model the distribution of genetic recombination. The crossover interference parameter for a gamma model of recombination is used with a value of v = 2.6 (Broman and Weber 2000), which approximates the degree of crossover interference implied by the Kosambi mapping function (Gaynor et al. 2021). Therefore, the random shuffling of genes created each individual’s genome in the simulated progeny test. In this process, Mendelian sampling variance is generated by the process of randomly sampling parental chromosomes during meiotic division at gametogenesis and estimated from the difference between the individual’s predicted transmission ability and its parent mean (Cole and VanRaden 2011).
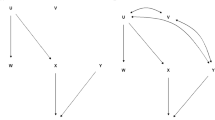
The process of creating a new population (progeny test) from the founders (breeding population) described above started with a given pedigree, i.e., a pair of crosses from the breeding population that will produce the offspring for the progeny test. The simulation for the OP population considered common issues that occur in improved seed orchards. Due to the timing of flowering (the probability of all trees in the improved seed orchard flowering at the same time is very low) and collection strategies, only a sample of trees become seed sources. In this case, we randomly selected a hypothetical set of 100/1000 trees to become seed sources and generated 20 progenies from each. From the genomic-based simulations, the relationship between progenies within families were half-sib (HS), full-sib (FS), self-half-sibs (SHS), and self-full-sibs (SFS) (Squillace 1974); however, in the pedigree-based model, all individuals within families are HS, as it assumes panmictic mating.
Therefore, the progeny test for OP populations has 2000 trees distributed across 100 families, four replications/blocks, and single-tree plots. During the crosses, we simulated self-crosses at the following selfing rates: 0, 5, 10, 25, 50, 75, and 95% and replicated each scenario 100 times, for a total of 700 simulations.
Inbreeding depression
We simulated self-crosses (selfing) for the OP population. In eucalypts, selfing is expected to cause a reduction in field growth compared to outcrossed trees (Hardner and Potts 1995), a phenomenon known as inbreeding depression (I). Inbreeding depression can be understood as the expected progressive decrease of the genotypic value as the inbreeding coefficient increases from zero to one (Wellmann and Bennewitz 2011). We simulated the expectation of inbreeding depression (E(I)) as follows (Wellmann and Bennewitz 2011):
Where, \({\mu }_{\delta }\) is the mean of the dominance effect; \({\sigma }_{a}\) is the additive genetic standard deviation; \({n}_{{QTN}}\) is the number of quantitative trait nucleotides (QTN; all QTN are biallelic and are assumed to have additive (a) and dominance (d) effects); \({p}_{i}\) is the frequency of allele 1 (the mutant allele) at the i-th locus; \({q}_{i}=1-{p}_{i}\) is the frequency of allele 0 (the ancestral allele) at the i-th locus; and \({\sigma }_{\delta }^{2}\) is the variance of the dominance effect. Therefore, the dominance effect was simulated as \(\sim N({\mu }_{\delta },\,{\sigma }_{\delta }^{2})\).
Relative phenotypic inbreeding depression due to self-pollination (ID) was calculated as (Hardner and Potts 1995):
Statistical models for genetic evaluation
To evaluate genetic parameters and predict breeding values, we applied a series of linear mixed models incorporating different sources of genetic information. Initially, we used pedigree-based models (ABLUP), which rely on the numerator relationship matrix (A) constructed from known pedigree records. These models estimate breeding values based on expected additive genetic relationships and are traditionally used in tree breeding programs.
We also employed genomic BLUP models, (GBLUP) models that use realized genomic relationship matrices (G) derived from SNP data. Two forms were tested:
-
An additive-only GBLUP, using the G matrix to model additive genetic effects
-
A genomic additive + dominance GBLUP (GDBLUP), which includes both additive (G) and dominance (D) genomic relationship matrices, thus capturing non-additive variance relevant for traits influenced by dominance.
To integrate all individuals, genotyped and non-genotyped, we used single-step GBLUP with dominance effect (ssGDBLUP). This model combines pedigree and genomic information in the H matrix and adds a dominance genomic matrix (D) to account for dominance deviations. In this model, the inverse of the H matrix was computed following (Legarra et al. 2009), and all models were fitted using ASReml-R. This modeling framework is especially relevant in open-pollinated populations where selfing and dominance effects can bias additive-only evaluations. Our approach aligns with findings in Eucalyptus pellita that emphasize the importance of dominance modeling for growth traits (Thavamanikumar et al. 2020).
Parameter estimates
We estimated the genetic parameters and genetic values (Breeding Values (BV) = additive effect; and Genotypic Values (GV) = additive + dominance effect) for the different simulated scenarios. First, analysis of variance using pedigree and marker-based models were performed to obtain the genetic and non-genetic estimates and to calculate the narrow-sense (\({h}_{a}^{2}\)) and broad-sense heritabilities (\({h}_{g}^{2}\)) for the different simulated scenarios (Table 1). The coefficient of dominance effect (\({h}_{d}^{2}\)) was also calculated.
The following general model was used for analysis based on REML/ABLUP and REML/GBLUP procedures:
Where, Y is the vector of individual observations; X is the known design matrix for fixed effects; b is the vector of fixed effects (overall mean and blocks). Zζ is the known design matrix for the additive genetic random effects; a is the vector of the additive genetic random effect with \(a \sim {N}(0,\,{\boldsymbol{\zeta }}{\sigma }_{a}^{2})\), where \({\sigma }_{a}^{2}\) is the additive genetic variance and \(\zeta\) is equal to:
-
i.
the additive genetic relationship matrix for pedigree-based models (HS and FAM models); the \({\sigma }_{a}^{2}\) in the FAM model was estimated as \(4{\sigma }_{p}^{2}\) (\({\sigma }_{p}^{2}\) is the variance between families), following the classical approach described by Falconer and Mackay (1996).
-
ii.
the marker-based additive genomic relationship matrix (VanRaden 2008) for marker-based models (GBLUP and GDBLUP models);
\({Z}_{{\boldsymbol{\tau }}}\) is the known design matrix of the dominance random effects; d is the dominance random effect vector with \({d} \sim {N}(0,\,{\boldsymbol{\tau }}{\sigma }_{d}^{2})\), where \({\sigma }_{d}^{2}\) is the dominance variance and τ is:
-
i.
the marker-based dominance genomic relationship matrix (Vitezica et al. 2013) for the marker-based model (GDBLUP model);
-
ii.
the combined pedigree and genomic-based dominance relationship matrix (HD matrix; Aliloo et al. 2017; Vitezica et al. 2013; Zhang et al. 2019) for the ssGDBLUP model.
e is the random residue vector with \({e} \sim {N}(0,{I}{\sigma }_{e}^{2})\), and \({\sigma }_{e}^{2}\) as the residual variance. The models that omitted dominance effects used the described model without the \({Z}_{\tau }d\) term. These models were applied to the data sets generated for each selfing rate.
The distribution of \({h}_{a}^{2}\) from the HS, GBLUP, GDBLUP, and ssGDBLUP models were compared with the true simulated \({h}_{a}^{2}\) for each selfing rate. We also performed a regression analysis for the simulated true breeding value as a function of the breeding value estimated from each model described and selfing rate.
The calculation of \({h}_{a}^{2}\), \({h}_{d}^{2}\), and \({h}_{g}^{2}\) were as follows:
Selective accuracy (raa)
The raa for breeding values were obtained as the correlation (“Pearson”) between the predicted BV from each model described in Table 1 and the true simulated BV.
Selection gain
We investigated the effect on selection gain for each progeny test structure and model class using the true simulated BV and GV as a baseline. Two different deployment strategies were applied: (1) SGBV – selection of trees for BV to become parents of the next generation of the breeding population; (2) SGGV – selection of trees for GV for cloning. Different levels of selfing rates and different selection intensities (SI) were considered.
For each deployment strategy, we calculated the “genetic gain gap” as the difference between selection based on each model described in Table 1 and the true BV and GV. To do so, the predicted breeding values of the best individual trees from each model (described in Table 1) were assumed to be indirectly selected and compared with the genetic gain from direct selection using the true BV and GV for each selfing rate. The following selection intensities (SI) were assumed: 1, 5, 10, 25, 50%. Table 2 describes the formulas used to calculate genetic gain for each deployment strategy.
Genetic and genotypic gain gap
The genetic gain gap quantifies the difference between the maximum achievable genetic gain (when selection is based on the true breeding values) and the gain achieved when individuals are ranked using breeding values (BV) or genotypic values (GV) estimated by different models (Table 1). For each selection intensity (SI%), the genetic gain gap was calculated as:
Where:
-
μ is the population mean.
-
\(\bar{B{V}_{{True},{SI} \% }}\) is the mean true breeding value of the top individuals selected based on their true BV at the given selection intensity.
-
\(\bar{B{V}_{{Model},{SI} \% }}\) is the mean true breeding value of individuals ranked using BV estimated by the models described in Table 1 (e.g., Half-Sib, GBLUP, GDBLUP, or ssGDBLUP).
The genotypic gain gap, which measures the difference in genotypic gains (additive + dominance effects), was similarly calculated as:
Where:
-
\(\bar{G{V}_{{True},{SI} \% }}\) is the mean true genotypic value of the top individuals selected based on their true GV.
-
\(\bar{G{V}_{{Model},{SI} \% }}\) is the mean true genotypic value of individuals ranked using BV (for HS model) or GV (GBLUP, GDBLUP and ssGDBLUP models) estimated by the models in Table 1.
The models used to estimate BV and GV (Half-Sib, GBLUP GDBLUP and ssGDBLUP) are detailed in Table 1, which provides a description on the kinship structures and deployment strategies for genetic and genotypic gains. By using the true BV or GV to calculate gains for both the True BV SI% and the Model and SI%, we ensure that the comparison reflects only the differences caused by ranking inaccuracies of the models.
Results
Genetic parameter estimates
In general, the pedigree-based models resulted in greater deviation from the true values of narrow-sense heritabilities (\({h}_{a}^{2}\)) (Fig. 1). It is important to note that by estimating the additive variance as \(4{\sigma }_{p}^{2}\), the Half-Sib (HS) and the Family (FAM) models have equal estimates of \({h}_{a}^{2}\). The median bias (HS/FAM model) was low up to a selfing rate of 10% and increased with greater inbreeding in the population (95% selfing rate), reaching more than 50% of median bias. Even in cases where the median bias was low, the pedigree-based models had more spread distribution around the true \({h}_{a}^{2}\) compared to the marker-based models.
Estimates are shown for the True \({h}_{a}^{2}\), Half-Sib/Family model, GBLUP model, GDBLUP model, and ssGDBLUP model (50% genotyped). Vertical dashed lines indicate median estimates; “±” values represent the standard deviation (SD), reflecting variability in \({h}_{a}^{2}\) under each selfing rate.
The ssGBLUP model, which considers that 50% of individuals are genotyped and accounts for the dominance effect (ssGDBLUP model), showed better performance with less bias than the pedigree-based model (Fig. 1). At a lower selfing rate (up to 50%), the ssGDBLUP model was intermediate between the pedigree and marker-based models to estimate \({h}_{a}^{2}\); however, at a higher selfing rate (greater than 50%) it was similar to the marker-based models, especially the model that does not consider the dominance effect (GBLUP model).
The marker-based model had a low standard deviation (2–5%) for all levels of selfing, and showed limited bias in estimates of dominance effect (\({h}_{d}^{2}\)) at low (up to 10%) and high (95%) selfing rates (Fig. 2). However, the bias was high at an intermediate selfing rate, with the highest bias at a 50% selfing rate. Despite high variability along the simulations, the ssGDBLUP model showed less bias than the marker-based model.
The marker-based models were more efficient than pedigree-based models in predicting the breeding values, showing higher accuracy regardless of selfing rate (Fig. 3). In the OP population, the pedigree-based model (Half-Sib) had stable accuracy across the selfing rate. The accuracy of the ssGDBLUP model was intermediate between pedigree and marker-based models (Fig. 3). The accuracy of ssGDBLUP increased with the selfing rate.
Selection gain
The highest genetic gain was achieved with the marker-based models at all levels of selfing and selection intensity (Fig. 4). In addition, the genetic gain was similar in the marker-based models with and without the dominance effect only at low (up to 10%) and high (95%) selfing rates. Smaller genetic gains were obtained when selection was based on the phenotypic value. The genotypic gain was also larger with the marker-based model, with selection based on phenotypic values showing the poorest results.
The ssGDBLUP model was intermediate between the marker-based and the pedigree-based models at all selection intensities for genetic gain (Fig. 4). For the genotypic gain, the ssGDBLUP model was similar to the pedigree-based model at high selfing rates (Fig. 5). The inferiority of the pedigree-based model and selection based on phenotypic values was clear and even stronger at lower selection intensities.
Genetic and genotypic gain gap
The genetic gain gap was greater with higher selection intensities and increased with an increase in selfing rate for all models (Fig. 6). The marker-based model, considering the dominance effect (GDBLUP), showed the lowest genetic gain gap among the selection models (Fig. 6). Nevertheless, it differed only slightly from the model that does not consider the dominance effect (GBLUP) at low selfing rates. The ssGDBLUP was intermediate between the marker-based and pedigree-based models, and selection based on phenotypic values resulted in a high genetic gain gap (Fig. 6).
Similar to the genetic gain gap, the genotypic gain gap was greater at higher selection intensities and increased with higher selfing rates. When taking the dominance effect into account, the marker-based model (GDBLUP model) resulted in a smaller genotypic gain gap compared to the other selection models (Fig. 7). The ssGDBLUP model showed a smaller genotypic gain gap than the pedigree-based and GBLUP model. Selection based on phenotypic value resulted in a greater genotypic gain gap at all selfing rates and selection intensities.
The average inbreeding depression across all simulations was 31% (supplementary material, Fig. S1). In summary, the mean DBH of crossed individuals was 10.5 cm, while the mean DBH of selfed individuals was 7 cm.
Discussion
Genetic parameter estimates
Compared to the pedigree-based model for open-pollinated populations, our results showed greater precision (lower standard deviation) and accuracy (lower bias) for heritability estimates using the marker-based model and the model that considers dominance effect. In fact, the marker-based model provided better estimates of variance and genetic gains because the markers correctly captured the relationship between individuals within the population (Powell et al. 2010; VanRaden 2008). Furthermore, with sufficient marker density, genomic models can also capture the variation in progeny due to Mendelian segregation of alleles at a locus during meiosis (Avendaño et al. 2005). In effect, the use of markers enabled us to estimate the realised, rather than the expected, proportion of the genome that dyads share identically by descent (IBD) (Visscher 2008).
Pedigree-based models do not take into account contemporary and historical kinship, which is usually unknown in the first generation of the breeding population. In open-pollinated populations with mixed mating systems, when breeders assume a half-sib structure, they neglect other types of relatedness, such as self-crosses and full-sibs, which undoubtedly exist in the population (Namkoong 1966; Tambarussi et al. 2018). Consequently, when applying the breeder’s equation (Lush 1937), the expectation of genetic gain from selection is overestimated as the selfing rate in the population with a mixed mating system increases. Even at low selfing rates, there is uncertainty about the true genetic transfer and therefore more noise in the selection of superior trees. Quezada et al. (2022) indicated the effectiveness of including selfing rate to reduce the upward bias of heritability estimates by between 7 and 30% in open-pollinated populations of Eucalyptus globulus.
As observed in this study, there is improved accuracy of the marker-based model when the dominance effect is included (Almeida Filho et al. 2016; Nadeau et al. 2023; Thavamanikumar et al. 2020). The models that consider non-additive genetic effects also predicted genetic values more accurately than models without non-additive genetic effects (Tan et al. 2018). When selfing or some level of inbreeding occurs in breeding populations, there is a partial transmission of non-additive genetic variance across generations (Charlesworth 2006; He et al. 2023). This is because there is a certain probability that the same combination of genotypes at multiple genetic locations can occur in the next generation (Lynch and Walsh 1998). Estimates of genetic parameters, such as heritability, typically assume random mating in OP populations (as defined by breeders to exclude selfing in the model), which is not the case in breeding populations with partial selfing. Finally, the prediction of genetic gain can be improved by considering non-additive effects resulting from selfing (He et al. 2023).
As the selfing rate increases from low to moderate levels, the dominance variance initially becomes more important in the response to selection in the population, while additive variance becomes less important (Heywood 2005; Kelly 1999). Indeed, the composition of genetic variation differs as a function of the mating system; outcrossing species have more heritable additive effects, whereas selfing species show greater variation in non-additive components (Clo et al. 2019; Clo et al. 2020). This suggests that the fraction of additive genetic variance is negatively correlated with the selfing rate (Clo et al. 2019), and the bias in genetic parameter estimates depends on how much information is available about the relationship between individuals. It is important to point out that dominance effects may contribute significantly to phenotypic variance for growth in Eucalyptus (Denis and Bouvet 2013; Klápště et al. 2017; Tan et al. 2017; Tan et al. 2018) and other forest species (Almeida Filho et al. 2016; Shalizi and Isik 2019). Therefore, for species with a mixed mating system, such as E. pellita, considering the open-pollinated population under study as half-sibs or controlled-pollinated, without properly considering the dominance effect, leads to errors in the expectation of genetic gains from selection.
The marker-based model that accounts for dominance effect was highly efficient in estimating the dominance effect at low (0 to 10%) and high (95%) selfing rates. However, the results showed bias at intermediate selfing rates (25% - 75%), especially 50%. At this level, the distribution of homozygosity becomes bimodal, as observed in our simulations (see Supplementary Fig. S2), with one group of individuals exhibiting high homozygosity (from selfing) and another group showing lower homozygosity (from outcrossing). This mixture of selfed and outcrossed individuals leads to a distortion in the population’s genetic structure, which can interfere with the accurate estimation of genetic variance components. The increase in homozygosity can mask dominance effects because dominance only manifests in heterozygous conditions (Billiard et al. 2021; Xu 2022). At very high selfing rates (75% and 95%), nearly all individuals become homozygous, reducing the role of dominance variance. Consequently, the model might more accurately estimate the remaining additive genetic effects, thus reducing the bias seen at 50% selfing, where heterozygosity (and thus dominance interactions) is more prevalent. The bias observed at 50% selfing can be attributed to complex interactions between additive and dominance effects. At this level of selfing, the population has a mix of selfing (Higher homozygosity) and non-selfing (Lower homozygosity) homozygous individuals, which affects the model’s ability to distinguish between additive and dominance effects.
The complex interplay between additive and dominance genetic variances, as well as their covariance, has been demonstrated in other species, underscoring the variability and challenges in dominance effect estimation across different models and populations (Fernández et al. 2017). Indeed, the application of genome-wide SNP information enhances the accuracy of genetic variance estimates by effectively separating genetic and non-genetic effects, reducing bias compared to pedigree-based methods. This approach proves particularly beneficial for traits characterized by significant dominance variance (Lee et al. 2010). In tree breeding, marker-based models have shown a more precise separation of additive and non-additive genetic variance components, for example, improving breeding value prediction for tree height in Pinus taeda (Muñoz et al. 2014). Therefore, our findings further indicate that GDBLUP models are more effective in estimating additive and dominance effects. However, we suggest that when the selfing rate is intermediate (around 50%), it is advisable to incorporate pedigree information with GDBLUP to improve accuracy. In other scenarios, GDBLUP is strongly recommended for use in open-pollinated populations such as E. pellita, as it effectively estimates genetic variance.
The ssGDBLUP model was intermediate between the marker-based and pedigree-based models. This highlights the fact that as more information is used to estimate the genetic parameters, the more effective it will be. It also indicates that despite full genomic information being the best scenario, the ssGDBLUP with 50% of individuals genotyped can be a cost-effective strategy to estimate genetic parameters and select individuals either to become parents of the next generation of the breeding population or deployed as clones in a Eucalyptus breeding program.
Selection gain
Marker-based models provide greater accuracy than pedigree-based models even in a single generation and this may increase over multiple generations (Grattapaglia 2022; Jurcic et al. 2023). The results simulated here corroborate previously reported empirical studies indicating that neglecting the kinship relationships between individuals leads to an overestimation of genetic variance (Beaulieu et al. 2022; Bush et al. 2015; Bush et al. 2011; Klápště et al. 2017; Nadeau et al. 2023; Tambarussi et al. 2018). Consequently, this can result in erroneous expectations of selection gains in tree breeding.
In tree breeding programs, the choice of appropriate breeding models and selection methods is crucial to achieve maximum genetic gains and improve desirable traits in the target species. The results obtained from the analysis of open-pollinated (OP) E. pellita populations provide valuable insights into the implications of different breeding models on genetic and genotypic gain. Our results suggest that incorporating molecular marker information into breeding models can enhance the efficiency of selection and improve genetic and genotypic gain. These results align with previous studies that have demonstrated the advantages of genomic selection in tree breeding programs (Beaulieu et al. 2022; El-Kassaby et al. 2024; Grattapaglia 2022; Klápště et al. 2022; Sharma et al. 2023). Furthermore, the inclusion of dominance effects in the marker-based models significantly increased genetic gain. Thus, considering dominance effects is important to capture the full potential of selection, especially when selfing occurs in the population. The positive impact of incorporating dominance effects in breeding models has also been reported for other tree species (Calleja-Rodriguez et al. 2021; Muñoz et al. 2014; Tan et al. 2018; Tan and Ingvarsson 2022; Thumma et al. 2022).
Our analysis indicates that selection based on phenotypic values alone results in smaller genetic gains compared to marker-based and pedigree-based models. This highlights the limitations of relying solely on phenotypic information for selection in tree breeding programs. Phenotypic selection can be influenced by environmental factors and may not accurately reflect the underlying genetic potential. Therefore, integrating marker information into breeding models provides a more reliable and efficient approach to genetic improvement.
In terms of genotypic gain, the model with half of individuals genotyped (ssGDBLUP models) showed intermediate efficiency between non-genotyped individuals (Half-Sib model) and fully genotyped individuals (GDBLUP model), especially at higher selection intensities. This suggests that although fully genotyping all individuals is desirable to maximize genotypic gain, genotyping a proportion of individuals can still yield comparable results in terms of genetic improvement and is preferable to no genotyping. This finding is consistent with studies emphasizing the cost-effectiveness and practicality of genotyping a proportion of individuals in tree breeding programs (Ratcliffe et al. 2017; Thavamanikumar et al. 2020).
Genetic and genotypic gain gap and implications for Eucalyptus breeding program
The “gap” we refer to is computed based on these true values and serves solely as a benchmark to evaluate how accurately different models (e.g., pedigree-based, genomic-based, and combined) recover the simulated genetic signal under varying conditions (e.g., selfing rates, marker density). The results obtained from the analysis of open-pollinated Eucalyptus pellita populations provide important insights for eucalypt breeding programs, particularly in terms of genetic gain and deployment strategies for the next generation of breeding and cloning. Our findings demonstrate that the genetic and genotypic gain gap—representing the difference between the true genetic and genotypic gain (respectively) and those estimated using different breeding models—increases with higher selection intensities. This suggests that selection intensity plays a crucial role in the ability of breeding models to achieve genetic and genotypic gains. In practice, lower selection intensities should be used if the breeding program relies solely on phenotypic or pedigree-based information. Conversely, selection intensities can be increased when additional information, such as genomics, are available.
Marker-based models, particularly models that consider dominance effect (GDBLUP), consistently outperformed the pedigree-based model in terms of reducing the genetic and genotypic gain gap. This highlights the superiority of marker-based models that incorporate molecular marker information over models that only consider pedigree relationships (Nadeau et al. 2023; Varshney et al. 2017). Marker-based models have been shown to enhance selection accuracy and increase genetic gains for various tree species, including Eucalyptus (Anders et al. 2023; Grattapaglia 2022; Harfouche et al. 2012; Klápště et al. 2020; Walker et al. 2022). The results found in the present study suggest that the use of ssGBLUP models in tree breeding programs, which combine pedigree and genomic data, provide an intermediate level of accuracy between marker-based and pedigree-based models. While ssGBLUP models offer improved accuracy over pedigree-based models, they are generally less accurate than marker-based models. This is supported by studies demonstrating the effectiveness of ssGBLUP in tree breeding programs such as C. africana, where ssGBLUP improved the accuracy of predicted breeding values but with less investment and effort required for genotyping (Ousmael et al. 2024).
For deployment in the next generation of breeding, the results suggest that marker-based models, especially when considering the dominance effect, offer significant advantages in terms of reducing the genetic gain gap and maximizing genetic improvement. The results demonstrate that the use of certain breeding models, such as the half-sib model in the OP population, can lead to significant genetic gain gaps, with clear consequences for forest companies. A larger genetic gain gap implies missed opportunities for selecting and deploying superior genotypes, leading to reduced productivity and financial losses. Meanwhile, the use of suboptimal genotypes results in lower-quality trees, slower growth rates, and decreased overall productivity (Fischer et al. 2017). Moreover, the inefficient allocation of resources, including land, labor, and capital, to these suboptimal genotypes further contributes to lost revenue.
Our study has implications not only for genetic gain and productivity, but also for eucalypt clone selection and deployment. The use of genomic models and a consideration of dominance effects in Eucalyptus breeding programs improves selection accuracy and increases genotypic gain (Denis and Bouvet 2013). In fact, cloning allows breeders to explore and multiply both additive and non-additive effects in their breeding strategy (Falconer and Mackay 1996). In the context of clone deployment, this advantage becomes even more significant when clones with superior traits can be identified using marker-based models, allowing the best performing clones to be selected for deployment.
The results of this study suggest that the dominance effect plays a crucial role in reducing the genotypic gain gap. Specifically, the GDBLUP model, which includes dominance effects, and the ssGDBLUP model, which combines pedigree and genomic data, are more efficient as the selfing rate increases. These models accurately capture both additive and dominance variances, providing improved prediction accuracy and reducing the genotypic gain gap. This efficiency is particularly pronounced at higher selfing rates where the genetic structure is more complex (Vencovsky et al. 2001). Conversely, selection based on phenotypic values results in the highest genotypic gain gap across all selection intensities and selfing rates. Therefore, breeders should avoid this method when selecting trees for cloning, as it leads to less accurate genetic improvement.
The forest industry relies heavily on the production of uniform and high-quality wood to meet the demands of the pulp and paper sector (Liu et al. 2025; Rezende et al. 2013; Wu 2019). By selecting and deploying eucalypt clones, forest companies can enhance wood productivity, improve fiber quality, and achieve greater operational efficiency (Wu 2019).
Reducing the genotypic gain gap is critical because it allows breeders to develop clones based on predicted genotypic values that are similar to the true genotypic value, and account for the significant influence of the dominance effect. By minimizing the genotypic gain gap, breeders can confidently select and propagate clones with superior genetic traits, ensuring the consistent and predictable performance of these individuals in commercial plantations. The similarity between estimated and true genotypic values leads to improved selection efficiency and increased genetic improvement. Meanwhile, the precise selection and deployment of clones contributes to the development of high performing and genetically superior eucalypt stands, ultimately resulting in greater productivity and profitability for the forestry sector.
Conclusion
This study highlights the impact of incorporating inbreeding and dominance effects into genetic parameter estimates for Eucalyptus pellita, a representative species with a mixed mating system. Our simulations clearly demonstrate that marker-based genomic models, particularly those integrating dominance effects (GDBLUP and ssGDBLUP), significantly enhance the accuracy of genetic predictions and markedly reduce genetic and genotypic gain gaps compared to traditional pedigree-based models. Conventional approaches that assume half-sib relationships substantially overestimate genetic gains by neglecting critical dominance and inbreeding variances, resulting in missed opportunities for optimal selection and deployment of superior genotypes. Consequently, adopting dominance-informed genomic selection is essential for breeders aiming to fully exploit genetic variation, improve selection efficiency, and achieve sustainable genetic improvement. This approach not only facilitates better management of genetic resources but also drives the development of genetically superior and highly productive Eucalyptus plantations, ensuring long-term sustainability and enhanced productivity in forestry operations.
Data availability
Data supporting the findings of this study have been deposited in the Dryad Digital Repository: https://doi.org/10.5061/dryad.nzs7h4544. The archived files include .qs datasets generated from the simulations of breeding population and progeny test scenarios across all selfing rates described in the manuscript.
Code availability
All R scripts used in this study, including the main custom simulation function Tree_breeding_simulator.R—developed specifically for generating the simulated open-pollinated breeding populations and results described here—are publicly available at the GitHub repository: https://github.com/AraujoMJ/Eucalyptus_Gain_Gap_Simulation
References
Aliloo H, Pryce JE, González-Recio O, Cocks BG, Goddard ME, Hayes BJ (2017) Including nonadditive genetic effects in mating programs to maximize dairy farm profitability. J Dairy Sci 100:1203–1222
Almeida Filho JE, Guimarães JFR, Silva FF, Resende MDV, Muñoz P, Kirst M et al. (2016) The contribution of dominance to phenotype prediction in a pine breeding and simulated population. Heredity 117:33–41
Anders C, Hoengenaert L, Boerjan W (2023) Accelerating wood domestication in forest trees through genome editing: advances and prospects. Curr Opin Plant Biol 71:102329
Araújo JM, Borralho N, Dehon G (2012) The importance and type of non-additive genetic effects for growth in Eucalyptus globulus. Tree Genet Genomes 8:327–337
Avendaño S, Woolliams JA, Villanueva B (2005) Prediction of accuracy of estimated Mendelian sampling terms. J Anim Breed Genet 122:302–308
Beaulieu J, Lenz P, Bousquet J (2022) Metadata analysis indicates biased estimation of genetic parameters and gains using conventional pedigree information instead of genomic-based approaches in tree breeding. Sci Rep 12:1–10
Billiard S, Castric V, Llaurens V (2021) The integrative biology of genetic dominance. Biol Rev 96:2925–2942
Bouvet J-M, Makouanzi G, Cros D, Vigneron PH (2015) Modeling additive and non-additive effects in a hybrid population using genome-wide genotyping: prediction accuracy implications. Heredity 116:146–157
Brawner JT, Bush DJ, Macdonell PF, Warburton PM, Clegg PA (2010) Genetic parameters of red mahogany breeding populations grown in the tropics. Aust Forestry 73:177–183
Broman KW, Weber JL (2000) Characterization of human crossover interference. Am J Hum Genet 66:1911–1926
Burgess IP, Williams ER, Bell JC, Harwood CE, Owen JV (1996) The effect of outcrossing rate on the growth of selected families of Eucalyptus grandis. Handlenet 45:2–3
Bush D, Kain D, Matheson C, Kanowski P (2011) Marker-based adjustment of the additive relationship matrix for estimation of genetic parameters—an example using Eucalyptus cladocalyx. Tree Genet Genomes 7:23–35
Bush DR, Kain D, Kanowski P, Matheson C (2015) Genetic parameter estimates informed by a marker-based pedigree: a case study with Eucalyptus cladocalyx in southern Australia. Tree Genet Genomes 11:1–14
Byers DL, Waller DM (1999) Do plant populations purge their genetic load? effects of population size and mating history on inbreeding depression. Annu Rev Ecol Syst 30:479–513
Calleja-Rodriguez A, Chen Z, Suontama M, Pan J, Wu HX (2021) Genomic predictions with nonadditive effects improved estimates of additive effects and predictions of total genetic values in pinus sylvestris. Front Plant Sci 12:666820
Charlesworth D (2006) Evolution of plant breeding systems. Curr Biol 16:R726–R735
Chen GK, Marjoram P, Wall JD (2008) Fast and flexible simulation of DNA sequence data. Genome Res 19:136–142
Clo J, Gay L, Ronfort J (2019) How does selfing affect the genetic variance of quantitative traits? An updated meta‐analysis on empirical results in angiosperm species. Evolution 73:1578–1590
Clo J, Ronfort J, Abu Awad D (2020) Hidden genetic variance contributes to increase the short‐term adaptive potential of selfing populations. J Evolut Biol 33:1203–1215
Coelho ASG, Vencovsky R (2003) Intrapopulation fixation index dynamics in finite populations with variable outcrossing rates. Sci Agric 60:305–313
Cole JB, VanRaden PM (2011) Use of haplotypes to estimate Mendelian sampling effects and selection limits. J Anim Breed Genet 128:446–455
Costa e Silva J, Borralho NMG, Potts BM (2004) Additive and non-additive genetic parameters from clonally replicated and seedling progenies of Eucalyptus globulus. Theor Appl Genet 108:1113–1119
Costa e Silva J, Hardner CM, Tilyard P, Potts BM (2011) The effects of age and environment on the expression of inbreeding depression in Eucalyptus globulus. Heredity 107:50–60
Costa e Silva J, Hardner C, Tilyard P, Pires AM, Potts BM (2010) Effects of inbreeding on population mean performance and observational variances in Eucalyptus globulus. Ann For Sci 67:605
Denis M, Bouvet J-M (2013) Efficiency of genomic selection with models including dominance effect in the context of Eucalyptus breeding. Tree Genet Genomes 9:37–51
Eldridge K, Davidson J, Harwood C, Van Wyk G (1993). Eucalypt domestication and breeding. Clarendon: London
El-Kassaby YA, Cappa EP, Chen C, Ratcliffe B, Porth IM (2024) Efficient genomics-based ‘end-to-end’ selective tree breeding framework. Heredity 132:98–105
Epperson BK (1992). Spatial structure of genetic variation within populations of forest trees. Population Genetics of Forest Trees 6:257–278.
Eyles A, Beadle C, Barry K, Francis A, Glen M, Mohammed C (2008) Management of fungal root-rot pathogens in tropical Acacia mangium plantations. Blackwell Verl 38:332–355
Fadwati AD, Hidayati F, Na’iem M (2023). Evaluation of Genetic Parameters of Growth Characteristics and Basic Density of Eucalyptus pellita Clones Planted at Two Different Sites in East Kalimantan, Indonesia. J Korean Wood Sci Technol 51:222–237.
Falconer DS, Mackay TC (1996) Introduction to quantitative genetics, 4th edn. Longman Scientific and Technical
Fernández EN, Legarra A, Martínez R, Sánchez JP, Baselga M (2017) Pedigree‐based estimation of covariance between dominance deviations and additive genetic effects in closed rabbit lines considering inbreeding and using a computationally simpler equivalent model. J Anim Breed Genet 134:184–195
Fischer DG, Wimp GM, Hersch‐Green E, Bangert RK, LeRoy CJ, Bailey JK et al. (2017) Tree genetics strongly affect forest productivity, but intraspecific diversity–productivity relationships do not (J Koricheva, Ed.). Funct Ecol 31:520–529
Gaynor RC, Gorjanc G, Hickey JM (2021) AlphaSimR: an R package for breeding program simulations (D-J de Koning, Ed.). G3 Genes|Genomes|Genetics 11
Gonzaga S, Manoel RO, Barbosa C, Pereira A, Luiz M, Luiz M et al. (2016) Pollen contamination and nonrandom mating in a Eucalyptus camaldulensis Dehnh seedling seed orchard. Silvae Genet 65:1–11
Goodwillie C, Kalisz S, Eckert CG (2005) The evolutionary enigma of mixed mating systems in plants: occurrence, theoretical explanations, and empirical evidence. Annu Rev Ecol Evol Syst 36:47–79
Grattapaglia D (2022) Twelve years into genomic selection in forest trees: climbing the slope of enlightenment of marker assisted tree breeding. Forests 13:1554
Grattapaglia D, Kirst M (2008) Eucalyptusapplied genomics: from gene sequences to breeding tools. N. Phytol 179:911–929
Griffin AR, Cotterill PP (1988) Genetic variation in growth of outcrossed, selfed and open-pollinated progenies of Eucalyptus regnans and some implications for breeding strategy. Silvae Genet 37:124–131
Griffin AR, Potts BM, Vaillancourt RE, Bell JC (2019) Life cycle expression of inbreeding depression in Eucalyptus regnans and inter-generational stability of its mixed mating system. Ann Bot 124:179–187
Griffin CAM, Eckert CG (2003) Experimental analysis of biparental inbreeding in a self-fertilizing plant. Evolution 57:1513–1519
Gusson E, Sebbenn AM, Kageyama PY (2006) Sistema de reprodução em populações de Eschweilera ovata (Cambess.) Miers. Rev Árvore 30:491–502
Hardiyanto EB (2003) Growth and genetic improvement of Eucalyptus pellita in South Sumatra, Indonesia. In: ACIAR PROCEEDINGS, Turnbull, J W: Zhanjiang, Guangdong, People’s Republic of China, pp. 82–88
Hardner C, Tibbits W (1998) Inbreeding depression for growth, wood and fecundity traits in Eucalyptus nitens. For Genet 5:11–20
Hardner CM, Potts B (1995) Inbreeding depression and changes in variation after selfing in Eucalyptus globulus ssp. Silvae Genet 14:46–54
Hardner CM, Potts BM (1997) Post dispersal selection following mixed mating in Eucalyptus regnans. Evolution 51:103–111
Harfouche A, Meilan R, Kirst M, Morgante M, Boerjan W, Sabatti M et al. (2012) Accelerating the domestication of forest trees in a changing world. Trends Plant Sci 17:64–72
Harwood CE, Alloysius D, Pomroy P, Robson KW, Haines NW (1997) Early growth and survival of Eucalyptus pellita provenances in a range of tropical environments, compared with E. grandis, E. urophylla and Acacia mangium. N. For 14:203–219
He Z-H, Xiao YU, Yan-Wen LV, Yeh FC, Wang X, Hu X-S (2023) Prediction of Genetic Gains from Selection in Tree Breeding. Forests 14:520–520
Heywood JS (2005) An exact form of the breeder’s equation for the evolution of a quantitative trait under natural selection. Evolution 59:2287–2298
Hickey JM, Gorjanc G (2012) Simulated data for genomic selection and genome-wide association studies using a combination of coalescent and gene drop methods. G3 Genes|Genomes|Genet 2:425–427
House APN, Bell JC (1996) Genetic diversity, mating system and systematic relationships in two red mahoganies, Eucalyptus pellita and E. scias. Aust J Bot 44:157
Hung TM, Brawner JT, Meder R, Lee DJ, Southerton SG, Thinh HH et al. (2015) Estimates of genetic parameters for growth and wood properties in Eucalyptus pellita F. Muell. to support tree breeding in Vietnam. Ann For Sci Vol 72:205–217
Ismail SA, Kokko H (2019) An analysis of mating biases in trees. Mol Ecol 29:184–198
Jones TH, Vaillancourt RE, Potts BM (2006) Detection and visualization of spatial genetic structure in continuous Eucalyptus globulus forest. Mol Ecol 16:697–707
Jurcic EJ, Dutour J, Villalba PV, Cantet R, Centurión C, Munilla S et al. (2023) Genomic-enhanced Markov causal models for predicting breeding values in forest trees. Agrociencia Urug 27:e1264
Kelly JK (1999) Response to selection in partially self-fertilizing populations. I. Selection on a single trait. Evolution 53:336–349
Kennington WJ, James SH (1997) Contrasting patterns of clonality in two closely related mallee species from Western Australia, Eucalyptus argutifolia and E. obtusiflora (Myrtaceae). Aust J Bot 45:679
Klápště J, Suontama M, Telfer E, Graham N, Low C, Stovold T et al. (2017) Exploration of genetic architecture through sib-ship reconstruction in advanced breeding population of Eucalyptus nitens. PloS one 12:e0185137
Klápště J, Dungey HS, Telfer EJ, Suontama M, Graham NJ, Li Y et al. (2020) Marker selection in multivariate genomic prediction improves accuracy of low heritability traits. Front Genet 11:499094
Klápště J, Ismael A, Paget M, Graham NJ, Stovold GT, Dungey HS et al. (2022) Genomics-enabled management of genetic resources in radiata pine. Forests 13:282–282
Lebedev VT, Lebedeva TN, Chernodubov A, Shestibratov KA (2020) Genomic selection for forest tree improvement: methods, achievements and perspectives. Forests 11:1190
Lee SH, Goddard ME, Visscher PM, van der Werf JH (2010) Using the realized relationship matrix to disentangle confounding factors for the estimation of genetic variance components of complex traits. Genet Select Evol 42:1–14
Legarra A, Aguilar I, Misztal I (2009) A relationship matrix including full pedigree and genomic information. J Dairy Sci 92:4656–4663
Leksono B, Kurinobu S, Ide Y (2006) Optimum age for selection based on a time trend of genetic parameters related to diameter growth in seedling seed orchards of Eucalyptus pellita in Indonesia. J For Res 11:359–364
Leksono B, Kurinobu S, Ide Y (2008) Realized genetic gains observed in second generation seedling seed orchards ofEucalyptus pellitain Indonesia. J For Res 13:110–116
Leksono B, Kurinobu S, Ide Y (2009) An optimum design for seedling seed orchards to maximize genetic gain: an investigation on seedling seed orchards of Eucalyptus pellita F. Muell. Indones J Forestry Res 6:85–95
Liu N, Van den Bulcke J, Van Acker J, Liu F, Gao C, Yu J et al. (2025) Enhancing large-diameter timber production: evaluating poplars by genotype and spacing. Ind Crops Products 223:120148
Lohr JN, Haag CR (2015) Genetic load, inbreeding depression, and hybrid vigor covary with population size: an empirical evaluation of theoretical predictions. Evolution 69:3109–3122
Lukmandaru G, Zumaini UF, Soeprijadi D, Nugroho WD, Susanto M (2016) Chemical properties and fiber dimension of Eucalyptus pellita from the 2nd generation of progeny tests in Pelaihari, South Borneo, Indonesia. J Korean Wood Sci Technol 44:571–588
Luo J, Arnold RJ, Aken K (2006) Genetic variation in growth and typhoon resistance in Eucalyptus pellita in south-western China. Aust Forestry 69:38–47
Lush JL (1937) Animal Breeding Plans, 1st edn. Iowa State Pr
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer Associates, Sunderland, Mass
McDonald MW, Rawlings M, Butcher PA, Bell JC (2003) Regional divergence and inbreeding in Eucalyptus cladocalyx (Myrtaceae). Aust J Bot 51:393
McPeek MS, Speed TP (1995) Modeling interference in genetic recombination. Genetics 139:1031–1044
Mendham D, Rimbawanto WA, Mohammed C, Glen M, Hardie M, Beadle C et al. (2015) Final report Increasing productivity and profitability of Indonesian smallholder plantations. ACIAR, Canberra ACT 2601 Australia
Muñoz PA, Deon M, Gezan SA, Deon M, de los Campos G, Kirst M et al. (2014) Unraveling additive from nonadditive effects using genomic relationship matrices. Genetics 198:1759–1768
Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J et al. (2014) The genome of Eucalyptus grandis. Nature 510:356–362
Nadeau S, Beaulieu J, Gezan SA, Perron M, Bousquet J, Patrick (2023) Increasing genomic prediction accuracy for unphenotyped full-sib families by modeling additive and dominance effects with large datasets in white spruce. Front Plant Sci 14:1137834
Nambiar S, Harwood C, Mendham D (2018) Paths to sustainable wood supply to the pulp and paper industry in Indonesia after diseases have forced a change of species from acacia to eucalypts. Aust Forestry 81:148–161
Namkoong G (1966) Inbreeding effects on estimation of genetic additive variance. For Sci 12:8–13
Nieto V, Giraldo-Charria D, Sarmiento M, Borralho N (2016) Effects of provenance and genetic variation on the growth and stem formation of Eucalyptus pellita in Colombia. J Trop For Sci 28:227–234
Ousmael KM, Cappa EP, Hansen JK, Hendre P, Hansen OK (2024) Genomic evaluation for breeding and genetic management in Cordia africana, a multipurpose tropical tree species. BMC Genom 25:1–16
Potts B, Savva M (1988) Self-incompatibility in Eucalyptus. In: Proceedings of the Pollination ’88 Symposium, School of Botany, University of Melbourne: Parkville, Victoria Vol 14, pp. 165–170
Powell JE, Visscher PM, Goddard ME (2010) Reconciling the analysis of IBD and IBS in complex trait studies. Nat Rev Genet 11:800–805
Pupin S, Sebbenn AM, Cambuim J, Silva AM, Zaruma DUG, Silva PHM et al. (2019) Effects of pollen contamination and non-random mating on inbreeding and outbreeding depression in a seedling seed orchard of Eucalyptus urophylla. For Ecol Manag 437:272–281
Quezada M, Aguilar I, Balmelli G (2022). Genomic breeding values’ prediction including populational selfing rate in an open-pollinated Eucalyptus globulus breeding population. Tree Genet Genomes 18:1–15
Ratcliffe B, El-Dien OG, Cappa EP, Porth I, Klápště J, Chen C et al. (2017) Single-step BLUP with varying genotyping effort in open-pollinated Picea glauca. G3 Genes|Genomes|Genet 7:935–942
Rezende GDSP, Resende MDV, Assis TF (2013). Eucalyptus Breeding for Clonal Forestry. In: Fenning T (ed) Challenges and opportunities for the World’s Forests in the 21st Century vol 81, Springer, Dordrecht, pp. 393–424
Ronfort J, Couvet D (1995) A stochastic model of selection on selfing rates in structured populations. Genet Res 65:209–222
Sampson JF, Byrne M (2008) Outcrossing between an agroforestry plantation and remnant native populations of Eucalyptus loxophleba. Mol Ecol 17:2769–2781
Sampson JF, Hopper SD, James SH (1995) The mating system and genetic diversity of the Australian Arid Zone Mallee, Eucalyptus rameliana. Aust J Bot 43:461
Shalizi MN, Isik F (2019) Genetic parameter estimates and GxE interaction in a large cloned population of Pinus taeda L. Tree Genet Genomes 15:1–13
Sharma U, Sankhyan HP, Kumari A, Thakur S, Thakur L, Mehta D et al. (2023) Genomic selection: a revolutionary approach for forest tree improvement in the wake of climate change. Euphytica 220:1–25
Squillace AE (1974) Average genetic correlations among offspring from open-pollinated forest trees. Silvae Genet 23:149–156
Tambarussi EV, Maria G, Engel M, Roque RH (2022) Estimation of the mating system of Eucalyptus benthamii Maiden at Cambage progeny. Rev do Inst Florest 34:163–171
Tambarussi EV, Boshier D, Vencovsky R, Luiz M, Sebbenn AM (2017) Inbreeding depression from selfing and mating between relatives in the Neotropical tree Cariniana legalis Mart. Kuntze. Conserv Genet 18:225–234
Tambarussi EV, Pereira FB, Silva PHM, Lee D, Bush D (2018) Are tree breeders properly predicting genetic gain? A case study involving Corymbia species. Euphytica 214:1–11
Tan B, Ingvarsson PK (2022) Integrating genome‐wide association mapping of additive and dominance genetic effects to improve genomic prediction accuracy in Eucalyptus. Plant Genome 15:e20208
Tan B, Grattapaglia D, Wu HX, Ingvarsson PK (2018) Genomic relationships reveal significant dominance effects for growth in hybrid Eucalyptus. Plant Sci 267:84–93
Tan B, Grattapaglia D, Martins GS, Ferreira KZ, Sundberg B, Ingvarsson PK (2017) Evaluating the accuracy of genomic prediction of growth and wood traits in two Eucalyptus species and their F1 hybrids. BMC Plant Biol 17:1–15
Tarigan M, Roux J, Van Wyk M, Tjahjono B, Wingfield MJ (2011) A new wilt and die-back disease of Acacia mangium associated with Ceratocystis manginecans and C. acaciivora sp. nov. in Indonesia. South Afr J Bot 77:292–304
Thavamanikumar S, Arnold RJ, Luo J, Thumma BR (2020) Genomic studies reveal substantial dominant effects and improved genomic predictions in an open-pollinated breeding population of Eucalyptus pellita. G3 Genes Genomes Genet 10:3751–3763
Thumma BR, Joyce K, Jacobs A (2022) Genomic studies with preselected markers reveal dominance effects influencing growth traits in Eucalyptus nitens. G3 Genes Genomes Genet 12:jkab363
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Varghese M, Kamalakannan R, Suraj PG (2024) Eucalyptus: taxonomy, geographic distribution, domestication, breeding, ecology and economic importance. Sustain Dev Biodivers 37:81–128
Varshney RK, Roorkiwal M, Sorrells ME (2017) Genomic selection for crop improvement: an introduction. In: Varshney RK, Roorkiwal M, Sorrells ME (eds) Springer eBooks, Springer Nature
Vencovsky R, Crossa J (1999) Variance effective population size under mixed self and random mating with applications to genetic conservation of species. Crop Sci 39:1282–1294
Vencovsky R, Pereira MB, Crisostomo JR, Ferreira MAJF (2001) Genética e melhoramento de populações mistas. In: Recursos genéticos e melhoramento - plantas, Fundação MT: Rondonópolis, pp. 231–282
Visscher PM (2008) Whole genome approaches to quantitative genetics. Genetica 136:351–358
Vitezica ZG, Varona L, Legarra A (2013) On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics 195:1223–1230
Walker TD, Cumbie WP, Isik F (2022) Single-step genomic analysis increases the accuracy of within-family selection in a clonally replicated population of Pinus taedaL. For Sci 68:37–52
Wellmann R, Bennewitz J (2011) The contribution of dominance to the understanding of quantitative genetic variation. Genet Res 93:139–154
Wright S (1921) Systems of mating. I. The biometric relations between parent and offspring. Genetics 6:111–123
Wright Stephen I, Ness Rob W, Foxe J, Barrett Spencer CH (2008) Genomic consequences of outcrossing and selfing in plants. Int J Plant Sci 169:105–118
Wu HX (2019) Benefits and risks of using clones in forestry – a review. Scand J For Res 34:352–359
Xu K (2022) The genetic basis of selfing rate evolution. Evolution 76:883–898
Yeh FC, Brune A, Cheliak WM, Chipman DC (1983) Mating system of Eucalyptus citriodora in a seed-production area. Can J For Res 13:1051–1055
Zhang H, Yin L, Wang M, Yuan X, Liu X (2019). Factors affecting the accuracy of genomic selection for agricultural economic traits in Maize, Cattle, and Pig Populations. Front Genet 10:1–10
Acknowledgements
We would like to thank Professor Roland Vencovsky (In Memoriam), a great supporter of studies of mixed species in Brazil. Evandro V. Tambarussi was supported by a CNPq research fellowship (grant no. 303789/2022-0). We also acknowledge AbacusBio for allowing the author Marcio J. Araujo to dedicate a few hours of personal development time to work on this manuscript.
Author information
Authors and Affiliations
Contributions
MJA: conceptualization, methodology, validation, formal code, analysis and writing - review & editing; DB: writing - review & editing; EVT: conceptualization, writing - review & editing of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Sebastián Ramos-Onsins.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g., a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Araujo, M.J., Bush, D. & Tambarussi, E.V. Quantifying genetic and genotypic gain gaps in Eucalyptus: the hidden cost of ignoring inbreeding and dominance. Heredity 134, 542–557 (2025). https://doi.org/10.1038/s41437-025-00792-8
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41437-025-00792-8