Abstract
Spastic paraplegia (SPG)10 is an autosomal dominant SPG caused by kinesin family member 5A (KIF5A) gene variants. We describe a Japanese patient with SPG whose deceased mother and maternal uncle also exhibited SPG. Exome analysis identified a rare KIF5A nonsense variant (NM_004984.4:c.2590C>T (p.Arg864Ter)) in the patient, regarded as pathogenic. As KIF5A mRNA expression was significantly decreased compared with that of a healthy control, the variant was deemed causative of SPG.
Similar content being viewed by others
Hereditary spastic paraplegia (SPG) comprises a group of clinically and genetically heterogeneous disorders, characterized mainly by progressive lower limb spasticity and weakness as a result of distal degeneration of corticospinal tract axons1. SPG10 (MIM; Mendelian Inheritance in Man: 604187) is an autosomal dominant hereditary SPG caused by variants in the kinesin family member 5A gene (KIF5A, MIM: 602821), also known as the kinesin heavy chain gene (KHC), which is located at chromosome 12q13 (refs. 2,3,4). KIF5A RNA is expressed exclusively in brain tissue5. KIF5A is associated with RNA transport in dendrites and consists of an N-terminal motor domain, a coiled-coil hinge domain and a C-terminal cargo-binding domain6.
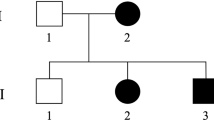
A 60-year-old male Japanese patient initially noticed muscle cramping in his calves while attending junior high school and exhibited pollakiuria while in high school. He subsequently experienced gait disturbance at the age of 20 and underwent catheter ablation for paroxysmal atrial fibrillation at 60 years of age. His mother and maternal uncle had spastic gaits. A neurological examination showed hyperreflexia, spasticity, weakness in his lower extremities and bilateral Babinski reflexes. He demonstrated mild impaired vibration sense with dominancy in the distal portion of the lower limbs. His superficial sensation and position sense were intact. Cognitive impairment was not observed. A cranial nerve examination found no abnormalities, and there was no sign of cerebellar ataxia. He did not have pes cavus. Serum anti-human T-lymphotropic virus-1 antibody levels were negative, and the standard cerebrospinal fluid examination was normal. Nerve conduction studies on the right median, ulnar, tibial and sural nerves revealed normal motor conduction velocities, distal compound muscle action potential amplitudes and sensory conduction velocities. The distal motor latency was slightly prolonged in the median nerve, and sensory nerve action potential amplitudes were mildly reduced in all nerves tested. Upon examination of motor evoked potentials, the bilateral central motor conduction time was normal, and no abnormalities were observed in brain or cervical magnetic resonance imaging. In summary, the patient showed slowly progressive SPG with bladder dysfunction and peripheral neuropathy, which is compatible with SPG107.
We first confirmed absence of an exonic copy number variant in SPAST (MIM: 604277) by fragment analysis using the SALSA MLPA Reagent Kit (MRC Holland) on the patient’s DNA. We detected 20,640 single-nucleotide variants by exome sequencing of the proband. We prioritized 17 genes known to be responsible for autosomal dominant SPG: ALDH18A1 (MIM: 138250), ATL1 (MIM: 606439), BSCL2 (MIM: 606158), CPT1C (MIM: 608846), HSPD1 (MIM: 118190), KIF1A (MIM: 601255), KIF5A (MIM: 602821), KPNA3 (MIM: 601892), NIPA1 (MIM: 608145), REEP1 (MIM: 609139), REEP2 (MIM: 609347), RTN2 (MIM: 603183), SLC33A1 (MIM: 603690), SPAST (MIM: 604277), UBAP1 (MIM: 609787), WASHC5 (MIM: 610657) and ZFYVE27 (MIM: 610243). Functional nonsynonymous, nonsense, frameshift, in-frame insertion/deletion and splice site variants with minor allele frequencies <0.0001 were selected using gnomAD (https://gnomad.broadinstitute.org), and a KIF5A nonsense variant, NM_004984.4:c.2590C>T (p.Arg864Ter) (rs1882578572), was identified in the patient. This was validated by Sanger sequencing using forward primer 5′-AGCTGGTACGTGACAATGCA-3′ and reverse primer 5′-CTCCTTGGCCTCCTTCAGTG-3′. The patient was confirmed to be heterozygous for the variant, consistent with the autosomal dominant mode of the predicted disease inheritance (Fig. 1).
A The genomic structure of KIF5A. Filled and open boxes represent coding and noncoding regions of transcript NM_004984.4, respectively. The nonsense variant NM_004984.4:c.2590C>T (p.Arg864Ter) is shown by a red arrow. KIF5A consists of 29 exons; exons 1–11, 11–23 and 24–28 encode the kinesin motor domain, coiled-coil hinge domain and cargo-binding domain, respectively. PTC, premature termination codon. B An electropherogram of the region of the variant NM_004984.4:c.2590C>T (p.Arg864Ter) in the patient and a healthy control. A heterozygous G-to-A substitution was detected at position 2590 in the patient, causing a premature termination at codon 864. The direction of translation is shown by the arrows.
This nonsense variant was previously reported in a Greek patient with sporadic SPG accompanied by sensorimotor axonal neuropathy, lower limb hemiatrophy and pes cavus8. It is located in KIF5A exon 24 and results in truncation of the KIF5A protein at the C-terminal cargo-binding domain. A significant decrease in KIF5A expression was detected in the patient’s peripheral blood, compared with that of a healthy control, probably resulting from nonsense-mediated decay (Fig. 2). According to ACMG–AMP (American College of Medical Genetics and Association of Molecular Pathology) guidelines9,10, the variant is classified as ‘pathogenic’, meeting PVS1, PS4, PM1, PM2 and PM4 criteria.
KIF5A mRNA was quantified by real-time quantitative PCR using the forward primer: 5′-CGGTGGCGCAATGGAGA-3′ and reverse primer: 5′-CCTCGTATTTCTGCCGCTCC-3′. The experiment was performed in triplicate. Values represent the mean ± s.d. normalized to GAPDH mRNA levels. Relative expression levels were calculated by the ΔΔCT method because of the quantitative limitation of the patient sample. *P = 0.0135.
The variant is expected to be translated into a truncated protein, lacking the C-terminal cargo-binding domain that is known to be essential for sliding and bundling of microtubule, and autoinhibition as well as cargo binding11,12,13. Although functional variants located in this domain are frequently associated with amyotrophic lateral sclerosis and neonatal intractable myoclonus syndrome (MIM: 617235)14, both our patient and the previous patient with the nonsense variant exhibited SPG with neuropathy, which is compatible with SPG10. The dominant negative effect of the truncated protein product and/or haploinsufficiency of the functional protein caused by nonsense-mediated decay are likely to be involved in the pathogenesis of SPG in the pedigree.
In this study, we conclude that the KIF5A nonsense variant NM_004984.4: c.2590C>T (p.Arg864Ter) is causative of SPG10.
HGV datbase
The relevant data from this Data Report are hosted at the Human Genome Variation Database at: https://doi.org/10.6084/m9.figshare.hgv.3500.
References
Panza, E., Meyyazhagan, A. & Orlacchio, A. Hereditary spastic paraplegia: genetic heterogeneity and common pathways. Exp. Neurol. 357, 114203 (2022).
Lawrence, C. J. et al. A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22 (2004).
Reid, E., Dearlove, A. M., Rhodes, M. & Rubinsztein, D. C. A new locus for autosomal dominant “pure” hereditary spastic paraplegia mapping to chromosome 12q13, and evidence for further genetic heterogeneity. Am. J. Hum. Genet. 65, 757–763 (1999).
Reid, E. et al. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10). Am. J. Hum. Genet. 71, 1189–1194 (2002).
Niclas, J., Navone, F., Hom-Booher, N. & Vale, R. D. Cloning and localization of a conventional kinesin motor expressed exclusively in neurons. Neuron 12, 1059–1072 (1994).
Kanai, Y., Dohmae, N. & Hirokawa, N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 (2004).
Goizet, C. et al. Complicated forms of autosomal dominant hereditary spastic paraplegia are frequent in SPG10. Hum. Mutat. 30, E376–E385 (2009).
Lynch, D. S. et al. Hereditary spastic paraplegia in Greece: characterization of a previously unexplored population using next-generation sequencing. Eur. J. Hum. Genet 24, 857–863 (2016).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Jarvik, G. P. & Browning, B. L. Consideration of cosegregation in the pathogenicity classification of genomic variants. Am. J. Hum. Genet. 98, 1077–1081 (2016).
Hirokawa, N., Noda, Y., Tanaka, Y. & Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 (2009).
Nakajima, K. et al. Molecular motor KIF5A is essential for GABAA receptor transport, and KIF5A deletion causes epilepsy. Neuron 76, 945–961 (2012).
Randall, T. S., Moores, C. & Stephenson, F. A. Delineation of the TRAK binding regions of the kinesin-1 motor proteins. FEBS Lett. 587, 3763–3769 (2013).
Brenner, D. et al. Hot-spot KIF5A mutations cause familial ALS. Brain 141, 688–697 (2018).
Acknowledgements
This work was supported by the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University, Japan. The study was approved by the Ethics Committees of Ehime University Graduate School of Medicine (#31-1-R3-K1 and #31-15-R3-K2) and Kyushu University, Faculty of Medicine (#818 and #819). We thank S. Williams from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miura, S., Suenaga, S., Goto, H. et al. A Japanese patient with hereditary spastic paraplegia with a rare KIF5A nonsense variant. Hum Genome Var 12, 11 (2025). https://doi.org/10.1038/s41439-025-00313-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41439-025-00313-3