Abstract
Photosystem I (PSI) from Fittonia albivenis, an Acanthaceae ornamental plant, is notable among green plants for its red-shifted emission spectrum. Here, we solved the structure of a PSI–light harvesting complex I (LHCI) supercomplex from F. albivenis at 2.46-Å resolution using cryo-electron microscopy. The supercomplex contains a core complex of 14 subunits and an LHCI belt with four antenna subunits (Lhca1–4) similar to previously reported angiosperm PSI–LHCI structures; however, Lhca3 differs in three regions surrounding a dimer of low-energy chlorophylls (Chls) termed red Chls, which absorb far-red beyond visible light. The unique amino acid sequences within these regions are exclusively shared by plants with strongly red-shifted fluorescence emission, suggesting candidate structural elements for regulating the energy state of red Chls. These results provide a structural basis for unraveling the mechanisms of light harvest and transfer in PSI–LHCI of under canopy plants and for designing Lhc to harness longer-wavelength light in the far-red spectral range.
Similar content being viewed by others
Introduction
Oxygenic photosynthesis is driven by photosystems I and II (PSI and PSII) in land plants, algae and cyanobacteria. In green plants, both photosystems have a common general organization comprising two moieties: (i) a peripheral antenna complex (light-harvesting complex I (LHCI) for PSI and LHCII for PSII) that has dual roles in light harvesting and dissipation of excess excitation energy and (ii) a reaction center complex (PSI or PSII core) that converts the excitation energy from the antenna system into chemical energy. In addition, both photosystems bind chlorophyll a (Chl a) and chlorophyll b (Chl b) as their light-harvesting pigments. Importantly, the two photosystems differ in their light-absorption range: the PSI–LHCI supercomplex can absorb some far-red light (longer than 700 nm) via a few low-energy Chl molecules (so-called red Chls), whereas the PSII–LHCII supercomplex cannot1. The red shift observed in the light-absorption range of the PSI–LHCI complex relative to that of PSII–LHCII reflects pigment properties, pigment–pigment and pigment–protein interactions, and differences in Chl arrangement, which depend on the structures of the two supercomplexes. The structures of the PSI–LHCI supercomplex have been resolved from pea (Pisum sativum, Ps) and maize (Zea mays, Zm) with improving resolution in the past two decades and have helped shape our understanding of the spectral properties and energy transfer mechanisms of PSI–LHCI in angiosperms2,3,4,5,6,7.
Based on the PSI–LHCI structures of flowering plants, the PSI core contains 14 photosystem subunits (PsaA–L, PsaN and PsaO) along with 98 Chl a and 22 β-carotene molecules; LHCI forms an arc-like belt composed of four light-harvesting antenna proteins, Lhca1, Lhca4, Lhca2 and Lhca3, in series when viewed from the stromal side (Supplementary Fig. 1). LHCI is anchored to the PSI core mainly through Lhca1–PsaG and Lhca3–PsaK interactions, with the two interacting regions being the main excitation energy transfer (EET) pathways from LHCI to the PSI core. The interactions between Lhca4 or Lhca2 and the PSI core are relatively weaker, which is consistent with the finding that EET from Lhca4 and Lhca2 to the PSI core relies on Lhca1 and the non-transmembrane PsaN subunit, respectively5,8,9. All four Lhcas share a similar protein structure characterized by three transmembrane helices (TMHs), A, B and C, forming 13 conserved Chl-binding sites and 1–2 unique Chl-binding sites. Notably, the four Lhcas do differ in their spectral properties, with reconstituted (r)-Lhca3 and r-Lhca4 showing more red-shifted absorption and fluorescence emission spectra than r-Lhca1 and r-Lhca210,11,12,13, suggesting that the energy state of red Chls in each rLhca can be different. The far-red spectral forms of PSI–LHCI were thought to originate from a mixing between the excited and charge-transfer states within tightly packed Chl dimers in early spectroscopic studies14,15,16,17,18, however, the precise mechanisms of the formation of these forms and the factors modulating their properties remain elusive. The Chl dimers were defined as Chls a3–a9 locating at the interfaces between each of the four Lhcas and the PSI core complex, and the special chlorophyll-protein interactions making the two Chl molecules within each dimer have a close distance and some overlap between their C and E rings5. It is important to understand the relationship between the environment surrounding Chls a3–a9 and their energy states. Indeed, Chl a9 binds to Glu in all four Lhcas, whereas the central Mg atom of Chl a3 is coordinated by His in Lhca1 and Lhca2 but by Asn in Lhca3 and Lhca4, as determined from structural comparisons. In vitro and more recent in vivo studies confirmed that Asn is necessary for maintaining the right geometry between the two packed Chls within the dimer to allow strong interaction15,16,19,20,21. However, other structural reasons likely regulate the extent of spectral red shift, as some flowering plants differing in the wavelength at which they show maximal fluorescence emission (λmax) share the same ligands for the Chl dimer22.
In agriculture, the far-red spectral forms of PSI–LHCI are associated with the utilization ratio of far-red light, which is usually very low. The top layer of a canopy generally absorbs more red light than far-red light, resulting in far-red light enrichment within or under a canopy23,24. The feasibility of increasing crop biomass by expanding the absorption spectrum into far-red wavelengths is receiving increasing attention25,26,27,28, along two possible lines of attack: borrowing unique pigments from prokaryotes, and modulating pigment–pigment and pigment–protein interactions without changing pigment types. Although some photosynthetic prokaryotes can synthesize Chl d and Chl f to capture a large amount of far-red light26,29,30,31,32,33,34, production of these pigments, which differ chemically from Chl a, in flowering plants is difficult and has never been successfully reported. To explore the second strategy, scientists have been searching for green plants with more prominent absorption of far-red light than most plants by collecting low-temperature (77-K) fluorescence emission spectra, a very sensitive method that reveals even small changes in the energy state of red Chls. Using λmax of Arabidopsis thaliana (At) as a baseline, λmax of the crop species barley (Hordeum vulgare) and the houseplant Calathea roseopicta exhibit a red shift of 6 nm and 9 nm, respectively, with the latter being the longest λmax reported to date. However, these two species have identical absorption spectra22,35. Therefore, it is necessary to find other flowering plants with longer λmax to exploit these natural resources to better use far-red light.
We posited that plants with strong far-red absorption may grow in shaded locations, as far-red light is enriched under canopy. In a broad search of shade-tolerant plants, we discovered that the houseplant known as nerve plant (Fittonia albivenis), in the Acanthaceae family, has a λmax of 753 nm and marked far-red absorption; ten Acanthaceae plants tested in this study also possessed λmax longer than 748 nm. We determined that purified photosynthetic complexes from F. albivenis (Fa) with long-wavelength 77-K fluorescence emission correspond to PSI–LHCI and analyzed their structure using single-particle cryo-electron microscopy (cryo-EM) to an overall resolution of 2.46 Å. Our structure not only revealed the detailed organization and pigment arrangement of Fa PSI–LHCI, a unique PSI–LHCI structure with strong far-red shift in fluorescence spectra of red Chls, but also provided important clues to the underlying structural reasons for red-shift in absorption and fluorescence spectra of Chls. These results also offer valuable suggestions to help redesign Lhc proteins to modulate the light-absorption range of plants.
Results
Several Acanthaceae species exhibit a far-red shift in their fluorescence spectra
We obtained plants from three understory traditional Chinese medicine species, Panax ginseng, Panax notoginseng and Dendrobium nobile, and two houseplants, Epipremnum aureum and Fa, to test their ability to use far-red light for photosynthesis by measuring their 77-K fluorescence emission spectra. Whereas At has a λmax of 735 nm, we observed a mean red shift for λmax of ~11.2 nm, ranging from 3.7 nm (D. nobile) to 18 nm for Fa, with a peak of 753 nm (Fig. 1a, b). We further determined the absorption spectra of two fluorescence red-shifted leaves, Fa and Epipremnum aureum, and the results showed that both had significant red-shifts, with Fa being the more red-shifted. The difference spectra between Fa and At showed that Fa absorption more light longer than 694 nm (Fig. 1c). To avoid possible interference from chlorophyll concentration on fluorescence spectroscopy, we repeated the 77-K fluorescence emission spectra using isolated chloroplasts and thylakoid membranes from Fa leaves. We detected one main red-shifted peak at 753.6 nm from Fa (Fa) chloroplasts, together with additional peaks at 685 nm and 695 nm (Fig. 1d). Purified Fa thylakoid membranes showed a similar fluorescence emission pattern but with a more moderate red shift at 750.4 nm (Fig. 1e). We also determined the absorption spectrum of thylakoids from Fa and At at room temperature to mimic normal growth conditions: the difference between the two spectra revealed a higher absorption spectrum for Fa thylakoids compared to At thylakoids in four main wavelength ranges, 690–750 nm, 645–655 nm, 465–500 nm and 440-465 (Fig. 1f). Specifically, we observed two major peaks at 488.5 nm and 650.5 nm, suggesting that Fa thylakoids contain more Chl b molecules than At thylakoids, as well as a peak at ~693.5 nm and a shoulder at 718 nm, which could be caused by the presence of more red-shifted Chls (red Chls) in Fa thylakoids than in At thylakoids. These results suggest that Fa chloroplasts absorb more far-red light than those of At, offering a possible explanation for the strong red-shifted fluorescence emission spectrum obtained in this species.
a Low-temperature (77-K) fluorescence emission spectra of Arabidopsis thaliana (At), three understory traditional Chinese medicine plants (P. ginseng, P. notoginseng and D. nobile) and two houseplants (E. aureum and F. albivenis [Fa]). b Wavelength of maximum fluorescence emission (λmax) of the plant species from (a) Error bars (standard deviation) were calculated from three different experimental units (n = 3 independent experiments), and data were presented as mean values ± SD. c The absorption spectra of leaves from At, Ea and Fa. The absorption spectra were normalized to the maximum in the red region, which was set to 1. The differential absorption spectrum between Fa and At is shown as a dashed line. d, e 77-K fluorescence emission spectra of chloroplast (d) and thylakoid membranes (e) from Arabidopsis and Fa. f Absorption spectra at room temperature of At thylakoids and Fa thylakoids. The differential absorption spectrum between Fa thylakoids and At thylakoids (multiplied by 5) is shown as a dashed line, with peaks at 456.5 nm, 488.5 nm, 650 nm, and 693.5 nm and the shoulder at 718 nm indicated by arrows. g 77-K fluorescence emission spectra of the ten Acanthaceae plants Fa, S. cusia, J. brandegeeana, P. curviflorus, B. cristata, R. tuberosa, C. infundibuliformis, H. phyllostachya, A. squarrosa and A. paniculata. h Wavelength of maximum fluorescence emission for plants shown in g Error bars (standard deviation) were calculated from three different experimental units (n = 3 independent experiments), and data were presented as mean values ± SD. The fluorescence emission spectra were collected following excitation at 440 nm and normalized with respect to their wavelength at maximal emission in the far-red region (750 nm), which was set to 1. The data are based on three independent experiments, each producing similar results.
The strongly red-shifted fluorescence emission detected using leaves, chloroplasts and thylakoid membranes of Fa (Fig. 1a–e) is unique among land plants. Fa is a native of the rainforests of South America that belongs to the family Acanthaceae. We therefore measured the 77-K fluorescence emission spectra of nine other Acanthaceae species. Notably, all 10 species showed enhanced far-red fluorescence emission, ranging from 748 nm in C. infundibuliformis to over 750 nm in the other species (Fig. 1g, h). We conclude that the pronounced red shift in the fluorescence emission spectra may be an important feature of the Acanthaceae.
Fluorescence emission red shift is associated with the PSI–LHCI in Fa
As Fa has soft, thick leaves that are easily ground for isolation of chloroplasts and thylakoid membranes, we chose this species as the source from which to isolate photosynthetic pigment–protein complexes and explore the reason for the red-shifted fluorescence (Supplementary Fig. 2a). As control, we purified pigment–protein complexes from At thylakoids. Sucrose density ultracentrifugation of At thylakoids treated with the detergent n-dodecyl-β-d-maltoside (β-DDM) resulted in four pigment-containing bands (Supplementary Fig. 2b), whose polypeptide composition we analyzed by SDS-PAGE. Band 1At and Band 2At were close together in the sucrose gradient and were assigned as LHCII monomers and trimers, respectively36. Band 3At contained mainly the PSI and PSII cores, whereas Band 4At comprised mainly PSI–LHCI (Supplementary Fig. 2c). The same isolation procedure applied to β-DDM-treated Fa thylakoids returned three main pigment-containing bands. Band 1Fa was much lighter than Band 2Fa, suggesting a low ratio of LHCII monomers to trimers. The lowest band in the gradient (Band 3Fa) migrated at the same position as Band 4At and contained mainly PSI–LHCI and a small fraction of PSII core. Finally, Fa thylakoids lacked a band corresponding to Band 3At (Supplementary Fig. 2b, c), suggesting differences in the organization of pigment–protein complexes between At and Fa thylakoid membranes.
We subjected all bands isolated from Fa thylakoid membranes to 77-K fluorescence emission spectroscopy (Supplementary Fig. 2d). Bands 1Fa and 2Fa each had one main emission peak at 680 nm, suggesting that LHCII does not contribute to the strongly red shift in chlorophyll fluorescence. In sharp contrast, Band 3Fa showed a major emission peak at 745.4 nm (F745) and a broad secondary peak at 685–695 nm (F685/695). As a peak around 690 nm is characteristic of PSII upon 77-K fluorescence spectrometry37, the F685/695 secondary peak is consistent with the presence of a small fraction of PSII core. The peak at 745 nm can thus be attributed to the PSI–LHCI supercomplex, suggesting that this supercomplex plays a role in the red shift in chlorophyll fluorescence in Fa. As λmax was 753 nm in leaves and chloroplasts but only 750 nm in purified thylakoid membranes and 745 nm for the PSI–LHCI supercomplex (Fig. 1 and Supplementary Fig. 2d), we suspect that additional factors may stabilize the far-red forms of PSI–LHCI in Fa. In addition, we determined the pigment composition of Fa PSI–LHCI by high-performance liquid chromatography (HPLC) analysis, resulting in the detection of Chl a, Chl b, violaxanthin, lutein and β-carotene (Supplementary Fig. 3), which are also present in the PSI–LHCI supercomplexes of At38, spinach (Spinacia oleracea)39, pea (Pisum sativum, Ps)5 and maize (Zea mays, Zm) thylakoid membranes8.
Despite the similar pigment composition of PSI–LHCI of Fa (Fa PSI–LHCI) and PSI–LHCI of At (At PSI–LHCI), their 77-K fluorescence emission spectra differed markedly, with λmax of 745.4 nm and 731.8 nm, respectively (Supplementary Fig. 4a), prompting us to measure their absorption spectra at room temperature. Unexpectedly, although Fa thylakoids displayed greater absorption than At thylakoids at wavelengths >700 nm, Fa PSI–LHCI and At PSI–LHCI had nearly identical absorption spectra at room temperature in the red and far-red regions, with small differences around 450–500 nm due to Chl b and carotenoids (Supplementary Fig. 4b). The loss of far-red absorption by PSI–LHCI suggests that the acting force of the thylakoid membrane helps control the red shift in fluorescence. Once PSI–LHCI particles dissociate from the thylakoid membrane, the spectral properties of red Chls from Fa and At are very similar based on absorption spectroscopy at room temperature. We therefore turned to 77-K fluorescence emission spectroscopy to characterize the spectral properties of PSI subfractions below.
The far-red fluorescence emission of Fa PSI–LHCI depends on interactions between the PSI core and LHCI
To further characterize the PSI core complex and its bound LHCI, we treated the PSI–LHCI sample with a combination of β-DDM and the zwitterionic detergent 3-(N,N-dimethylpalmitylammonio) propanesulfonate (zw 3-16) and then subjected the samples to sucrose density gradient ultracentrifugation. We detected three dark green bands (Bands 1, 2 and 3) and then assessed their polypeptide composition by SDS-PAGE (Supplementary Fig. 4c,d). Bands 1 and 2 from the sucrose density gradient resolved as two main bands of Lhca subunits, suggesting that Band 1 is a mixture of Lhca monomers (LHCI-1 thereafter), whereas Band 2 may be a polymer of Lhca subunits (LHCI-2 thereafter). We assigned Band 3 to the PSI core complex, as it mainly contained the largest subunits, PsaA and PsaB, and several small subunits of the PSI core. We determined the 77-K fluorescence emission spectra of Bands 1–3 (Supplementary Fig. 4e). The PSI core (Band 3) showed a main peak at 721.6 nm (F721.6), similar to the emission peak of the PSI core complex from some algae and flowering plants39,40,41,42,43. LHCI-1 (Band 1) displayed a main peak at 679.4 nm (F679.4) and a minor broad peak with a maximum between 730 nm and 740 nm, similar to the emission peak of LHCI-680, a LHCI fraction isolated from some flowering plants39,40,41,44,45. LHCI-2 (Band 2) showed a main peak at 686 nm (F686), as well as a secondary peak at 737.8 nm (F737.8) with an 8-nm red shift relative to the emission peak of LHCI-730, the other isolated LHCI fraction in some flowering plants39,40,41,44,45. Notably, the red-shifted F737.8 peak was still blue-shifted by ~8 nm compared to the main fluorescence peak of Fa PSI–LHCI at 745.4 nm (Supplementary Fig. 4e).
Thus, the fluorescence emission peaks of red Chls in Fa undergo a gradual blue shift from PSI–LHCI within the thylakoid membrane, to PSI–LHCI isolated from the thylakoid membrane, and to LHCI. We hypothesize that some intrinsic structural elements of Fa LHCI, absent from other plant LHCI, enable its red emission shift (737.8 nm). Looking at the whole PSI–LHCI supercomplex, the interactions of LHCI with the PSI core complex may enhance the extent of the red shift to reach 745.4 nm; finally, within thylakoids and chloroplasts, the binding of the PSI–LHCI supercomplex to the thylakoid membrane further increases the red shift to 750 nm and 753 nm, respectively. Thus, Fa likely possesses the strongest far-red-shifted fluorescence emission among land plants due to three consecutive enhancements of red-shift levels.
Architecture of the PSI–LHCI supercomplex from Fa
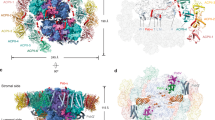
To reveal the detailed structure of Fa PSI–LHCI and explore the relationship between its structure and its observed red shift in fluorescence emission, we determined the structure of Fa PSI–LHCI by cryo-EM. Briefly, automated particle picking selected 3.7 million positions from 12,023 cryo-EM micrographs. Through multiple rounds of 2D classification, heterogeneous refinement and 3D variability analysis, we identified the most representative and highest-quality particles, from which we used a subset of 157,342 particles to reconstruct a cryo-EM map of Fa PSI–LHCI at a global resolution of 2.46 Å (Supplementary Fig. 5; Supplementary Table 1). We identified 18 protein subunits, 162 Chls, 37 carotenoids, three iron-sulfur (Fe4S4) clusters and nine lipids in the structure of the Fa PSI–LHCI supercomplex (Fig. 2, Supplementary Fig. 6 and Supplementary Table 2). We describe the structural features of the Fa PSI–LHCI supercomplex and their functional implications below.
a, b View along the membrane normal from the stromal side (a) and along the membrane plan from the LHCI side (b). Protein subunits are shown as ribbon models and colored differently. c Pigment arrangement in the Fa PSI–LHCI supercomplex. Chls are shown as stick models in the same color as their interacting protein subunits in a, and carotenoids are shown as stick models in light blue. d Distribution of lipids in Fa PSI–LHCI. Five phosphatidyl glycerol molecules (PG, green), three monogalatosyl diglyceride molecules (MGDG, blue) and one digalactosyl diacylglycerol molecule (DGDG, magenta) are depicted as stick models. e, f Overlay of the Fa PSI–LHCI structure (PDB code 8WGH, red) with that of maize PSI–LHCI (Zm PSI–LHCI, PDB code 5ZJI, light blue), At PSI–LHCI (PDB code 8J7B, smudge) and pea PSI–LHCI (Ps PSI–LHCI, PDB code 4XK8, gray; 5L8R, tv_blue; 7DKZ, yellow) based on PsaA viewed along the membrane normal from the stromal side, with the protein structure and the chlorophyll arrangement shown in (e) and f, respectively. In c and f, the phytol chains of all Chls have been deleted for clarity.
The overall structure of Fa PSI–LHCI consists of two moieties—the PSI core complex and its peripheral LHCI belt (Fig. 2a, b)—that are very similar to the structures reported for PSI–LHCI from land plants4,5,7,46. As the Fa genome sequence is not currently available, we determined the sequences of the genes encoding four Lhca subunits and some PSI core subunits (Supplementary Fig. 7 and Supplementary Fig. 8). The Fa PSI core complex is composed of nine transmembrane subunits (PsaA, PsaB, PsaF, PsaG, PsaH, PsaI, PsaJ, PsaK and PsaL), three extrinsic subunits (PsaC, PsaD and PsaE) at the stromal side and one (PsaN) at the lumenal side, and the LHCI belt composed of four Lhca subunits (Lhca1 to Lhca4) binds to the PSI core complex at the PsaG-PsaF-PsaJ-PsaK side (Fig. 2a, b). Notably, we observed PsaO and PsaN subunits in the Fa PSI–LHCI structure (Fig. 2a, b and Supplementary Fig. 9a). PsaO, which is critical for associating LHCII with the PSI core complex to assemble the PSI–LHCI–LHCII supercomplex and to functionally favor energy transfer from LHCII to the PSI core complex in green algae and land plants during state 2 (refs. 47,48,49,50), which with the similar function as the subunit of PsaK, PsaL, PsaH and possibly PsaP51,52,53,54, is located at the outermost edge of the PSI core complex, and is thus typically absent in previously reported purified PSI–LHCI structures. Fa PsaO has a structure similar to those of PsaO from the moss Physcomitrium patens (Pp)50, the green alga Chlamydomonas reinhardtii (Cr)48,49, and the red alga Cyanidioschyzon merolae (Cm)55, which are composed of a shorter TMH B, an amphipathic helix C at the lumenal side, and a longer TMH A from the N-terminal to its C-terminal end, although we noticed some differences in loops AC and BC between Fa PsaO, Pp PsaO and Cm PsaO (Supplementary Fig. 9b). Fa PsaO binds three Chls (a201, a202 and a203) and two transmembrane β-carotenes (BCR204 and BCR205), forming two pigment clusters residing at either side of the crossed TMHs A and B. One cluster is composed of two Chls (a201 and a203) close to the lumenal side and of one β-carotene (BCR204); the other cluster comprises one Chl (a202) close to the stromal side, whose phytol tails protrude into PsaA, and one β-carotene (BCR205) (Supplementary Fig. 9b). Comparison of the pigment arrangement in Fa PsaO and that of land plants (Zm and Pp), a green alga (Cr) and a red alga (Cm) revealed extensive similarity, although with fewer pigments were assigned in the structures of Zm PsaO (two Chls), Cm PsaO (three Chls) and Pp PsaO (three Chls and one β-carotene)8,50,55, and an almost identical pigment pattern in Cr PsaO and Fa PsaO (Supplementary Fig. 9b). These results suggest that Fa PsaO may have functions similar to those previously reported for other plant PsaO proteins.
The PsaN subunit is easily detached from the PSI core due to its loose association, explaining how it is missing from structures containing the PSI core, with the exception of the maize (Zm) PSI–LHCI–LHCII structure8. Importantly, we resolved PsaN in the Fa PSI–LHCI structure. Superposition of the Fa PSI–LHCI structure with that of Zm PSI–LHCI revealed that Fa PsaN is in the same location within Fa PSI–LHCI as Zm PsaN is within Zm PSI–LHCI (Supplementary Fig. 9c). Fa PsaN has two directly connected helices (A and B) at its N terminus, a long loop BC and a short helix C followed by a long C-terminal loop, similar to the structure of maize PsaN (Supplementary Fig. 9d). Helices A and B bind to the lumenal region of PsaA, PsaF and PsaJ, while loops as well as two Chls (a201 and a202) fill in a large gap between PsaA and Lhca2 at the lumenal side (Supplementary Fig. 9e), supporting a role for PsaN in building direct EET pathways from Lhca2 to the PSI core complex, which was suggested for PsaN in Zm PSI–LHCI8.
An amino acid sequence alignment showed high conservation for each of the four groups of Lhca subunits (Lhca1, Lhca2, Lhca3 and Lhca4) among Fa, At, pea and maize (Supplementary Fig. 7). Based on their predicted amino acid sequences and relative positions around the PSI core, we assigned the four Lhca subunits in Fa PSI–LHCI as Lhca1, Lhca4, Lhca2, and Lhca3 from PsaG to PsaK when viewed from the stromal side (Fig. 2a). The four Lhca subunits are in the same order in Ps PSI–LHCI4,5,7 and Zm PSI–LHCI8. Although there is a possibility of relative movement of antennae relative to the core complex, such as in the PSI-IsiA complex56, the results of the 3D variability analysis showed that there were no obvious variances in the positions of Lhca relative to the PSI core in Fa PSI-LHCI (Supplementary Movie 1).
When the Fa PSI–LHCI structure and the five PSI–LHCI structures from maize (PDB code 5ZJI), At (PDB code 8J7B)57 and pea (PDB codes: 4XK8, 5L8R and 7DKZ) are superposed based on PsaA, we discovered that their PSI core complexes as well as the chlorophyll molecules they are associated with are in similar positions, while their LHCI belts and associated chlorophyll molecules show some shifts (Fig. 2e,f). To assess the extent of these shifts, we measured the inter-Mg distance (Mg-to-Mg distance) between Chl a801PsaA, one of the special pair (the primary electron donor of photosystem I), and Chl a603 in each Lhca subunit in these PSI–LHCI structures (Supplementary Table 3). We obtained average distances across all five published structures of 63.7 Å from Chl a603Lhca1, 55.7 Å from Chl a603Lhca2, 63.3 Å from Chl a603Lhca3 and 58.4 Å from Chl a603Lhca4. The Fa LHCI belt was slightly farther away from the PSI core complex than that of other structures, with distances of 64.6 Å from Chl a603Lhca1, 56.0 Å from Chl a603Lhca2, 64.0 Å from Chl a603Lhca3 and 59.3 Å from Chl a603Lhca4. These results suggest that the interactions between the PSI core complex and the LHCI belt might be different among the six PSI–LHCI structures.
Structure of Fa LHCI
All Fa Lhca subunits comprise three major TMHs B, C and A and a short amphipathic helix D from their N termini to C termini, with three loop regions: an N-terminal loop, loop BC and loop AC (Fig. 3a). Each Fa Lhca subunit binds 14 or 15 Chl molecules, which are distributed into two layers, one close to the stromal side and the other close to the lumenal side. At the stromal layer, eight Chl molecules (a/b301, a302, a303, a/b308, a309, a310, a311 and a312) are located around TMHs A and B; at the lumenal layer, three Chl molecules (a304, a/b306 and a/b307) are located at the interface between TMHs B and C and two Chl molecules (a313 and a314) are located at the helix A-turn-helix D region. Besides these conserved Chl-binding sites, each Lhca binds 1–2 unique Chl molecules and 3–4 carotenoids (Fig. 3a).
a Structure of Lhca1, Lhca2, Lhca3 and Lhca4 from F. albivenis. Each Lhca is shown as ribbon model with the same color code as in Fig. 2a. Color codes: gray, Chls a; blue, Chls b; orange, carotenoids. b, c Overlay of each of the four Fa Lhca apoprotein structures (PDB code 8WGH, red) (b) and pigment arrangement (c) and the corresponding structures from maize PSI–LHCI (PDB code 5ZJI, light blue) and pea PSI–LHCI (PDB code 4XK8, gray; 5L8R, tv_blue; 7DKZ, yellow). d Superposition of the LHCI belt based on Lhca3 between the six land plants PSI–LHCI structures. Shifts between helix CFa Lhca1 and helix Cmaize/pea/At Lhca1 are enlarged. The structure of PSI–LHCI from Arabidopsis thaliana is also included (PDB code 8J7B, smudge).
To look for structural differences between Fa Lhcas and Lhcas from other flowering plants, we compared the structures of each Lhca group individually and within the entire LHCI belt by using the five PSI–LHCI structures (PDB codes: 8WGH, 5ZJI, 4XK8, 5L8R and 7DKZ) (Fig. 3b–d). For each Lhca group, structural superposition of each member showed a very high conservation in the three TMHs and loop regions, although Fa Lhca1 and Fa Lhca3 differed in their C-terminal loop regions (Fig. 3b). In addition, all Chl-binding sites are conserved between Fa, maize and pea, as are all amino acids coordinating to the central Mg atoms of Chl molecules, with the exception of one additional Chl a315 binding to the outer margin of Fa Lhca3 and Zm Lhca3 (Figs. 2f and 3c, Supplementary Fig. 7). These results indicate that the structure of each Lhca is highly conserved between different flowering plants.
Furthermore, we superposed the Fa LHCI belt structure with those of LHCI belts from the other five published PSI–LHCI structures based on the Lhca3 subunit, which revealed some shifts between the six LHCI structures, with At LHCI exhibiting the greatest shifts, followed by Fa LHCI (Fig. 3d). To quantify these shifts, we measured the distance between helix CFa Lhca1 and helix Cothers Lhca1, as helix CLhca1 is the farthest helix from Lhca3 and will reflect the cumulative effect of any shift affecting each Lhca. We detected a 3 Å distance between helix CFa Lhca1 and helix CAt Lhca1, and a 4–5 Å distance between helix CFa Lhca1 and each of helix C from other Lhca1s (Fig. 3d). These shifts may be related to a variety of factors, including the PSI-LHCI structure itself, the technical approach used, and whether or not it is combined with LHCII. however, here we focus on the possible relationship between shifts and the PSI-LHCI structure. As each Lhca can interact with adjacent Lhcas within the LHCI belt and with the PSI core complex, the different shifts observed here suggest differences in these interactions, in turn regulating the three-dimensional PSI–LHCI structure. Whether these differences affect the spectral red shift is unknown. LHCI can exist in three possible states: (i) in PSI–LHCI particles embedded in the thylakoid membrane; (ii) in isolated PSI–LHCI particles; and (iii) dissociated from the PSI core. Compared to PSI–LHCI particles in situ, isolated PSI–LHCI particles lose the acting force from the thylakoid membrane; similarly, separated LHCI no longer interacts with the PSI core. In each state, the local environment of red Chls and their conformation may thus differ, which may contribute to their spectral differences in all plant species, but not to great differences in chlorophyll fluorescence between Fa and other plant species. To investigate possible structural changes related to the energy state of red Chls, we compared Fa Lhca structures to those of other Lhcas in more detail below.
Structure elements possibly enhancing far-red forms in Fa Lhca3
To explore structural elements in Lhcas that may lead to the red-shifted spectral forms in Fa, we examined the coordination of the red Chl dimer (a303 and a309) in each Fa Lhca subunit. In all Fa Lhcas, Chl a309 is coordinated by a glutamate residue in TMH C (GluTMH C), while the coordinating amino acid for Chl a303 varied, with HisTMH B in Fa Lhca1 and Fa Lhca2 and AsnTMH B in Fa Lhca3 and Fa Lhca4 (Fig. 4). The coordination pattern of the red Chl dimer in Fa was identical to that in pea and maize4,5,7,8, ruling out coordinating amino acids as causal structural elements for the red shift seen in Fa compared to common flowering plants.
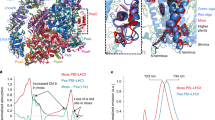
We then focused on the local environment of Chl a303 and Chl a309 in each Fa Lhca and compared them to that in the corresponding pea and maize Lhca. Although Lhca1, Lhca2 and Lhca4 differed little between the three plant species (Supplementary Fig. 10), Fa Lhca3 displayed three regions that were distinct relative to Lhca3 from pea and maize (Fig. 5). First, four continuous amino acid residues (88–91, Phe-Trp-Phe-Gln) located in the N-terminal loop of Fa Lhca3 and next to TMH B, form a large canopy for Chl a303 and Chl a309 due to the large benzene ring side chains of Phe and Trp; by contrast, the corresponding amino acids in pea and maize Lhca3 are Gly-Phe-Ile-Glu, with smaller side chains than in Fa Lhca3, thus forming a much smaller canopy for Chl a303 and Chl a309 (Fig. 5b, c, region I). The porphyrin rings of Chl a303 and Chl a309 in all Lhca3 structures have two sides—one facing TMHs B and C, whose two residues provide ligands to the central Mg atoms of the two Chls, and the other facing the PSI core complex; the long phytyl tails of Chl a303 and Chl a309 protrude towards the stromal side and become bent to the core side once touch the middle two residues of the canopy region, causing steric hindrance for the phytyl tails (Fig. 5b, region I). Therefore, the size of the canopy, and especially the size of the middle two residues, may affect the conformation of Chl a303 and Chl a309. To quantify the steric hindrance effect, we compared the structure of Chl a303Fa Lhca3 to that of Chl a303Ps Lhca3 and Chl a303Zm Lhca3: the phytyl chain of Chl a303Fa Lhca3 was bent closer to the core side (Supplementary Fig. 11), which might be caused by a larger steric hindrance from the stromal canopy in Fa Lhca3.
a Structure of Fa Lhca3 showing the extent of surface hydrophobicity. The degree of hydrophobicity is shown from white (no hydrophobicity) to red (high hydrophobicity). The black dashed boxes indicate three regions (I, II and III) surrounding Chl a303 and Chl a309, with different amino acid residues between Fa Lhca3, pea (Pisum sativum, Ps) Lhca3 and maize (Zea mays, Zm) Lhca3. b Enlarged views of boxed areas I–III) in Fa Lhca3 and the structure of the corresponding area in Ps Lhca3 and Zm Lhca3. c Sequence alignment of regions I–III from Fa Lhca3, Ps Lhca3 and Zm Lhca3. Color codes: Chl a303 and Chls a309, green; other Chl molecules, purple. PDB codes: Fa PSI–LHCI, 8WGH; Ps Lhca3, 4XK8; Zm Lhca3, 5ZJI.
Second, an amino acid residue surrounding Chl a309, polar Tyr-170 in TMH C, is located within 4 Å from Chl a309 in Fa Lhca3; in other species, this residue is replaced by nonpolar Phe (pea Lhca3) or Leu (maize Lhca3), leading to differences in hydrophobicity (Fig. 5b, c, region II). Third, there is a large gap between TMHs B and C in the neighborhood of Chl a309 in Fa Lhca3, with the nearest distance between Thr-105TMH B and Val-162TMH C being up to 10.3 Å. Notably, the corresponding gap was much smaller in pea and maize Lhca3, with the shortest distance between PheTMH B and MetTMH C of pea Laca3, 4.1 Å, and between TyrTMH B and MetTMH C in maize Lhca3, 3.3 Å (Fig. 5b, c, region III). Therefore, amino acids in the three regions constitute candidate structural elements for the strong red shift in Fa.
To delineate the amino acids critical to the red shift in Fa, we compared the sequence of Fa Lhca3 to two available Lhca3 sequences from Acanthaceae plants, Chinese rain bell (Strobilanthes cusia) and green chiretta (Andrographis paniculata), with a comparable red shift in their λmax (Fig. 1g, h). We also compared the sequence of Fa Lhca3 to the Lhca3 sequence from 15 plant species belonging to different families outside the Acanthaceae (Supplementary Fig. 12). The sequence in region I of Lhca3 in Acanthaceae is Phe-Trp-Phe-Gln (in Fa and S. cusia) or Met-Trp-Phe-Gln (in A. paniculata), with the middle two residues having benzene ring side chains forming a large canopy; in all non-Acanthaceae species except P. notoginseng, the sequence is Gly-Phe-Ile-Glu, forming a smaller canopy as in pea and maize (Fig. 5b, region I). Region I in P. notoginseng has the sequence Ile-Phe-Phe-Glu; the third residue Phe has a larger side chain than Ile (the counterpart of non-Acanthaceae Gly-Phe-Ile-Glu), while the second residue Phe has a smaller side chain than Trp (the counterpart of Acanthaceae Phe-Trp-Phe-Gln or Met-Trp-Phe-Gln), thus likely forming a larger canopy than that in pea and maize but smaller than that in Fa. Notably, P. notoginseng had a fluorescence emission peak of 747 nm, intermediate between non-Acanthaceae land plants and Fa (Fig. 1a, b), suggesting that residues with larger side chains in region I may lead to a more red-shifted fluorescence emission spectrum.
In region II, the amino acid is polar Tyr in the three Acanthaceae plant species and P. notoginseng, and nonpolar Phe or Leu in the 14 non-Acanthaceae, suggesting that polar Tyr in region II may favor the red shift. The sequence in region III does not clearly distinguish between Acanthaceae and non-Acanthaceae, as the two non-Acanthaceae plants pineapple (Ananas comosus) and Boxelder maple (Acer negundo) have the same residues as the three Acanthaceae plants, with a Thr in TMH B and a Val in TMH C (Supplementary Fig. 12). However, these residues in region III may contribute to the energy state of red Chl dimers, as evidenced by the 77-K fluorescence emission spectra of A. comosus and A. negundo, with λmax values of 747.2 nm and 744.4 nm, representing red shifts of 12 nm and 10 nm compared to At, respectively (Supplementary Fig. 13). Therefore, all three regions likely contain structural elements that explain the strong red shift in fluorescence emission.
Discussion
Expanding the photosynthetic absorption spectrum into the far-red region is important for agricultural production to feed the growing human population and achieve food security. However, uneven light distribution is an important challenge to increasing cropping density. Visible light intensity can exceed 1000 μmol m−2s−1 at the top of the canopy on a sunny day, while being only a few μmol m−2s−1 at the bottom of the canopy. Although the canopy undergrowth is rich in far-red light, this wavelength is of poor quality for photosynthetic utilization. It has been estimated that the full use of the entire 700–750 nm light range for photosynthesis could increase the light energy absorbed by crops by 19%24. Considering that in actual agricultural production, solar energy conversion utilization is only ~1%58, increased utilization of far-red light will provide new ideas for improving crop yields. Therefore, it is of great significance to search for examples of far-red light utilization in extant land plants and to analyze the structural causes that promote a red shift in the chlorophyll a absorption spectrum.
A screen of shade plants identified the Acanthaceae plant Fa as being able to capture more far-red light, with its thylakoid membrane mediating greater absorption of far-red light, as evidenced by a fluorescence emission peak at 753 nm, ~20 nm longer than that of At. In Fa, the enhanced far-red spectral forms originated from PSI–LHCI, prompting us to resolve the spatial structure of the Fa PSI–LHCI supercomplex at 2.46 Å.
The overall structure of Fa PSI–LHCI is very similar to previously reported PSI–LHCI structures from angiosperms. Their subunit compositions are similar, although Fa PSI–LHCI was unique in retaining the PsaN and PsaO subunits easily lost in other crystal structure preparations. The Chl a/b-binding sites are also highly conserved, suggesting that the greater red shift seen in Fa is not due to the addition of new binding sites for Chl dimers or trimers in Fa PSI–LHCI, but rather to a change in the interaction between the red Chl dimers and their surrounding protein environments.
We established that the ligand amino acids of red Chls are identical between Fa Lhcas and Lhcas from pea and maize: His for Lhca1 and Lhca2, and Asn for Lhca3 and Lhca4. Previous reports have suggested that the red-shifted state of red Chls is better maintained by Asn rather than His as the ligand amino acid15,20,21,59. We conclude that the red shift of Fa LHCI is not due to a change in the coordination of the red Chl of Lhca1 and/or Lhca2 from His to Asn. According to the published structures of PSI–LHCI from land plants4,5,7,8 and that reported in this article, Lhca1 and Lhca3 at the two ends of the LHCI belt directly bind to the core complex, while Lhca4 and Lhca2 in the center of LHCI connect with the core complex through lipid molecules distributed at their interface, making the overall set of interactions complex and flexible. We determined that the relative positions between the LHCI antenna and the core complex differ slightly among all the structures examined, with LHCI of Fa PSI–LHCI being the furthest away from the reaction center, suggesting that the interaction between LHCI and the core complex in Fa is somehow different compared to those in other species. However, how this interaction may affect Chl energy levels is not known.
The comparison of the Fa PSI–LHCI structure to that of PSI–LHCI from other land plants indicated several amino acid changes in three regions that might contribute to the enhanced red shift: Trp-89–Phe-90 in region I with large benzene ring side chains, polar Tyr-167 in region II, and Thr-102TMHB and Val-159TMHC in region III, located around the red Chls of Fa Lhca3. Notably, these unique amino acids are conserved among Fa and two other Acanthaceae (S. cusia and A. paniculata) with a red-shifted fluorescence emission, as well as in non-Acanthaceae plants that exhibit some degree of red-shifted fluorescence features. Changing these amino acids in the Lhca sequences from model plants and crops might produce more red-shifted spectral forms, and thus further studies are needed to test the possible mechanisms regulating the energy level of chlorophylls.
Modulation of the red Chl energy level affects not only the absorption spectral range in the far-red region, but also EEC from the LHCI to the core. Most of the excitation energy absorbed by LHCI is transferred to red Chls before it can be handed to the core complex60, such that a lower energy level for red Chl makes it more difficult to overcome the energy barrier for energy transfer. Although the physiological significance of red Chls in EEC is not well understood, the energy gradient where red Chls is at the lowest value may be related to plant evolutionary adaptation, as it becomes more and more pronounced across plant evolution61. Indeed, the λmax PSI–LHCI is ~710 nm in green algae42, 725 nm in bryophytes46 and generally >730 nm in angiosperms. The effect of the lower red Chl energy level in Fa PSI–LHCI on plant photosynthetic light reaction needs to be investigated in the future.
Plants can be categorized into two types based on their adaptation to the light environment: sun plants, which grow robustly under strong light intensity but poorly in low light and in the shade; and shade plants, which grow better in low light than in strong light. Based on our results, we propose that shade plants can be further divided into two categories as a function of their degree of far-red light utilization. Fa and other species can increase their utilization of far-red light; other species cannot, making them more similar to sun plants and suggesting that they may trap more light by increasing LHCII contents or through changes to leaf shape62,63. Future physiological and ecological studies of shade plants should distinguish between these two different types of shade plants, which may employ different mechanisms.
In conclusion, the cryo-EM structure of PSI–LHCI from Fa solved in this study provides new insights into the structure of a land plant PSI–LHCI supercomplex with strongly red-shifted fluorescence emission. This structure adds to the list of land plant PSI–LHCI supercomplexes solved with different λmax values (727 nm in P. patens, 730 nm in pea, 745 nm in Fa), reflecting the diversity in the energy level of red Chls, which may be a result of changes in the antenna protein microenvironment due to amino acid polymorphisms during evolution to adapt to different light environments. These findings will shed light on the design of light-harvesting antenna proteins that improve light trapping capacity and increase biomass production.
Methods
Plant materials
Three understory traditional Chinese medicine plants (Asian ginseng [Panax ginseng], Chinese ginseng [Panax notoginseng] and noble dendrobium [Dendrobium nobile]), the houseplant Devil’s ivy (Epipremnum aureum) and ten species belonging to the Acanthaceae family (nerve plant [Fittonia albivenis], Chinese rain bell [Strobilanthes cusia], shrimp plant [Justicia brandegeeana], curved-flower flaming-Acanthus [Phlogacanthus curviflorus], Philippine violet [Barleria cristata], minnieroot [Ruellia tuberosa], firecracker flower [Crossandra infundibuliformis], polka dot plant [Hypoestes phyllostachya], zebra plant [Aphelandra squarrosa] and bitterweed [Andrographis paniculata]) were purchased from a local market or from Taobao, the biggest online shopping website in China. All plants were then grown in a growth chamber (20 μmol photons m−2s−1, 16-h light/8-h dark photoperiod, 23 °C) for 2 days before harvesting leaves. Arabidopsis thaliana (Columbia 0 [Col-0] accession) plants were grown in a plant cabinet set to 45% humidity and a temperature of 23 °C, with a 16-h light/8-h dark photoperiod under 130 μmol photons m−2s−1 for 6 weeks.
Isolation of thylakoids and purification of PSI–LHCI
Thylakoid membranes of Fa and At were isolated under dim green light as described previously40 with slight modifications. Plants were grown under normal white light conditions with no special light treatments to induce the state transition. Their leaves were homogenized in prechilled buffer I (20 mM Tricine-NaOH, pH 7.8, 0.3 M sucrose, 5 mM NaCl). To isolate chloroplasts, the slurry was filtered through 8 layers of gauze and the filtered solution was centrifuged at 7000 × g for 10 min at 4 °C. The pellet was resuspended in buffer II (20 mM Tricine-NaOH, pH 7.8, 5 mM NaCl) to break chloroplasts, and then centrifuged at 20,000 × g for 10 min at 4 °C in a JA14 rotor (Beckman, USA) to obtain the thylakoid membranes. Chlorophyll concentrations were determined by extraction in 80% (v/v) acetone using the method of Arnon64. Purification of PSI–LHCI was performed as previously described19. Freshly prepared thylakoids (1 mg Chl/mL equivalent) were solubilized by the addition of 10% (w/v) n-dodecyl-β-d-maltoside (β-DDM) to a final concentration of 1% (w/v). After stirring for 30 min in an ice bath, the sample was centrifuged at 40,000 × g for 10 min at 4 °C before the supernatant were loaded on top of ultracengtrifuge tubes containing 12 mL of a 0.3–0.9 M sucrose gradient containing 20 mM Tricine-Tris, pH 7.5, 0.015% (w/v) β-DDM. After centrifugation at 243,500 × g for 16 h at 4 °C in an SW40Ti rotor (Beckman, USA), three chlorophyll-containing bands were resolved, and the lowest one contained PSI–LHCI.
Isolation of the PSI core complex and LHCI from PSI–LHCI
The PSI–LHCI sample obtained above was further separated into the PSI core and LHCI following a procedure similar to that previously reported43,44. The purified PSI–LHCI supercomplex (0.5 mg Chl/mL equivalent) was solubilized with β-DDM at a final concentration of 0.75% (w/v) and 3-(N,N-dimethylpalmitylammonio) propanesulfonate (zw 3-16) at a final concentration of 0.75% (w/v) for 30 min at 4 °C. An aliquot of 0.5 ml detergent-treated PSI–LHCI was applied to a 0.3–0.9 M sucrose density gradient tube containing 20 mM Tricine-Tris, pH 7.5, 0.05% (w/v) β-DDM. Following centrifugation at 165,000 × g for 16 h at 4 °C in a Beckman SW40Ti rotor, three chlorophyll-containing bands were obtained.
Gel electrophoresis
To analyze the polypeptide composition, samples were treated with lithium dodecyl sulfate (LDS) sample buffer containing 2% (w/v) LDS, 60 mM dithiothreitol and 60 mM Tris-HCl (pH 8.5) at 40 °C for 10 min, before being subjected to SDS-PAGE as described by Ikeuchi and Inoue65 using a 16% gel (acrylamide:bisacrylamide = 75:1 [w/w]) containing 7.5 M urea. Samples corresponding to 2.5 µg Chl were loaded in each lane.
Absorption and fluorescence spectroscopy
Absorption spectra of leaves and isolated samples were recorded at room temperature using a UV-3600 Plus spectrophotometer (Shimadzu, Japan) and a U-3900H spectrophotometer (Hitachi, Japan) in the 350–800 nm range, respectively. Isolated samples were diluted in a buffer containing 10 mM Tricine-NaOH (pH 7.5) and 0.03% (w/v) β-DDM to a Chl concentration with a maximal absorption of about 0.7. Absorption at 800 nm was set to zero and the spectra were normalized with respect to their maximal absorption peak in the red region. Low-temperature (77-K) fluorescence emission spectra were recorded using a F-4700 instrument (Hitachi, Japan) with 440 nm as the excitation wavelength. A small piece of leaf or 300 μL of liquid samples were placed into an nuclear magnetic resonance (NMR) tube in a glass Dewar filled with liquid nitrogen. For preparation of liquid samples, the chlorophyll concentration was adjusted to about 5 μg/mL with a buffer containing 50% (v/v) glycerol, 10 mM Tricine-NaOH, pH 7.5, and 0.015% (w/v) β-DDM.
Pigment analysis
The pigments were extracted in 80% (v/v) cold acetone from purified Fa PSI–LHCI and analyzed by high-performance liquid chromatography (HPLC) as described66 with slight modifications. HPLC was performed with a C-18 reversed-phase column (4.6 × 250 mm, 5-μm particle size, Grace, USA) in a Thermo U-3000 separation module equipped with an Ultimate 3000 diode array detector. The pigments were eluted at a flow rate of 1 mL/min using 100% solvent A (acetonitrile:methanol:tetrahydrofuran, 81:14:5 [v/v/v]) for the first 9 min, followed by a 3-min linear gradient with 0–100% solvent B (methanol:ethyl acetate, 70:30 [v/v]), which was continuously used until the end of the 30-min separation. The column was re-equilibrated in solvent A for 10 min prior to the next injection and the loading amount for each sample is 40 μL. Pigments were detected by their absorbance at 445 nm. Pigment identification was achieved by comparing their retention times and absorption spectra to those of authentic standards and pigments extracted from At PSI–LHCI.
Sequence analysis of PSI–LHCI from Fa
To obtain amino acid sequences of the PSI subunits, total RNA was extracted from Fa leaves using the reverse transcription kit and subjected to nonparametric transcriptome deep sequencing by Beijing Novogene Bioinformatics Technology Company. The sequence analysis of genes encoding subunits of the PSI–LHCI complex was performed using Novomagic, an online platform for data analysis (https://magic.novogene.com). Specifically, the assembled coding sequences (CDSs) encoding PSI core subunits and Lhcas were identified via comparison to related sequences in the National Center for Biotechnology Information (nih.gov). To confirm the sequences of Lhca genes, total RNA (about 2 μg) was converted to first-strand cDNA using the reverse transcription kit (vazyme), and each predicted CDS was amplified by PCR using gene-specific designed primers. Each PCR product was cloned into the pMD™ 19-T vector; five independent colonies per PCR product were picked and the sequence of their insert was determined by Sanger sequencing. All resulting sequences were aligned with CLUSTALW and ENDscript 3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) to assess consistency among clones.
Cryo-EM sample preparation and data collection
The purified PSI–LHCI complex was concentrated to 4 mg Chl/mL. A 4-μL aliquot of the protein sample was dropped onto glow-discharged holey carbon grids (Quantifoil Cu R1.2/1.3, 400 mesh). The grids were then blotted using a Vitrobot Mark IV (Thermo Fisher Scientific) with the following settings: a 10-s wait time, a 4-s blot time, 100% humidity at 8 °C and blotting force of 0; and finally the grids were plunge-frozen in liquid ethane cooled by liquid nitrogen.
The cryo-EM grids were imaged using a Titan Krios transmission electron microscope (FEI) operating at 300 kV with a magnification of 81,000×. The images were recorded using a Gatan K3 Summit direct electron detector by AutoEMation67. An energy filter with a slit width of 20 eV was used, and the preset defocus ranged from −1.5 to −1.0 μm. Each image was dose-fractionated to 32 frames with a total electron dose of 50 e−/Å2 and a total exposure time of 3.2 s. The data collection statistics are summarized in Supplementary Table 1.
Cryo-EM image processing
All movie stacks were motion-corrected using MotionCor268, and all motion-corrected images were processed using cryoSPARC v4.4.169. The overall workflow is shown in Extended Data Fig. 1. Following estimating the CTF (contrast transfer function) parameters using Patch CTF estimation, 11,517 micrographs with a CTF fit resolution better than 5 Å were selected. Around 2159 thousand particles were picked using the blob picker, with a diameter ranging from 50 to 300 Å. After several rounds of 2D classification, 468 thousand particles were chosen as a training dataset for Topaz70, resulting in the identification of 3737 thousand particles from all micrographs. Following multiple rounds of 2D classification, 1148 thousand particles were kept and subjected to ab initio reconstruction to generate three initial models for heterogenous refinement, which yielded a high-quality 3D volume containing 718,766 particles. To analyze conformational or compositional heterogeneity, 3D variability analysis was employed. From this analysis, 157,342 particles with the PsaN subunit bound were selected. After performing stepwise global CTF refinement, local CTF refinement, non-uniform refinement71 and local refinement, a final map with a resolution of 2.46 Å was obtained. The local resolution map was generated from a local resolution estimation job in cryoSPARC where the FSC threshold parameter used 0.5.
Considering the possibility of relative movement of the antennae relative to the core complex, another round of 3D variability analysis was performed. The 3D variability display was conducted in simple mode with a filter resolution of 3 Å, and a movie was created using the output of 20 frames (Supplementary Movie 1).
Modeling and refinement
The atomic model of maize PSI–LCHI/LHCII (PDB code: 5ZJI) was docked into the EM map using UCSF ChimeraX v1.2.572 and the LHCII subunits were manually removed. The amino acid sequences of all maize subunits were then changed to the corresponding sequences of Fa (Supplementary Fig. 8) using Coot v.0.8.9.273. Meanwhile, the cofactors, including Chl a and Chl b, were assigned based on the model of maize PSI-LHCI, and manually checked their match with the EM map (Extended Data Fig. 2). Specifically, the amino acid sequences of PsaA, PsaD, PsaE, PsaH, PsaL and four Lhca subunits were changed to the corresponding sequences from Fa, which were determined in this study as described above. For those subunits whose amino acid sequences were not indentified in Fa, a model (model-1) was built using their amino acid sequences substituted by amino acid sequences from Strobilanthes cusia (S. cusia), which belongs to the same family as Fa. Meanwhile, a model (model-2) was automatically built using ModelAngelo74 in no fasta file mode to aid in identifying amino acid sequences. Upon careful comparison focused on side chains, we found that for most of subunits, the model-1 shows a better side chain match with the EM map than the model-2. Therefore, model-1 was adopted. However, For the PsaF subunit, the amino acid sequence was replaced by that from At, which shows a better match than both the sequence from S. cusia and the one generated by ModelAngelo.
Then, the side chain of residues and the cofactors were manually adjusted and optimized based on the EM density using Coot v.0.8.9.2. Finally, real-space refinement was performed with Phenix v1.20.1-448775. The final statistics for data processing and structure refinement are summarized in Supplementary Table 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding authors upon request. The cryo-EM map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-37513. Atomic model coordinates have been deposited in the Protein Data Bank (PDB) under accession code 8WGH. Source data are provided with this paper.
References
Croce, R. & van Amerongen, H. Light-harvesting in photosystem I. Photosynth. Res. 116, 153–166 (2013).
Ben-Shem, A., Frolow, F. & Nelson, N. Crystal structure of plant photosystem I. Nature 426, 630–635 (2003).
Amunts, A., Drory, O. & Nelson, N. The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature 447, 58–63 (2007).
Mazor, Y., Borovikova, A. & Nelson, N. The structure of plant photosystem I super-complex at 2.8 Å resolution. eLife 4, e07433 (2015).
Qin, X., Suga, M., Kuang, T. & Shen, J.-R. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348, 989–995 (2015).
Mazor, Y., Borovikova, A., Caspy, I. & Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 3, 17014 (2017).
Wang, J. et al. Structure of plant photosystem I-light harvesting complex I supercomplex at 2.4 Å resolution. J. Integr. Plant Biol. 63, 1367–1381 (2021).
Pan, X. et al. Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II. Science 360, 1109–1113 (2018).
Novoderezhkin, V. I. & Croce, R. The location of the low-energy states in Lhca1 favors excitation energy transfer to the core in the plant PSI-LHCI supercomplex. Photosynth. Res. 156, 59–74 (2023).
Schmid, V. H., Cammarata, K. V., Bruns, B. U. & Schmidt, G. W. In vitro reconstitution of the photosystem I light-harvesting complex LHCI-730: heterodimerization is required for antenna pigment organization. Proc. Natl Acad. Sci. USA 94, 7667–7672 (1997).
Schmid, V. H., Paulsen, H. & Rupprecht, J. Identification of N- and C-terminal amino acids of Lhca1 and Lhca4 required for formation of the heterodimeric peripheral photosystem I antenna LHCI-730. Biochemistry 41, 9126–9131 (2002).
Castelletti, S. et al. Recombinant Lhca2 and Lhca3 subunits of the photosystem I antenna system. Biochemistry 42, 4226–4234 (2003).
Croce, R. Structural biology. A close view of photosystem I. Science 348, 970 (2015).
Ihalainen, J. A. et al. Red spectral forms of chlorophylls in green plant PSI - a site-selective and high-pressure spectroscopy study. J. Phys. Chem. B 107, 9086–9093 (2003).
Morosinotto, T., Breton, J., Bassi, R. & Croce, R. The nature of a chlorophyll ligand in Lhca proteins determines the far red fluorescence emission typical of photosystem I. J. Biol. Chem. 278, 49223–49229 (2003).
Morosinotto, T., Mozzo, M., Bassi, R. & Croce, R. Pigment-pigment interactions in Lhca4 antenna complex of higher plants photosystem I. J. Biol. Chem. 280, 20612–20619 (2005).
Croce, R. et al. The low-energy forms of photosystem I light-harvesting complexes: spectroscopic properties and pigment-pigment interaction characteristics. Biophys. J. 93, 2418–2428 (2007).
Romero, E. et al. The origin of the low-energy form of photosystem I light-harvesting complex Lhca4: mixing of the lowest exciton with a charge-transfer state. Biophys. J. 96, L35–L37 (2009).
Ballottari, M., Govoni, C., Caffarri, S. & Morosinotto, T. Stoichiometry of LHCI antenna polypeptides and characterization of gap and linker pigments in higher plants Photosystem I. Eur. J. Biochem. 271, 4659–4665 (2004).
Wientjes, E., Roest, G. & Croce, R. From red to blue to far-red in Lhca4: how does the protein modulate the spectral properties of the pigments? Biochim Biophys. Acta 1817, 711–717 (2012).
Li, X. et al. LHCA4 residues surrounding red chlorophylls allow for fine-tuning of the spectral region for photosynthesis in Arabidopsis thaliana. Front. Plant Sci. 13, 1118189 (2023).
Chukhutsina, V. U., Liu, X., Xu, P. & Croce, R. Light-harvesting complex II is an antenna of photosystem I in dark-adapted plants. Nat. Plants 6, 860–868 (2020).
Andrea et al. The importance of PS I chlorophyll red forms in light-harvesting by leaves. Photosynth. Res. 60, 209–215 (1999).
Chen, M. & Blankenship, R. E. Expanding the solar spectrum used by photosynthesis. Trends Plant Sci. 16, 427–431 (2011).
Lee, M.-J., Son, K.-H. & Oh, M.-M. Increase in biomass and bioactive compounds in lettuce under various ratios of red to far-red LED light supplemented with blue LED light. Hortic. Environ. Biotechnol. 57, 139–147 (2016).
Wolf, B. M. & Blankenship, R. E. Far-red light acclimation in diverse oxygenic photosynthetic organisms. Photosynth. Res. 142, 349–359 (2019).
Legendre, R. & van Iersel, M. W. Supplemental far-red light stimulates lettuce growth: disentangling morphological and physiological effects. Plants 10, 166 (2021).
Kim, D., Moon, T., Kwon, S., Hwang, I. & Son, J. E. Supplemental inter-lighting with additional far-red to red and blue light increases the growth and yield of greenhouse sweet peppers (Capsicum annuum L.) in winter. Hortic. Environ. Biotechnol. 64, 83–95 (2022).
Miyashita, H. et al. Chlorophyll d as a major pigment. Nature 383, 402–402 (1996).
Miyashita, H. et al. Pigment composition of a novel oxygenic photosynthetic prokaryote containing chlorophyll d as the major chlorophyll. Plant Cell Physiol. 38, 274–281 (1997).
Kuhl, M., Chen, M., Ralph, P. J., Schreiber, U. & Larkum, A. W. D. Ecology: a niche for cyanobacteria containing chlorophyll d. Nature 433, 820 (2005).
Chen, M. et al. A red-shifted chlorophyll. Science 329, 1318–1319 (2010).
Behrendt, L. et al. Endolithic chlorophyll d-containing phototrophs. ISME J. 5, 1072–1076 (2011).
Chen, M., Li, Y. Q., Birch, D., & Willows, R.D. A cyanobacterium that contains chlorophyll f - a red-absorbing photopigment.FEBS Lett. 586, 3249–3254 (2012).
Bos, P. R. et al. Spectral diversity of photosystem I from flowering plants. Photosynth. Res. 155, 35–47 (2023).
Bos, I. et al. Multiple LHCII antennae can transfer energy efficiently to a single Photosystem I. Biochim Biophys. Acta Bioenerg. 1858, 371–378 (2017).
Satoh, K. Photosystem II particles largely depleted in the two intrinsic polypeptides in the 30 kDa region from Synechococcus sp. FEBS Lett. 204, 357–362 (1986).
Klimmek, F. et al. Structure of the higher plant light harvesting complex i: in vivo characterization and structural interdependence of the LHCA proteins. Biochem. 44, 3065–3073 (2005).
Qin, X. et al. Rapid purification of photosystem I chlorophyll-binding proteins by differential centrifugation and vertical rotor. Photosynth. Res. 90, 195–204 (2006).
Bassi, R. & Simpson, D. Chlorophyll-protein complexes of barley photosystem I. Eur. J. Biochem. 163, 221–230 (1987).
KNOETZEL, J., Svendsen, I. & SIMPSON, D. J. Identification of the photosystem I antenna polypeptides in barley: Isolation of three pigment‐binding antenna complexes. Eur. J. Biochem. 206, 209–215 (1992).
Qin, X. et al. Isolation and characterization of a PSI-LHCI super-complex and its sub-complexes from a siphonaceous marine green alga, Bryopsis Corticulans. Photosynth. Res. 123, 61–76 (2015).
Tian, L. et al. Isolation and characterization of PSI-LHCI super-complex and their sub-complexes from a red alga Cyanidioschyzon merolae. Photosynth. Res. 133, 201–214 (2017).
Pålsson, L., Tjus, S. E., Andersson, B. & Gillbro, T. Ultrafast energy transfer dynamics resolved in isolated spinach light-harvesting complex I and the LHC I-730 subpopulation. Biochem Biophys. Acta Bioenerg. 1230, 1–9 (1995).
Schmid, V. H. et al. Pigment binding of photosystem I light-harvesting proteins. J. Biol. Chem. 277, 37307–37314 (2002).
Yan, Q. et al. Antenna arrangement and energy-transfer pathways of PSI-LHCI from the moss Physcomitrella patens. Cell Discov. 7, 10 (2021).
Jensen, P. E., Haldrup, A., Zhang, S. & Scheller, H. V. The PSI-O subunit of plant photosystem I is involved in balancing the excitation pressure between the two photosystems. J. Biol. Chem. 279, 24212–24217 (2004).
Pan, X. et al. Structural basis of LhcbM5-mediated state transitions in green algae. Nat. Plants 7, 1119–1131 (2021).
Huang, Z. et al. Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2. Nat. Commun. 12, 1100 (2021).
Zhang, S. et al. Structural insights into a unique PSI-LHCI-LHCII-Lhcb9 supercomplex from moss. Nat. Plants 9, 832–846 (2023).
Galka, P. et al. Functional analyses of the plant photosystem I-light-harvesting complex II supercomplex reveal that light-harvesting complex II loosely bound to photosystem II is a very efficient antenna for photosystem I in state II. Plant Cell 24, 2963–2978 (2012).
Lunde, C., Jensen, P. E., Haldrup, A., Knoetzel, J. & Scheller, H. V. The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408, 613–615 (2000).
Zhang, S. & Scheller, H. V. Light-harvesting complex II binds to several small subunits of photosystem I. J. Biol. Chem. 279, 3180–3187 (2004).
Kouril, R. et al. Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana. Biochem. 44, 10935–10940 (2005).
Pi, X. et al. Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga. Proc. Natl Acad. Sci. USA 115, 4423–4428 (2018).
Harris, D. et al. Energetic robustness to large scale structural fluctuations in a photosynthetic supercomplex. Nat. Commun. 14, 4650 (2023).
Wu, Jianghao et al. Regulatory dynamics of the higher-plant PSI–LHCI supercomplex during state transitions. Mol. Plant 16, 1937–1950 (2023).
Zhu, X.-G., Long, S. P. & Ort, D. R. What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr. Opin. Biotechnol. 19, 153–159 (2008).
Sláma, V. et al. Origin of Low-Lying Red States in the Lhca4 Light-Harvesting Complex of Photosystem I. J. Phys. Chem. Lett. 14, 8345–8352 (2023).
Croce, R., Zucchelli, G., Garlaschi, F. M., Bassi, R. & Jennings, R. C. Excited state equilibration in the photosystem I−light-harvesting I complex: P700 Is Almost Isoenergetic with Its Antenna. Biochemistry 35, 8572–8579 (1996).
Jian-Ren Shen, Kimiyuki Satoh and Suleyman I. Allakhverdiev. Photosynthesis: Molecular approaches to solar energy conversion. (Springer, New York, 2021).
Feng, L. et al. The influence of light intensity and leaf movement on photosynthesis characteristics and carbon balance of soybean. Front. Plant Sci. 9, 1952 (2019).
Hu, C., Nawrocki, W. J. & Croce, R. Long‐term adaptation of Arabidopsis thaliana to far‐red light. Plant Cell Environ. 44, 3002–3014 (2021).
Arnon, D. I. Copper enzymes in isolated chloroplasts. polyphenoloxidase in beta vulgaris. Plant Physiol. 24, 1–15 (1949).
Ikeuchi, M. & Inoue, Y. A new photosystem II reaction center component (4.8 kDa protein) encoded by chloroplast genome. FEBS Lett. 241, 99–104 (1988).
Val, J., Monge, E. & Baker, N. R. An Improved HPLC Method for Rapid Analysis of the Xanthophyll Cycle. Pigments J. Chromatogr. Sci. 32, 286–289 (1994).
Lei, J. & Frank, J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai. electron Microsc. J. Struct. Biol. 150, 69–80 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T., Morin, A., Rapp, M., Brasch, J. & Berger, B. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Nature 628, 450–457 (2024).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
We thank J. Lei and the staff at the Tsinghua University Branch of the National Center for Protein Sciences Beijing for providing facility support, and the “Explorer 100” cluster system of the Tsinghua National Laboratory for Information Science and Technology for providing computation resources. The project was funded by the National Natural Science Foundation of China (32270260, 32070267, 32241030 and 32271245); State Key Laboratory of Membrane Biology, Taishan Scholars Project (Xiaochun Qin), Shandong Provincial Natural Science Foundation, China (ZR2019ZD48).
Author information
Authors and Affiliations
Contributions
X.Q. and S.F.S. planned and supervised the project, analyzed the data, interpreted the structures, and wrote the paper; X.L. and L.Z. prepared the sample, analyzed the data, interpreted the structures, prepared the figures, and wrote the paper; G.H. processed the cryo-EM data, built and refined the structure model, analyzed the data, prepared the figures and wrote the paper; C.H. assisted in sample preparation. All authors discussed and commented on the results and the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Dmitry Semchonok and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, X., Huang, G., Zhu, L. et al. Structure of the red-shifted Fittonia albivenis photosystem I. Nat Commun 15, 6325 (2024). https://doi.org/10.1038/s41467-024-50655-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-50655-9
This article is cited by
-
Structure, regulation and assembly of the photosynthetic electron transport chain
Nature Reviews Molecular Cell Biology (2025)