Abstract
The meninges are critical for the brain functions, but the diversity of meningeal cell types and intercellular interactions have yet to be thoroughly examined. Here we identify a population of meningeal lymphatic supporting cells (mLSCs) in the zebrafish leptomeninges, which are specifically labeled by ependymin. Morphologically, mLSCs form membranous structures that enwrap the majority of leptomeningeal blood vessels and all the mural lymphatic endothelial cells (muLECs). Based on its unique cellular morphologies and transcriptional profile, mLSC is characterized as a unique cell type different from all the currently known meningeal cell types. Because of the formation of supportive structures and production of pro-lymphangiogenic factors, mLSCs not only promote muLEC development and maintain the dispersed distributions of muLECs in the leptomeninges, but also are required for muLEC regeneration after ablation. This study characterizes a newly identified cell type in leptomeninges, mLSC, which is required for muLEC development, maintenance, and regeneration.
Similar content being viewed by others
Introduction
The meninges provide mechanical support and protective barriers for the central nervous system (CNS) but also play indispensable roles in brain development, neuroinflammation, and CNS immunity1,2,3. Along the axis from the skull to the brain parenchyma, the meninges consist of the external dura mater attached to the skull, the arachnoid mater in the middle, and the inner pia mater covering the parenchyma1,2,3. The arachnoid, the subarachnoid space filled with cerebrospinal fluid, and the pia mater are referred to as the leptomeninges1. The leptomeninges of mammals are composed of fibroblasts, neural progenitor cells, immune cells, perivascular cells, and meningeal blood vessels connected to the parenchymal vasculature1,4,5,6. Although multiple meningeal cells have been known to be important for the proper functioning of the brain, the constitutions and functions of meningeal cell types remain to be more thoroughly understood.
A network of lymphatic vessels consisting of a single layer of blind-ended lymphatic endothelial cells (LECs) is distributed throughout most body tissues and is responsible for maintaining interstitial fluid homeostasis, immune surveillance, and lipid absorption7. The meningeal lymphatic vessels in the dura mater of mice drain cerebrospinal fluid, macromolecules, and immune cells from the CNS to the peripheral lymphatic network, thus participating in the regulation of immune surveillance and inflammatory responses in the brain8,9,10. The presence of meningeal lymphatics has also been found in the meninges of nonhuman primates and humans11, and has recently been detected in the juvenile and adult zebrafish12. Furthermore, a population of perivascular LECs have been revealed to reside in the membranes attached to the zebrafish brain parenchyma13,14,15, and named as mural lymphatic endothelial cells (muLECs)15. These muLECs are transcriptomically characterized as an LEC population and express canonical LEC markers including prox1a, lyve1b, vegfr3, and mrc1a, and muLEC development is dependent on the vegfc/vegfr3/ccbe1 signaling axis13,14,15. Notably, under physiological conditions, muLECs do not form lymphatic vessels but consistently remain as dispersed single-cell communities13,14,15. The characteristics of non-tubular muLECs are distinct from those of tubularized peripheral and intracranial LECs. Apart from eliminating various metabolic wastes from the brain14,16, muLECs, as a type of perivascular cells, have also been found to be involved in regulating the development of the meningeal vasculature15 and assisting the post-injured cerebrovascular regeneration17,18. However, mechanisms underlying the development of muLECs as well as the formation and maintenance of their dispersed distributions in the leptomeninges remain unclear.
In this study, we identified a population of meningeal lymphatic supporting cells (mLSCs) specifically labeled by ependymin (epd) in zebrafish leptomeninges. These flat and irregular spindle-shaped mLSCs accumulate at 2 days post-fertilization (dpf) in a partially overlapping manner, forming a membranous structure that covers the brain and maintains throughout adulthood. mLSCs exhibit a unique transcriptomic profile and express unique markers that distinguish them from other known meningeal cells. muLECs always locate on the mLSC-composed membrane. Functionally, at early developmental stages, mLSCs provide migratory tracks and produce pro-lymphangiogenic factors to activate the germination of muLECs. Later, mLSCs are essential for maintaining the morphologies and dispersed distributions of non-tubular muLECs as well as muLEC regeneration after ablation. Collectively, these findings improve understanding of cellular constitutions and interactions of vertebrate meninges.
Results
epd is expressed in the larval zebrafish brain
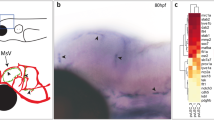
Ependymin, encoded by gene epd, is a secretory glycoprotein first discovered in the extracellular fluid and cerebrospinal fluid of the goldfish brain19. According to previous reports showing specific expression of epd mRNA in the zebrafish leptomeninges20, we examine whether epd is generally expressed in the whole leptomeninges or exclusively labeled a group of meningeal cells. Whole-mount in situ hybridization (WISH) on zebrafish larvae at 5 dpf showed that epd was exclusively expressed over the brain (Supplementary Fig. 1a), accumulated at the dorsal side of the brain mainly in symmetrical circles over the optic tectum (TeO) and in horizontal lines over the cerebellum (Supplementary Fig. 1a, b). This expression pattern of epd was reminiscent of muLECs, which similarly form bilateral circular structures on the TeO at this developmental stage13,14,15. However, fluorescence in situ hybridization (FISH) combined with antibody staining (FISH-antibody staining) in the Tg(lyve1b:EGFP) transgenic line18,21 showed that epd was expressed around muLECs rather than in muLECs (Supplementary Fig. 1c). Furthermore, epd appeared to be around the tubular structures that were assumed as mesencephalic vein (MsV) (Supplementary Fig. 1c, arrowheads), which was confirmed by the FISH-antibody staining under the Tg(kdrl:GFP) transgenic background (Supplementary Fig. 1d, arrowheads).
epd-positive cells enwrap meningeal blood vessels and muLECs
To further explore the nature of the epd-positive cells, we generated a Tg(epd:EGFP) transgenic line, in which EGFP was driven by the epd promoter. The distributions of EGFP-positive cells in the brain (Supplementary Fig. 1e–h) were highly similar to the location of epd mRNA expression, suggesting that the 3.9 kb of epd promoter could drive its endogenous expression. DAPI staining under the Tg(epd:EGFP) background at 5 dpf showed the nuclei of epd-positive cells as well as the adjacent nuclei of epd-negative cells (Supplementary Fig. 1i). In the Tg(epd:EGFP; kdrl:mCherry-Ras) and Tg(epd:EGFP; lyve1b:DsRed) zebrafish brains from larvae to adults, the epd-positive cells, although widely distributed on TeO, were more clustered around muLECs and meningeal blood vessels at multiple stages (Fig. 1a–f). Higher magnification images revealed the presence of visible cavities of muLECs/blood vessels among epd-positive cells (Fig. 1a–d, arrowheads), implicating that the epd-positive cells were different from either muLECs or meningeal blood vessels.
a, c, e Dorsal confocal images of epd-positive cells and blood vessels on Tg(epd:EGFP; kdrl:mCherry-Ras) brains at 5 dpf (a n = 20), 7 mm (c n = 20) and 4 mpf (e n = 12). White arrowheads indicate the cavities of mural lymphatic endothelial cells (muLECs). b, d, f Dorsal confocal images of epd-positive cells and muLECs on Tg(epd:EGFP; lyve1b:DsRed) brains at 5 dpf (b n = 20), 7 mm (d n = 20) and 4 mpf (f n = 12). White arrowheads indicate the cavities of meningeal blood vessels. g A representative confocal image of the meningeal lymphatics in the dura mater beneath the skull at 4 mpf. n = 12. h Illustration of cross-sections (horizontal line) and sagittal sections (vertical line) of adult zebrafish brain in (i–l). i–l Confocal images of brain sections showing the locations of epd-positive cells in the leptomeninges at 4 mpf. White arrowheads indicate the distributions of epd-positive cells. n = 12 adult brains per experiment. Each experiment was repeated three times independently with similar results. The white dashed boxes outline the enlarged areas. Scale bars: 100 µm.
Detailed examination of the epd-positive cells, muLECs, and meningeal vasculature on the surface of the brains of 5-dpf larval and 7-mm juvenile zebrafish revealed the constant presence of epd-positive cells around muLECs (Fig. 1a–d), but the blood vessels were not always around muLECs (Fig. 1aI–dI, and Supplementary Fig. 2a–d). These results were further validated by the intracerebroventricular (ICV) injection of fluorescent macromolecule Alexa647-lgG (150 kDa) into the Tg(epd:mCherry-NTR; fli1:GFP) transgenic larvae (Supplementary Fig. 2e–g). Although fli1 labels both muLECs and blood vessel endothelial cells (BECs), only muLECs can take up Alexa647-lgG14. Thus, BECs were only labeled by GFP, while muLECs were double-labeled by GFP and Alexa647-lgG. In some brain areas in particular the posterior half of the midbrain and the midbrain-hindbrain boundary, muLECs were always enwrapped by the epd-positive cells, but not adjacent to blood vessels (Supplementary Fig. 2f–h). These data suggest that muLECs are more closely associated with epd-positive cells than with blood vessels during development.
epd-positive cells present in zebrafish leptomeninges
In adults at 4 months post-fertilization (mpf), the epd-positive cells were shown to be absent in the dura mater attached to the skull (Fig. 1g), whereas were widely distributed in the midbrain and hindbrain and still featured by encompassing blood vessels and muLECs (Fig. 1e, f). Sagittal sections of the adult zebrafish brain showed that the epd-positive cells were localized in the olfactory bulb, the base of the forebrain, midbrain, and hindbrain, the regions where muLECs settled, and were absent in the brain parenchyma (Fig. 1i, k). Cross-sections of the adult brain demonstrated that the epd-positive cells were only detected in the meninges and further illustrated that the meningeal vasculature and muLECs were embedded in the membranes formed by the epd-positive cells (Fig. 1j, l). Altogether, these meningeal epd-positive cells are exclusively resident in the leptomeninges throughout zebrafish lifespan and consistently around muLECs and most of meningeal blood vessels.
epd-positive cells are not other known meningeal cell types
The mammalian leptomeninges contain a variety of cell types, including fibroblasts, neural progenitor cells, LECs, BECs, perivascular cells, and various immune cells1,4,5,6,22,23,24. To investigate whether the epd-positive cell belongs to one of the common meningeal cell types, we first analyzed the brains of Tg(epd:H2B-mCherry; fli1:nEGFP) zebrafish at 5 dpf and 4 mpf. Co-localization between the H2B-mCherry-labeled nuclei and nEGFP-labeled nuclei was never observed (Fig. 2a, b), further indicating that the epd-positive cell did not belong to endothelial cell types. The fibroblasts labeled by Collagen I are the main component of mouse brain membranes6. The sections of adult zebrafish brain at 6 mpf displayed that the Collagen I-labeled fibroblasts located at a layer different from, but immediately beneath, the epd-positive cell layer (Fig. 2c), suggesting that the epd-positive cells are not fibroblasts, but form a membranous structure similar to fibroblasts. In multiple transgenic lines including Tg(abcc9BAC:Gal4ff; UAS:GFP) labeling pericytes25, Tg(pdgfrbBAC:EGFP) labeling pericytes and/or vascular smooth muscle cells26, Tg(acta2:GFP) labeling vascular smooth muscle cells27, Tg(coro1a:Kaede) labeling macrophages and neutrophils28, Tg(lyz:GFP) labeling neutrophils29, Tg(mpeg1:GFP) labeling macrophages30, Tg(prox1aBAC:KalTA4; UAS:TagRFP) labeling LECs and neuronal cells31, Tg(nkx2.2a:GFP) labeling neuronal and/or oligodendrocyte cells32, and Tg(elavl3:GFP) labeling neurons33, the epd-positive cells did not share any overlap or morphological similarities with these cell types (Fig. 2d–l). Hence, the epd-positive cell is anatomically distinct from the currently known meningeal cell types.
a, b Dorsal confocal images of epd:H2B-mCherry-positive nuclei and fli1:nEFP-positive nuclei in Tg(epd:H2B-mCherry; fli1:nEFP) brains at 5 dpf (a n = 20) and 4 mpf (b n = 8). c Confocal image of a cross-section of an adult brain showing collagen I-labeled fibroblasts at a different layer to epd-positive cells at 6 mpf. A 2D view of the enlarged area is shown. n = 8 adult brains. The experiment was repeated three times independently with similar results. d–l Confocal images showed epd-positive cells did not co-stain with abcc9BAC:Gal4ff; UAS:GFP-positive pericytes (d), pdgfrbBAC:EGFP-positive pericytes (e), acta2:GFP-positive smooth muscle cells (f), coro1a:Kaede-positive immune cells (g), lyz:GFP-positive neutrophils (h), mpeg1:GFP-positive macrophages (i), prox1aBAC:KalTA4; UAS:TagRFP-positive lymphatic endothelial cells and neuronal cells (j), nkx2.2a:GFP-positive neuronal and/or oligodendrocytes (k), and elavl3:GFP-positive neurons (l) at 8 dpf. Dorsal and lateral views of the larval brains were shown. n = 20 per experiment. Each experiment was repeated three times independently with similar results. The white dashed boxes outline the enlarged areas. Scale bars: 100 µm.
The previously reported single-cell RNA-sequencing (scRNAseq) data from whole zebrafish embryos and larvae as well as adult telencephalon revealed that epd is specifically expressed in a group of cells34,35,36. Here, we re-analyzed the scRNAseq data from whole zebrafish embryos and larvae by Farnsworth et al.34. On the basis of marker genes and other zebrafish scRNAseq results1,4,6,23,34,35, we picked out epd-positive cells and common meningeal cell types (including fibroblasts, neural progenitor cells, endothelial cells, mural cells, and immune cells) for analyses (Supplementary Data 1). We found that each cluster was featured by distinctive gene expression patterns (Supplementary Fig. 3a–c). Particularly, the epd-positive cells possessed their exclusively highly expressed genes that have previously not been reported as marker genes for other meningeal cell populations (Supplementary Fig. 3d). Taken together, these data suggest that epd-positive cells represent a meningeal cell type with unique anatomical and transcriptomic characteristics.
epd-positive cells produce pro-lymphangiogenic factors
To further understand the molecular characteristics of the early-stage epd-positive cells, we isolated the epd-positive cells from zebrafish brains at 55 h post-fertilization (hpf) and 5 dpf by fluorescence-activated cell sorting (FACS) (Fig. 3a, b) and carried out transcriptomic profiling by RNA-seq analyses. To efficiently separate the epd-positive cells from adjacent muLECs, FACS was performed using the Tg(epd:EGFP-NTR; lyve1b:DsRed) transgenic line (Fig. 3c, d). The sorted epd:EGFP+ cells were applied for RNA-sequencing analyses, using whole embryos/larvae at 55 hpf and 5 dpf as controls. Principal component analysis verified the similarities between our RNA-seq data and the published scRNAseq data34 of epd-positive cells (Supplementary Fig. 3e). Then, the expressions of various meningeal cell type marker genes were analyzed in the sorted epd-positive cells and in whole fish. The makers of immune cells (ImCs), glial and neural cells (GAN), BECs, pericytes (PCs), LECs, and fibroblasts (FBs) failed to express or expressed at very low levels in the epd-positive cells (Fig. 3e; Supplementary Data 2). By contrast, the epd-positive cells expressed high levels of their unique markers such as slc13a4, soul5, slc7a2, igfbp2a, nid1b, apof, ggctb, and epd (Fig. 3e; Supplementary Data 2), which were validated by WISH and FISH-antibody staining (Supplementary Fig. 4).
a, b Dorsal confocal images of fluorescence-negative wild-type and Tg(epd:EGFP-NTR; lyve1b:DsRed) embryonic/larval brains at 55 hpf (a) and 5 dpf (b). n = 30 embryos/larvae per panel. Scale bars: 200 µm. c, d Representative plots for fluorescence-activated cell (FAC) sorted EGFP-NTR-positive cells in brains of fluorescence-negative wild-type and Tg(epd:EGFP-NTR; lyve1b:DsRed) at 55 hpf (c) and 5 dpf (d). e Heatmap for differential expression levels of epd-positive cells and other selected known meningeal cell marker genes between whole fish (n = 100 fish per replicate) and FAC sorted epd-positive cells (EGFP-NTR-positive, n = 100 cells per replicate). ImC immune cell, GAN glial and neural cells, BEC blood vessel endothelial cell, PC pericytes, LEC lymphatic endothelial cell, FB fibroblasts, epd+ cells epd-positive cells. Scale bar represents relative expression by log2(FPKM + 1), from 0 (lowest) to 15 (highest). f Heatmap for differential expression levels of neurotrophic, pro-angiogenic, and pro-lymphangiogenic factors between whole fish (n = 100 fish per replicate) and FAC sorted epd-positive cells (EGFP-NTR-positive, n = 100 cells per replicate). Scale bar represents relative expression by log2(FPKM + 1), from 0 (lowest) to 7.55 (highest).
Notably, we revealed that although the epd-positive cells did not express neurotrophic factors or pro-angiogenic factors, they expressed high levels of pro-lymphangiogenic factors such as vegfc, vegfd, ccbe1 and mmp2 (Fig. 3f). Analysis of Enriched Ontology Clusters for highly expressed genes in the epd-positive cells relative to whole fish showed that the epd-positive cell-enriched genes have terms associated with the SLC-mediated transmembrane transport, import into the cell, mesenchyme development, and lymphangiogenesis (Supplementary Fig. 5; Supplementary Data 3, 4). Thus, the epd-positive cells represent a previously unidentified leptomeningeal cell population that produce pro-lymphangiogenic factors.
A portion of muLECs migrate along epd-positive cells
Since the epd-positive cells produce pro-lymphangiogenic factors, we first track their development to see whether they develop prior to the muLEC formation. Live imaging of zebrafish embryos from 0 hpf was carried out. The appearance of epd-positive cells was first detected in the midbrain at the 18-somite-stage (ss) (Fig. 4a), and then the number of this cell population increased from 18 ss to 24 ss (Supplementary Movie 1). To display the morphologies of individual epd-positive cell, we generated the Tg(epd:H2B-GFP; epd:mCherry-Ras) transgenic strains, in which the membranes and nuclei of epd-positive cells were labeled by the membrane-targeted mCherry-Ras and the nuclear H2B-GFP, respectively (Fig. 4b). The epd-positive cells formed a membrane structure covering the TeO from 2 dpf (Fig. 4b). At 5 dpf, individual epd-positive cell in forebrain, midbrain, and hindbrain all exhibited a flattened irregular spindle cell shape, and multiple cells look like partially stacked rather than interconnected together in terms of the overlap between cell membranes (Fig. 4b, Supplementary Fig. 6a, b).
a A representative lateral confocal image of epd-positive cells first appeared in Tg(epd:EGFP; kdrl:mCherry-Ras) brains at 18-somite stage (ss). White arrowheads indicate the epd-positive cells. n = 10. The experiment was repeated three times independently with similar results. b Lateral confocal images of the development of epd-positive cells in Tg(epd:H2B-GFP; epd:mCherry-Ras) from 2 dpf to 5 dpf. The nuclei and membranes of epd-positive cells are labeled with H2B-GFP and mCherry-Ras, respectively. Enlarged area showing irregular spindle-shaped epd-positive single-cell. n = 15 per stage. c Schematic diagram showing the strategy for ablation of epd-positive cells. d, e Quantification of the number of epd-positive cells on the optic tectum (TeO) at 72 hpf/0 dpt (d) and the number of mural lymphatic endothelial cells (muLECs) in bilateral loop at 5 dpf/2 dpt (e) in the nonablation and epd-positive cell ablation group. 16 fish were observed in three independent experiments in each group. f, g, i, j Dorsal confocal images of epd-positive cells and muLECs in brains that did not induce muLEC or epd-positive cell injury at 72 hpf/0 dpt and 5 dpf/2 dpt. White arrowheads indicate muLECs. n = 34 per experiment. h Dorsal confocal images of ablation of epd-positive cells in brains at 72 hpf/0 dpt. n = 38. The experiment was repeated three times independently with similar results. k Dorsal confocal images of massive missing in muLECs after epd-positive cell ablation at 5 dpf/2 dpt. White arrowheads indicate where the muLECs should have been. n = 33. The experiment was repeated three times independently with similar results. Error bars, mean ± SEM. Unpaired two-tailed Student’s t-test. P values included in the graphs. Source data are provided as a Source Data file. The white dashed boxes outline the enlarged areas. Scale bars: 50 µm.
The muLECs originate in the optic choroidal vascular plexus (OCVP), then posteriorly migrate either along the medial side of MsV or along the ventral side of the midbrain, finally forming a classical bilateral symmetrical circular structure on the TeO at around 4 dpf13,14. Given the close proximity of epd-positive cells and muLECs, we next study whether muLECs migrate along the epd-positive cells during development. Time-lapse imaging from 60 hpf when muLECs begin to emerge to 106 hpf when typical muLEC loop structures are formed was carried out under the Tg(epd:mCherry-NTR; lyve1b:EGFP) transgenic background. Before the budding of muLECs from OCVP, the epd-positive cells already clustered (Supplementary Movie 2). Migration of muLECs along the ventral side of the midbrain turns out to follow a track provided by the epd-positive cells (Supplementary Movie 2, yellow arrowhead). The lateral framework of epd-positive cells at the midbrain-hindbrain junction formed at 60 hpf, and then muLECs sprouted out at about 77 hpf, again demonstrating guidance of muLEC migration by the epd-positive cells (Supplementary Movie 2, blue arrowhead). These results suggest that the epd-positive cells are potentially important for the development of muLECs.
epd-positive cells regulate the development of muLECs
In order to explore whether the epd-positive cells are required for the formation of muLECs, we generated the Tg(epd:EGFP-NTR) and Tg(epd:mCherry-NTR) transgenic lines to ablate the epd-positive cells based on the nitroreductase-metronidazole (NTR-Mtz) system37,38. Because muLECs sprout from OCVP at approximately 54–56 hpf, treatment with Mtz was applied to the Tg(epd:EGFP-NTR; lyve1b:DsRed) transgenic lines from 48 hpf to 72 hpf, which resulted in the ablation of epd-positive cells (Fig. 4c, d, h). The Tg(lyve1b:DsRed) line treated with Mtz (Fig. 4c, f) and the Tg(epd:EGFP-NTR; lyve1b:DsRed) line treated with DMSO (Fig. 4c, g) were used as controls. At 2 days post Mtz treatment (dpt), equivalent to 5 dpf, the muLECs were massively missing, including the typical bilateral circular structures (Fig. 4e, i–k). These results were validated by using prox1a to label LECs under the Tg(epd:EGFP-NTR; prox1aBAC:KalTA4; UAS:TagRFP) transgenic background (Supplementary Fig. 6c–e). Using the Alexa647-lgG macromolecule endocytosis function to label muLECs, intracranial injection of Alexa647-lgG further confirmed the loss of muLECs after ablation of the epd-positive cells (Supplementary Fig. 6f–h). Time-lapse imaging from 60 hpf to 106 hpf using the Tg(epd:mCherry-NTR; lyve1b:EGFP) transgenic lines revealed that after ablation of the epd-positive cells, muLECs failed to sprout from OCVP (Supplementary Movie 3). Taken together, all these data demonstrate that the epd-positive cell is an essential supportive cell type for muLEC sprouting and development, thereafter we name it as a meningeal lymphatic supporting cell (mLSC).
Ablation of mLSCs causes muLECs to form lymphatic vessels
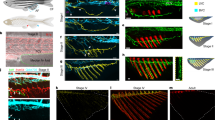
Although muLECs express multiple LEC markers, they do not form lymphatic vessels but maintain dispersed cell distributions13,14,15. Next, we examined whether mLSCs were responsible for the maintenance of muLECs at different stages. Mtz was applied to the Tg(epd:EGFP-NTR; lyve1b:DsRed) transgenic larvae to ablate mLSCs at 5 dpf, when the basic pattern of muLECs was initially established (Fig. 5a). At 6 dpf/0 dpt when Mtz was withdrawn after a 24-h-treatment, mLSCs were successfully ablated and the continuous endothelial loops of muLECs (Fig. 5b, c, Supplementary Fig. 7a) began to be fractured (Fig. 5d). Time-lapse live imaging of the Mtz-treated larvae from 5 dpf to 6.5 dpf illustrated that muLECs collapsed from elongated strips to ovals along with the ablation of mLSCs (Supplementary Movie 4). Although the morphologies of muLECs changed after mLSC ablation, the number of muLECs was maintained (Supplementary Fig. 7b). Moreover, TUNEL assays showed that muLECs did not undergo apoptosis (Supplementary Fig. 7c, d). After mLSC ablation, the adjacent muLECs converged to form cell clusters at 10 dpf/4 dpt (Fig. 5e–g), then protruded filopodium-like structures to interconnect at 14 dpf/8 dpt (Fig. 5h–j) and formed tubular-like structures at 17 dpf/11 dpt (Fig. 5k–m) (Supplementary Fig. 8a–d). Eventually, these muLECs became lumenized vessels at 20 dpf/14 dpt after mLSC ablation (Fig. 5n–p, Supplementary Fig. 8a–d).
a Schematic diagram showing experimental design for meningeal lymphatic supporting cell (mLSC) ablation. b, c, e, f, h, i, k, l, n, o Dorsal confocal images of mural lymphatic endothelial cells (muLECs) and mLSCs in brains that did not induce muLEC or mLSC injury at 6 dpf/0 dpt, 10 dpf/4 dpt, 14 dpf/8 dpt, 17 dpf/11 dpt and 20 dpf/14 dpt. White arrowheads indicate muLECs. Yellow arrowheads indicate meningeal lymphatic vessels (MLV). n = 20 per experiment. d, g, j, m, p Dorsal confocal images showing the progressive formation of lymphatic vessels from collapsed muLECs after inducing injury to mLSCs. White arrowheads in (d) indicate collapsed muLECs. White arrowheads in (p) indicate muLEC-derived lymphatic vessels. Yellow arrowheads indicate MLV. d n = 28; g n = 24; j n = 20; m n = 17; p n = 19. The experiments were repeated three times independently with similar results. The white dashed boxes outline the enlarged areas. Scale bars: 50 µm.
Then, we analyze whether the loss of mLSCs leads to the formation of functional lymphatic vessels by muLECs. At 14 dpt after Mtz treatment, some muLEC-derived lymphatic vessels were observed to connect to the developing meningeal lymphatic vessels (MLVs) sprouting from otolithic lymphatic vessels (Fig. 5pII, Supplementary Fig. 8e). MLVs play important roles in the immune surveillance of the brain and drainage of cerebrospinal fluid8,9,10,12. Time-lapse imaging under the Tg(epd:EGFP-NTR; lyz:GFP; lyve1b:DsRed) background at 20 dpf/14 dpt after mLSC ablation showed that the lyz:GFP+ lymphocytes slowly moved in the muLEC-derived lymphatic vessels (Fig. 6b, c). The muLECs in the physiological state at the same time do not carry lyz:GFP+ lymphocytes (Fig. 6a, c). The ICV injection of Alexa647-Dextran (10 kDa) into the brain of Tg(epd:EGFP-NTR; lyve1b:DsRed) juvenile at 20 dpf/14 dpt, which was capable of tracing cerebrospinal fluid (Fig. 6d), showed that the Alexa647-Dextran-labeled fluid filled the muLEC-derived vessels after mLSC ablation (Fig. 6h, n), in contrast to the muLECs under physiological conditions that endocytosed the dye (Fig. 6f, g, m). These results indicate that the mLSC ablation-induced, muLEC-derived lymphatic vessels obtain the functions of immune cell transport and cerebrospinal fluid drainage. Furthermore, the injection of Alexa647-Dextran into the dorsal aorta (DA) to image blood circulation (Fig. 6e) revealed that the dye was absent from the muLEC-derived vessels after mLSC ablation, suggesting separation of the muLEC-derived lymphatic vessels from blood vessels (Fig. 6i–k, m, n).
a Dorsal confocal images of lyz:GFP+ lymphocytes in DMSO-treated brains at 20 dpf/14 dpt. n = 6. b Time-lapse images of the flow of lyz:GFP+ lymphocytes in mural lymphatic endothelial cell (muLEC)-derived lymphatic vessels of Mtz-treated brains at 20 dpf/14 dpt. The duration of time-lapse imaging is represented in hours:minutes. n = 6. The experiment was repeated three times independently with similar results. c Percentage of fish whose muLEC-derived vessels with (green) and without (gray) immune cells. 10 fish were observed in three independent experiments in each group. d, e, l Illustrations of intracerebroventricular (ICV) and dorsal aorta (DA) injection points of Alexa647-dextran and image areas. f, g Confocal images of the uptake of Alexa647-dextran by muLECs in meningeal lymphatic supporting cell (mLSC)-uninjured brains after ICV injection. n = 20 per experiment. Each experiment was repeated three times independently with similar results. h Confocal images of the uptake of Alexa647-dextran by muLEC-derived lymphatic vessels in mLSC-injured brains after ICV injection. n = 17. The experiment was repeated three times independently with similar results. i, j Confocal images of the uptake of Alexa647-dextran by blood vessels in mLSC-uninjured brains after DA injection. n = 14 per group. Each experiment was repeated three times independently with similar results. k Confocal images of the flow of Alexa647-dextran in blood vessels but not muLEC-derived lymphatic vessels in mLSC-injured brains after DA injection. n = 14. The experiment was repeated three times independently with similar results. m, n Quantification of the number of muLECs endocytosed dye as a percentage of total muLECs (m) and the length of muLEC-derived vessels draining fluid as a percentage of total muLEC-derived vessels (n) after ICV or DA injection of Alexa647-dextran. 12 fish were observed in three independent experiments in each group. Error bars, mean ± SEM. Unpaired two-tailed Student’s t-test. P values included in the graphs. Source data are provided as a Source Data file. The white dashed boxes outline the enlarged areas. Scale bars: 50 µm.
mLSC ablation does not change vascular and pericyte morphology
Since mLSCs are also in close proximity to meningeal vessels, we therefore explored whether mLSC ablation affects the morphology of cerebral vasculature. After application of Mtz to the Tg(epd:mCherry-NTR; kdrl:GFP) larvae to ablate mLSCs, neither obvious alterations of MsV morphologies and cell number at 7 dpf/1 dpt nor brain BEC apoptosis at 6 dpf/0 dpt were observed (Supplementary Fig. 9a–f). Additionally, pericytes also remained normal at 7 dpf/1 dpt (Supplementary Fig. 9g, h). These results indicate that mLSCs are specific for the maintenance of muLECs, but not for brain vasculature.
mLSCs maintain muLEC morphology in juvenile zebrafish
The studies above ablate the mLSCs in zebrafish larvae at 5 dpf, we next examine the phenomenon in juvenile zebrafish at 25 dpf (Supplementary Fig. 10a). Again, the mLSC ablation-induced muLEC morphological change, lymphatic vessel formation and connection to MLVs were observed (Supplementary Fig. 10b–e), indicating that the maintenance of muLEC morphologies and distributions by mLSCs is conserved among different stages. Taken together, all these results demonstrate that the cell morphologies and dispersed distribution pattern of muLECs are supported by mLSCs, and loss of mLSCs will induce formation of authentic functional leptomeningeal lymphatic vessels by muLECs.
sox10-positive cell ablation does not alter muLEC morphology
To ensure the morphological changes of muLECs are specifically caused by ablation of mLSCs but not generally caused by massive cell death, we ablate oligodendrocytes and oligodendrocyte progenitor cells using the sox10:EGFP-NTR transgene to induce massive cell death in the brain. sox10 labels oligodendrocytes and oligodendrocyte progenitor cells in zebrafish39,40. Mtz was applied to the Tg(sox10:EGFP-NTR; lyve1b:DsRed) transgenic larvae at 5 dpf (Supplementary Fig. 11a). At 6 dpf/0 dpt after Mtz treatment, the sox10-positive cells were successfully ablated. However, the morphologies of muLECs remained unaffected at 6 dpf/0 dpt, 8 dpf/2 dpt, and 10 dpf/4 dpt (Supplementary Fig. 11b–g). These results suggest that the morphological change of muLECs after mLSC ablation is not an indirect effect of massive cell death.
mLSCs are required for muLEC regeneration
The muLECs are able to self-repair after partial injury by high-energy lasers15. Because of the pivotal roles of mLSCs in muLEC development and maintenance, the question whether mLSCs are involved in muLEC regeneration was investigated. The muLECs develop a lymphatic endothelial bilateral loop on TeO at 5 dpf (Fig. 1b), we chose this time point to induce muLEC ablation (Fig. 7a). Controlled by the Tg(lyve1b:DsRed) larvae treated with Mtz (Fig. 7b) and the Tg(epd:mCherry-Ras; lyve1b:EGFP-NTR) larvae treated with DMSO (Fig. 7c), application of Mtz to Tg(epd:mCherry-Ras; lyve1b:EGFP-NTR) larvae caused massive ablation of muLECs (Fig. 7d, o), which became partially regenerate at 11 dpf/5 dpt (Fig. 7e–g, p, q). By contrast, double ablation of muLECs and mLSCs could be successfully induced in the Tg(epd:mCherry-NTR; lyve1b:EGFP-NTR) transgenic larvae (Fig. 7h–k, o, r). But most muLECs failed to regenerate at 11 dpf/5 dpt (Fig. 7l–n, q). These evidence suggest that mLSCs are required for muLEC regeneration.
a, h Experimental design for ablation of meningeal lymphatic supporting cells (mLSCs) and mural lymphatic endothelial cells (muLECs). b, c, e, f, i, j, l, m Dorsal confocal images of muLECs and mLSCs in unablated brains at 6 dpf/0 dpt and 11 dpf/5 dpt. n = 37 per experiment. White arrowheads indicate muLECs. d Dorsal confocal images of muLEC ablation at 6 dpf/0 dpt. White arrowheads indicate muLECs after ablation. n = 37. The experiment was repeated three times independently with similar results. g Dorsal confocal image of partial regeneration of muLECs after injury at 11 dpf/5 dpt. n = 28. The experiment was repeated three times independently with similar results. k Dorsal confocal images of muLEC and mLSC ablation at 6 dpf/0 dpt. n = 39. The experiment was repeated three times independently with similar results. n Dorsal confocal images of less regeneration of muLECs after muLEC and mLSC ablation at 11 dpf/5 dpt. n = 34. The experiment was repeated three times independently with similar results. o, p Quantification of the number of muLECs in bilateral loop in the nonablation, muLEC ablation, and muLEC and mLSC double ablation larvae at 6 dpf/0 dpt (o) and at 11 dpf/5 dpt (p). 16 fish were observed in three independent experiments in each group. q Quantification of the recovery rate of muLECs in bilateral loop after muLEC ablation and double muLEC and mLSC ablation. 16 fish were observed in three independent experiments in each group. The same larvae as in (o, p) were used. r Quantification of the number of mLSCs on the optic tectum (TeO) for larvae with unablated and ablated mLSCs at 6 dpf/0 dpt. 16 fish were observed in three independent experiments in each group. Error bars, mean ± SEM. Unpaired two-tailed Student’s t-test. P values included in the graphs. Source data are provided as a Source Data file. The white dashed boxes outline the enlarged areas. Scale bar: 50 µm.
Discussion
Our study reveals a previously unidentified population of lymphatic support cells in the leptomeninges of zebrafish that are required for the development, maintenance, and regeneration of muLECs. Fibroblasts are known to provide microenvironmental cues and mechanical support to surrounding cells in multiple organs41. In zebrafish, mLSCs form interconnected membranous structures and exhibit similar cell morphologies and functions to fibroblasts. Previous scRNAseq data classified mLSCs as a subclass of mesenchyme35, and our RNA-sequencing analysis showed that the mLSC-enriched genes at 55 hpf also have a GO term of mesenchyme development, suggesting that mLSCs may be functional in supporting adjacent cells. Solute carriers (SLCs) mediate membrane transport of various substances and maintain the stability of the intracellular environment, and some of the SLCs have been identified to express in brain barrier cells42. The mLSC-enriched genes were shown to correlate with SLC-mediated transmembrane transport pathways, suggesting that mLSCs may carry out barrier functions in maintaining brain homeostasis.
The muLECs remain dispersed and relatively stationary throughout the zebrafish lifespan. They are invariably encapsulated by mLSCs and do not always coexist with meningeal blood vessels, suggesting a close association between muLECs and mLSCs. Previous studies have suggested that the early development of muLECs may migrate along blood vessels13,14. Based on our study, those muLECs migrating ventrally along the hindbrain and laterally along the midbrain-hindbrain junction where the meningeal blood vessel is absent should be directed and supported by mLSCs rather than by blood vessels. However, on the tracks of those muLECs migrating along MsV, both blood vessels and mLSCs are present. In these regions, the possibility that mLSCs and blood vessels cooperate to guide and support muLEC migration should not be excluded. The fundamental physiological function of muLECs under dispersed morphological conditions is their capacity to endocytose various types of substrates14,16. Previous studies have demonstrated distributions of tasks between muLECs and microglia in the endocytosis of extracellular cargo molecules from the brain16. After mLSC ablation, muLECs transform to vessels and no longer possess the capacity of endocytosis, but instead perform liquid transport function in a tubular form (Fig. 6f–h). Thus, mLSCs maintain the dispersed morphologies of muLECs under physiological conditions.
The membrane-like structures formed by mLSCs (Fig. 4b) are likely to be fundamental for them to support the development, maintenance, and regeneration of muLECs. The migration and growth of one cellular structure could be dependent on the guidance and support provided by another pre-formed cellular structure17,43,44. This mechanism could explain how the pre-formed framework of mLSCs provides support and guidance for the sprouting and post-injured regeneration of muLECs. The mLSCs express several pro-lymphangiogenic factors and their ablation severely impairs muLEC development. However, at later stages, many other cell types including BECs, smooth muscle cells, macrophages, and fibroblasts could also produce pro-lymphangiogenic factors45,46. Even muLECs themselves could express some pro-lymphangiogenic factors such as Vegfc, Vegfd, and Mmp215. After ablation of mLSCs at 5 dpf, the formation of lymphatic vessels by muLEC interconnection and lumenization (Fig. 5) is a relatively slow process compared to the outgrowth of muLECs, and this process may be assisted by non-mLSC-derived pro-lymphangiogenic factors. The process of muLEC interconnection and lumenization to form lymphatic vessels may involve more complex mechanisms, which need further investigation. Furthermore, after the ablation of mLSCs, the muLECs that lose support from mLSCs gradually transform into functional lymphatic vessels rather than undergo cell death. These observations indicate that the morphologies but not survival of muLECs are dependent on mLSCs, also suggesting the muLEC plasticity. These findings provide compelling evidence that muLECs represent a distinct LEC lineage and help to clarify the plasticity, heterogeneity, and special functions of LECs in the meninges.
Previous scRNAseq data of whole embryos/larvae and adult telencephalon showed that epd is specifically expressed in one-cell cluster34,35,36, which also expresses other mLSC markers used in our study. Here, we validated the transcriptomic specificity of mLSCs by re-analyzing scRNAseq data from Farnsworth et al.34. Besides, we verified that mLSCs in embryos/larvae did not express common meningeal cell markers by bulk RNA sequencing (Fig. 3e) and confirmed morphological differences between mLSCs and other common meningeal cells using multiple transgenic lines (Fig. 2). Furthermore, apoptosis assays confirmed that ablation of mLSCs do not lead to widespread cell death, especially not in BECs and muLECs (Supplementary Figs. 7 and 9). Thus, these findings provide evidence for the transcriptional and anatomical specificities of mLSCs.
Although Ependymin is a protein specific in fish, several molecular markers of mLSCs are conserved in mammals such as slc13a4, igfbp2a, and nid1b, providing preliminary evidence to implicate the existence of a similar cell type in mammals. It is important to note that the mammalian leptomeninges feature a more complex structure and cellular composition than those of zebrafish, therefore the mLSC marker-expressing cells in mammals could format a different structure and carry out functions in a more complex manner. After the discovery of meningeal lymphatics, recent findings of novel lymphatic functions in several tissues and identification of related cell types further enhance our understanding of lymphatic diversities and functional heterogeneities in different organs and tissues47,48,49,50. For example, recent findings that lymphatics in bone are important for bone regeneration have added new insights into the organ-specific functions and molecular specialization of lymphatics47.
Methods
Study approval
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Southwest University Laboratory Animal Center. All animal procedures followed standard conditions in accordance with the regulations of the Ethics Committee of Southwest University (Chongqing, China).
Zebrafish handling and strains
All zebrafish lines were maintained and raised under standard laboratory conditions. In order to inhibit pigmentation, zebrafish embryos were treated with 0.003% 1-phenyl-2-thiourea (PTU, Sigma–Aldrich) from 24 hpf, or alternatively, their parents were crossed into the casper background.
The transgenic zebrafish lines generated in this study were Tg(epd:EGFP)cq188, Tg(epd:EGFP-NTR)cq189, Tg(epd:mCherry-Ras)cq190, Tg(epd:mCherry-NTR)cq191, Tg(epd:H2B-GFP)cq192, Tg(epd:H2B-mCherry)cq193, Tg(lyve1b:EGFP-NTR)cq194, and Tg(sox10:EGFP-NTR)cq195. The previously published zebrafish lines used were Tg(lyve1b:DsRed)cq27 (refs. 17,21), Tg(lyve1b:EGFP)cq86 (ref. 18), Tg(abcc9BAC:Gal4ff)ncv34 (ref. 25), Tg(pdgfrbBAC:GFP)ncv22 (ref. 26), Tg(acta2:GFP)ca7 (ref. 27), Tg(coro1a:Kaede)cq22 (ref. 28), Tg(lyz:GFP)nz117 (ref. 29), Tg(mpeg1:GFP)gl22 (ref. 30), Tg(prox1aBAC:KalTA4; UAS:TagRFP)nim5 (ref. 31), Tg(nkx2.2a:GFP)ia3 (ref. 32), Tg(elavl3:GFP) knu3 (ref. 33), Tg(kdrl:mCherry-Ras)s896 (ref. 51), Tg(fli1:GFP)y1 (ref. 52), Tg(kdrl:GFP)s843 (ref. 53), Tg(fli1:nEGFP)y7 (ref. 54), and casper 55. All the lines used in this study were stable and breeding transgenic lines. The strains, numbers, and ages of zebrafish used in each experiment are described in the corresponding figures and legends. All zebrafish were used with an equal number of males and females when sex determination has occurred.
Generation of plasmids and transgenic lines
To generate reporter lines driven by the epd promoter, the ~3.9 kb upstream DNA sequence of the epd gene was amplified from the wild-type zebrafish genomic DNA. The ApaI and AgeI digest sites were added to the 5′ and 3′ end of the amplified sequence, respectively. The epd promoter DNA was digested with ApaI and AgeI enzymes, serving as the insert fragment. Additionally, the plasmid pBluescript2KS(-)_krt18:EGFP-NTR was used as the vector, which had been digested with ApaI, AgeI, and EcoRI enzymes. Subsequently, the digested fragments were ligated with T4 DNA ligase resulting in the formation of the plasmid pBluescript2KS(-)_epd:EGFP-NTR. The sequences encoding EGFP, mCherry-NTR, H2B-mCherry, H2B-GFP, and mCherry-Ras were amplified by PCR before being inserted into the 3′ end of epd promoter through subcloning, resulting in the generation of epd:EGFP, epd:mCherry-NTR, epd:H2B-mCherry, epd:H2B-GFP, and epd:mCherry-Ras constructs. These plasmids were respectively co-injected with I-SceI and 10× I-SceI buffer into one-cell-stage wild-type zebrafish embryos to obtain F0. F1 was screened from the descendant embryos generated from the cross of F0 and wild-type based on the correct expression position of the respective fluorescent proteins.
For the generation of Tg(lyve1b:EGFP-NTR) zebrafish line, the plasmid pT2KXIGDin_lyve1b:EGFP was digested with ClaI enzyme and the plasmid pBluescript2KS(-)_cldn15lb-1:EGFP-NTR was digested with NotI-HF enzyme. Then, the two digested DNA fragments were blunted with Klenow to eliminate the sticky ends created by the endonucleases. Subsequently, both blunted DNA fragments were subjected to AgeI-HF digestion. Finally, the produced vector fragment pT2KXIGDin_lyve1b and the insert fragment EGFP-NTR were ligated with T4 DNA ligase to obtain the plasmid pT2KXIGDin_lyve1b:EGFP-NTR. F0 was generated by co-injecting the plasmid pT2KXIGDin_lyve1b:EGFP-NTR with Tol2 transposase RNA (40–50 pg) into one-cell-stage wild-type zebrafish. F0 and wild-type zebrafish were crossed to select F1 possessing the correct fluorescence expression.
To generate the Tg(sox10:EGFP-NTR) zebrafish line, the ~5.1 kb sox10 promoter was amplified from the genomic DNA of wild-type zebrafish. The insert fragment was obtained by digesting the sox10 promoter with ApaI and AgeI enzymes, while the vector fragment was obtained by digesting the plasmid pBluescript2KS(-)_krt18:EGFP-NTR with ApaI, AgeI, and EcoRI enzymes. Subsequently, T4 DNA ligase was used to ligate the insert and vector fragments, resulting in the generation of the plasmid pBluescript2KS(-)_sox10:EGFP-NTR. F0 was generated by co-injecting the plasmid pBluescript2KS(-)_sox10:EGFP-NTR with I-SceI and 10× I-SceI buffer into wild-type zebrafish embryos at one-cell stage. F1 with correct fluorescence expression was selected from the offspring of the cross between F0 and wild-type zebrafish.
Information about primers for plasmid construction is provided in Supplementary Table 1.
Whole-mount in situ hybridization (WISH) and antibody staining
WISHs were performed as previously described56. Briefly, larval zebrafish were fixed in 4% paraformaldehyde (PFA) at 4 °C for 24 h. They were then dehydrated with 100% methanol and incubated at −30° overnight. The dehydrated larvae were rehydrated with methanol and 1× PBT (1× PBS with 0.1% Tween 20). Rehydrated larvae were then digested with proteinase K (10 µg/ml in 1× PBT) at room temperature (RT) for 30 min. Followed by incubation with 4% PFA again for half an hour at RT. Then digested larvae were prehybridized with 100% HYB at 68.5 °C for 5 h and hybridized with HYB-containing probe at 68.5 °C overnight. Subsequently, the larvae were washed at 68.5 °C with preheated HYB, 2× SSCT, and 0.2× SSCT and washed with 0.2× SSCT and 1× MABT at RT. Then they were blocked with blocking buffer for 2 h at RT and incubated with Anti-digoxigenin AP, Fab fragment (1:2000, 11093274910, Roche) at 4 °C overnight. Finally, the larvae were washed with 1× MABT and NTMT at RT and incubated with NBT/BCIP solution (11681451001, Roche) (0.2% NBT/BCIP stock solution in 1× MABT) at 37 °C. For probes (epd, ggctb, slc13a4, apof, nid1b, slc7a2, soul5, and igfbp2a), templates were acquired by PCR amplification of cDNA with a T7 promoter sequence from 3-dpf AB strains. Information about primers for probe synthesis is provided in Supplementary Table 1.
Antibody staining was performed following previously established protocols57,58,59. Briefly, zebrafish larvae and dissociated adult brains were fixed in 4% formaldehyde at 4 °C for 24 h. Larval zebrafish were dissociated to obtain brains after being fixed. Subsequently, the specimens were rinsed in 1× PT (1× PBS with 1% TrintonX-100) at RT and blocked with 1× PBTN (1× PT with 4% BSA) at 4 °C for 2 h. Then they were incubated with primary antibodies at 4 °C overnight. Primary antibodies used were: anti-GFP (1:2000, ab6658, Abcam), anti-mCherry (1:2000, ab125096, Abcam), anti-DsRed2 (1:2000, sc-101526, Santa Cruz), and Anti-Collagen I (1:1000, ab23730, Abcam). After that, the specimens were washed with 1× PT at RT and then stained with secondary antibodies at 4 °C overnight. Secondary antibodies used were: Donkey anti-goat IgG Alexa fluor 488-conjugated (1:2000, A11055, Invitrogen), Donkey anti-mouse IgG Alexa fluor 568-conjugated (1:2000, A10037, Invitrogen), and Donkey anti-rabbit IgG Alexa fluor 568-conjugated (1:2000, A10042, Invitrogen). Finally, the specimens were washed with 1× PT and then stained with DAPI (D8417, Sigma–Aldrich) at RT for 30 min. Information for the antibodies is summarized in Supplementary Table 2.
Fluorescence in situ hybridization (FISH) combined with antibody staining (FISH-antibody staining)
FISH-antibody stainings were performed as previously described60. In brief, after being fixed in 4% PFA for 24 h, the zebrafish larvae were dissected and their brains were obtained and dehydrated with 100% methanol. Subsequently, the brains were rehydrated with methanol and 1× PBT, then digested with proteinase K at RT for 5 min, followed by incubation with 4% PFA again at RT for 20 min. After that, the brains were prehybridized with 100% HYB at 65 °C for 5 h, and then hybridized with a HYB-containing probe at 65 °C overnight. Then, the brains were washed with preheated HYB, 2× SSCT, and 0.2× SSCT at 65 °C, and washed with 0.2× SSCT and 1× MABT at RT. Followed by blocked brains with blocking buffer and incubated with Anti-digoxigenin POD, Fab fragment (1:2000, 11207733910, Roche) at 4 °C overnight. The brains were then washed with 1× MABT and 1× PBS at RT and then incubated with Cyanine 3 in amplification diluent buffer (1:50, NEL701A001KT, PerkinElmer) at RT overnight. The subsequent procedures were performed according to the antibody staining procedure. The probes used were identical to those employed in WISHs. The antibodies utilized were anti-GFP (1:2000, ab6658, Abcam) and Donkey anti-goat IgG Alexa fluor 488-conjugated (1:2000, A11055, Invitrogen).
Vibratome section of zebrafish brains
The fixed adult brains of zebrafish were mounted in the 4% low melting agarose and sliced at 200 µm using LEICA VT1000S vibratome.
Tissue isolation, FAC sorting, and data analysis for RNA sequencing of mLSCs and zebrafish embryos/larvae
Heads of transgenic Tg(epd:EGFP-NTR; lyve1b:DsRed) zebrafish at 55 hpf and 5 dpf were separately dissected and placed in 1 mL 1× PBS on ice. They were then washed with 1 mL 1× PBS once and centrifuged at 4000 × g at 4 °C for 2 min. Next, the supernatant was removed and heads were dissociated with a mixture solution of 200 µL PBS-EDTA (1 mM EDTA in 1× PBS) and 50 µL 2.5% trypsin. The homogenized cell suspension was centrifuged at 4000 × g at 4 °C for 2 min before being washed with 1 mL 1× PBS twice. The cells were then resuspended in 200 µL 1× PBS within 5 min and the cell suspension was collected by filtering through a 40 µm cell strainer into a 2 mL EP tube. Wild-type zebrafish heads at 55 hpf and 5 dpf were operated using the same dissociation procedure as a fluorescent-negative control for FAC sorting. Subsequently, cell sorting was performed using flow cytometry (Moflo XDP, Beckman) to obtain 55 hpf and 5 dpf of EGFP-NTR (+) DsRed (−) cells as two biological replicates. Next, single-cell transcriptomic amplification was performed on the obtained cells. All tissue isolation procedures were carried out on ice. The sorted cells were harvested directly in a single-cell collection solution containing cell lysis components and RNase inhibitors. The 1st cDNA was generated via reverse transcription using a nucleic acid sequence with Oligo dT, followed by PCR amplification to enrich the nucleic acid and purify the amplified product for library construction. For control tissues of 55 hpf or 5 dpf, there were three biological replicates per period, each containing one hundred zebrafish. Total RNA was extracted from whole zebrafish by Trizol-Chloroform-Isopropanol isolation and then purified by magnetic beads with Oligo dT. The purified mRNA was fragmented by adding a fragmentation buffer to create short fragments, which were then used as templates for constructing cDNA libraries. Constructed libraries were sequenced using the Illumina platform with the sequencing strategy PE150. The raw data aligned to the zebrafish (Danio rerio) reference genome (GRCz11.96). Data analysis was carried out as previously described28. In brief, Clean Reads are obtained by processes such as removing Raw Reads from low-quality sequences to complete data processing, and all subsequent analysis is based on Clean Reads. Gene enrichment ontology analysis of highly expressed genes in epd-positive cells relative to whole fish was performed using Metascape (https://metascape.org/gp/index.html#/main/step1)61. The top 500 protein-coding genes in 55 hpf as well as 5-dpf epd-positive cells with padj <0.05 and ranked in descending order by Log2FoldChange were selected for gene enrichment ontology analysis.
Data analysis of single-cell RNA sequencing
The single-cell RNA-sequencing (scRNAseq) data analyzed in this paper were mined from Farnsworth et al.34. ScRNAseq data were preprocessed and normalized using the R package “Seurat v4.3.0”. The epd-positive cells, fibroblasts, mural cells, neural progenitor cells, immune cells, and endothelial cells were picked out for further analysis according to the classical marker genes of each cell type. The Uniform Manifold Approximation and Projection (UMAP) plot was obtained by dimensionality reduction analysis using the RunUMAP function (R package “Seurat v4.3.0”). The FindAllMarkers function (R package “Seurat” v4.3.0) was used to identify the highly expressed genes in each cluster, and the genes with padj <0.05 were taken to be sorted by avg_log2FC, and the genes in the top 50 avg_log2FC were taken to be plotted by heatmap (R package “pheatmap 1.0.12”), take the genes in the top 20 of avg_log2FC of epd-positive cell population and plot them by dotplot (R package “Seurat v4.3.0”). Violin plot of each cell-specific marker gene using the VlnPlot function in the R package “Seurat v4.3.0”. For Principle component analysis (PCA), the epd-positive cells in scRNAseq data were first split into 3 groups for pseudo-bulk process according to the mean value of expression, then ComBat_seq in R package “sva 3.42.0” was used to eliminate the batch effect on bulk RNA-sequencing data (epd-positive cells at 55 hpf and 5 dpf, and whole zebrafish at 55 hpf and 5 dpf) generated in this study and the epd-positive cells in single-cell RNA-sequencing data that were split into 3 groups, and finally PCA clustering analysis was performed with R package “ape 5.7.1”.
Imaging
Larvae in which whole-mount in situ hybridization was performed were imaged using a ZEISS SteREO Discovery.V20 microscope. Antibody-stained brains or brain sections, FISH-antibody-stained brains, live embryos, live larvae, or live juveniles were mounted in 1% low melting point agarose. They were then imaged with either a ×10 air objective or a ×20 water immersion objective installed in the ZEISS LSM780 or LSM 880 confocal microscope. Live embryos or larvae were mounted in 1% low melting point agarose and then time-lapse imaged using the Zeiss Lightsheet Z.1 or ZEISS LSM 880 confocal microscope.
Chemical treatment
In the treatment of metronidazole (Mtz, Sigma–Aldrich), the Mtz was completely dissolved in egg water that contained 0.2% dimethyl sulfoxide (DMSO). Within a Tg(epd:EGFP-NTR) or Tg(epd:mCherry-NTR) background, embryos at 48 hpf and larvae at 5 dpf were incubated with 10 mM Mtz for 24 h, while juveniles at 25 dpf were incubated with 5 mM Mtz for 22 h; within a single Tg(lyve1b:DsRed) background, embryos at 48 hpf were incubated with 10 mM Mtz for 24 h, while larvae at 5 dpf were incubated with 5 mM Mtz or 10 mM Mtz for 24 h; within a Tg(sox10:EGFP-NTR) background, larvae at 5 dpf were incubated with 5 mM Mtz for 24 h; within a Tg(lyve1b:EGFP-NTR) background, larvae at 5 dpf were incubated with 5 mM Mtz for 20 h. For muLEC ablation, larvae with a large number of muLECs ablated on TeO and some other muLECs retained need to be selected for analysis. Each ablated group was incubated with 0.2% DMSO in the egg water as a control. Subsequently, the embryos, larvae, or juveniles were washed three times with egg water and recovered in egg water, and marked as 0 dpt.
Dye injection
The dye injection was carried out as previously described14,17,18. Briefly, the dye is injected directly into the dorsal aorta or into the intracerebroventricular of zebrafish using a glass capillary needle. The dyes used were IgG-conjugated Alexa fluor 647 (150 kDa) (2 mg/ml, A31573, Invitrogen) and Alexa647-Dextran (10 kDa) (2 mg/ml, D22914, Invitrogen).
TUNEL assay
The TUNEL assay for the detection of cell apoptosis was performed as previously described17. In brief, zebrafish larvae underwent fixation, dehydration, rehydration, brain dissected, and digestion in accordance with the procedures of FISHs. Then the brains were treated with acetone at −20 °C for 1 h and subsequently were fixed with 4% PFA for 20 min at RT. The brains were sequentially incubated with buffer and enzyme from In Situ Cell Death Detection Kit (12156792910, Roche) at 37 °C for 2 h and then were washed with preheated 2× SSCT at 37 °C and washed with 2× SSCT and 1× PBT at RT.
Statistics and reproducibility
All confocal imaged pictures and WISHs represented at least 3 independent experiments with similar results. The numbers of animals represented by the images were indicated in the corresponding figure legends. The number of specimens used for RNA sequencing along with the number of biological parallels were described in the corresponding figure legends. The statistical calculations were performed using GraphPad Prism 8.0.1. The statistical data were analyzed by unpaired two-tailed Student’s t-test and two-way ANOVA Sidak’s multiple comparisons test. All “n” and “P” values as well as statistical tests are presented in the corresponding figures and legends. The results with P values less than 0.05 were considered to be statistically significant. With the exception of Fig. 7q, Supplementary Fig. 8a–d, each dot and number in the remaining statistics figures represents an independent fish. All movies were handled with Fiji (ImageJ) 2.14.0 and ZEN2010 Imaging software. The heatmaps in Fig. 3e, f were created using Heatmap Illustrator (HemI) version 1.0.3.7 software62.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-sequencing data generated in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center under the accession code GSA: CRA016778. Previous published scRNAseq data that were re-analyzed in this study are available in the National Centre for Biotechnology (NCBI) SRA: PRJNA56481034. All other relevant data that support the findings of this study are available within the article, its Supplementary Information, and its Supplementary Data, or from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Coles, J. A., Myburgh, E., Brewer, J. M. & McMenamin, P. G. Where are we? The anatomy of the murine cortical meninges revisited for intravital imaging, immunology, and clearance of waste from the brain. Prog. Neurobiol. 156, 107–148 (2017).
Rua, R. & McGavern, D. B. Advances in meningeal immunity. Trends Mol. Med. 24, 542–559 (2018).
Alves de Lima, K. A., Rustenhoven, J. & Kipnis, J. Meningeal immunity and its function in maintenance of the central nervous system in health and disease. Annu. Rev. Immunol. 38, 597–620 (2020).
Bifari, F. et al. Meninges harbor cells expressing neural precursor markers during development and adulthood. Front. Cell Neurosci. 9, 383 (2015).
Mastorakos, P. & McGavern, D. The anatomy and immunology of vasculature in the central nervous system. Sci. Immunol. 4, eaav0492 (2019).
DeSisto, J. et al. Single-cell transcriptomic analyses of the developing meninges reveal meningeal fibroblast diversity and function. Dev. Cell 54, 43–59 (2020).
Oliver, G., Kipnis, J., Randolph, G. J. & Harvey, N. L. The lymphatic vasculature in the 21st century: novel functional roles in homeostasis and disease. Cell 182, 270–296 (2020).
Aspelund, A. et al. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J. Exp. Med. 212, 991–999 (2015).
Louveau, A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015).
Louveau, A. et al. CNS lymphatic drainage and neuroinflammation are regulated by meningeal lymphatic vasculature. Nat. Neurosci. 21, 1380–1391 (2018).
Absinta, M. et al. Human and nonhuman primate meninges harbor lymphatic vessels that can be visualized noninvasively by MRI. eLife 6, e29738 (2017).
Castranova, D. et al. Live imaging of intracranial lymphatics in the zebrafish. Circ. Res. 128, 42–58 (2021).
Venero Galanternik, M. et al. A novel perivascular cell population in the zebrafish brain. eLife 6, e24369 (2017).
van Lessen, M. et al. Intracellular uptake of macromolecules by brain lymphatic endothelial cells during zebrafish embryonic development. eLife 6, e25932 (2017).
Bower, N. I. et al. Mural lymphatic endothelial cells regulate meningeal angiogenesis in the zebrafish. Nat. Neurosci. 20, 774–783 (2017).
Huisman, Y. et al. Meningeal lymphatic endothelial cells fulfill scavenger endothelial cell function and cooperate with microglia in waste removal from the brain. Glia 70, 35–49 (2022).
Chen, J. et al. Cerebrovascular injuries induce lymphatic invasion into brain parenchyma to guide vascular regeneration in zebrafish. Dev. Cell 49, 697–710.e695 (2019).
Chen, J. et al. Acute brain vascular regeneration occurs via lymphatic transdifferentiation. Dev. Cell 56, 3115–3127 e3116 (2021).
Shashoua, V. E. Ependymin, a brain extracellular glycoprotein, and CNS plasticity. Ann. N. Y. Acad. Sci. 627, 94–114 (1991).
Sterrer, S., Konigstorfer, A. & Hoffmann, W. Biosynthesis and expression of ependymin homologous sequences in zebrafish brain. Neuroscience 37, 277–284 (1990).
Okuda, K. S. et al. lyve1 expression reveals novel lymphatic vessels and new mechanisms for lymphatic vessel development in zebrafish. Development 139, 2381–2391 (2012).
Zhao, Z., Nelson, A. R., Betsholtz, C. & Zlokovic, B. V. Establishment and dysfunction of the blood-brain barrier. Cell 163, 1064–1078 (2015).
Vanlandewijck et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018).
Holm, A., Heumann, T. & Augustin, H. G. Microvascular mural cell organotypic heterogeneity and functional plasticity. Trends Cell Biol. 28, 302–316 (2018).
Ando, K. et al. Peri-arterial specification of vascular mural cells from naive mesenchyme requires Notch signaling. Development 146, dev165589 (2019).
Ando, K. et al. Clarification of mural cell coverage of vascular endothelial cells by live imaging of zebrafish. Development 143, 1328–1339 (2016).
Whitesell, T. R. et al. An α-smooth muscle actin (acta2/alphasma) zebrafish transgenic line marking vascular mural cells and visceral smooth muscle cells. PLoS ONE 9, e90590 (2014).
Liu, C. et al. Macrophages mediate the repair of brain vascular rupture through direct physical adhesion and mechanical traction. Immunity 44, 1162–1176 (2016).
Hall, C., Flores, M. V., Storm, T., Crosier, K. & Crosier, P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 7, 42 (2007).
Ellett, F., Pase, L., Hayman, J. W., Andrianopoulos, A. & Lieschke, G. J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56 (2011).
van Impel, A. et al. Divergence of zebrafish and mouse lymphatic cell fate specification pathways. Development 141, 1228–1238 (2014).
Pauls, S., Zecchin, E., Tiso, N., Bortolussi, M. & Argenton, F. Function and regulation of zebrafish nkx2.2a during development of pancreatic islet and ducts. Dev. Biol. 304, 875–890 (2007).
Park, H. C. et al. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol. 227, 279–293 (2000).
Farnsworth, D. R., Saunders, L. M. & Miller, A. C. A single-cell transcriptome atlas for zebrafish development. Dev. Biol. 459, 100–108 (2020).
Sur, A. et al. Single-cell analysis of shared signatures and transcriptional diversity during zebrafish development. Dev. Cell 58, 1–20 (2023).
Pandey, S., Moyer, A. J. & Thyme, S. B. A single-cell transcriptome atlas of the maturing zebrafish telencephalon. Genome Res. 33, 658–671 (2023).
Curado, S. et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev. Dyn. 236, 1025–1035 (2007).
He, J., Lu, H., Zou, Q. & Luo, L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 146, 789–800.e788 (2014).
Kirby, B. B. et al. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat. Neurosci. 9, 1506–1511 (2006).
Preston, M. A., Finseth, L. T., Bourne, J. N. & Macklin, W. B. A novel myelin protein zero transgenic zebrafish designed for rapid readout of in vivo myelination. Glia 67, 650–667 (2019).
Plikus, M. V. et al. Fibroblasts: origins, definitions, and functions in health and disease. Cell 184, 3852–3872 (2021).
Hu, C., Tao, L., Cao, X. & Chen, L. The solute carrier transporters and the brain: physiological and pharmacological implications. Asian J. Pharm. Sci. 15, 131–144 (2020).
Tsai, H. H. et al. Oligodendrocyte precursors migrate along vasculature in the developing nervous system. Science 351, 379–384 (2016).
Cattin, A. L. et al. Macrophage-induced blood vessels guide schwann cell-mediated regeneration of peripheral nerves. Cell 162, 1127–1139 (2015).
Petrova, T. V. & Koh, G. Y. Biological functions of lymphatic vessels. Science 369, eaax4063 (2020).
Wang, G. et al. Specific fibroblast subpopulations and neuronal structures provide local sources of Vegfc-processing components during zebrafish lymphangiogenesis. Nat. Commun. 11, 2724 (2020).
Biswas, L. et al. Lymphatic vessels in bone support regeneration after injury. Cell 186, 382–397 (2023).
Harrison, M. R. M. et al. Late developing cardiac lymphatic vasculature supports adult zebrafish heart function and regeneration. eLife 8, e42762 (2019).
Liu, X. et al. Lymphoangiocrine signals promote cardiac growth and repair. Nature 588, 705–711 (2020).
Goto, N. et al. Lymphatics and fibroblasts support intestinal stem cells in homeostasis and injury. Cell Stem Cell 29, 1246–1261.e1246 (2022).
Chi, N. C. et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 22, 734–739 (2008).
Lawson, N. D. & Weinstein, B. M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318 (2002).
Jin, S. W., Beis, D., Mitchell, T., Chen, J. N. & Stainier, D. Y. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209 (2005).
Roman, B. L. et al. Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development 129, 3009–3019 (2002).
White, R. M. et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189 (2008).
Liu, X. et al. NF-kappaB and Snail1a coordinate the cell cycle with gastrulation. J. Cell Biol. 184, 805–815 (2009).
Lu, H., Ma, J., Yang, Y., Shi, W. & Luo, L. EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev. Cell 24, 543–553 (2013).
Yang, Y. et al. A single-cell-resolution fate map of endoderm reveals demarcation of pancreatic progenitors by cell cycle. Proc. Natl Acad. Sci. USA 118, e2025793118 (2021).
Yang, Y. et al. Intestinal precursors avoid being misinduced to liver cells by activating Cdx-Wnt inhibition cascade. Proc. Natl Acad. Sci. USA 119, e2205110119 (2022).
He, J., Mo, D., Chen, J. & Luo, L. Combined whole-mount fluorescence in situ hybridization and antibody staining in zebrafish embryos and larvae. Nat. Protoc. 15, 3361–3379 (2020).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Deng, W. K., Wang, Y. B., Liu, Z. X., Cheng, H. & Xue, Y. HemI: a toolkit for illustrating heatmaps. PLoS ONE 9, e111988 (2014).
Acknowledgements
We thank J He, J Chen, Y Yang, X Wei, P Cai for helpful discussions. This work was supported by the National Key R&D Program of China (2021YFA0805000) to L.L., and the National Natural Science Foundation of China (32192400) to L.L.
Author information
Authors and Affiliations
Contributions
L.L., X.H., and D.X. designed the experimental strategies, analyzed data, and wrote the manuscript. L.Z. conducted analyses of RNA-sequencing data. J.F. generated the pericyte-labeling transgenic line. X.H. and D.X. performed all the other experiments in this study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, X., Xiong, D., Zhao, L. et al. Meningeal lymphatic supporting cells govern the formation and maintenance of zebrafish mural lymphatic endothelial cells. Nat Commun 15, 5547 (2024). https://doi.org/10.1038/s41467-024-49818-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-49818-5