Abstract
The ClC-3 chloride/proton exchanger is both physiologically and pathologically critical, as it is potentiated by ATP to detect metabolic energy level and point mutations in ClC-3 lead to severe neurodegenerative diseases in human. However, why this exchanger is differentially modulated by ATP, ADP or AMP and how mutations caused gain-of-function remains largely unknow. Here we determine the high-resolution structures of dimeric wildtype ClC-3 in the apo state and in complex with ATP, ADP and AMP, and the disease-causing I607T mutant in the apo and ATP-bounded state by cryo-electron microscopy. In combination with patch-clamp recordings and molecular dynamic simulations, we reveal how the adenine nucleotides binds to ClC-3 and changes in ion occupancy between apo and ATP-bounded state. We further observe I607T mutation induced conformational changes and augments in current. Therefore, our study not only lays the structural basis of adenine nucleotides regulation in ClC-3, but also clearly indicates the target region for drug discovery against ClC-3 mediated neurodegenerative diseases.
Similar content being viewed by others
Introduction
The ClC family of ion channels and exchangers mediates the transmembrane movements of the Cl- ion, which is the most abundant anion in human, so that they are critically involved in many biological processes1,2,3. The exchanger branch of ClC proteins, including the ClC-3 to ClC-7, mediates the counter exchange of two Cl- ions and one H+ ion1,2. As a member in the exchanger branch of ClC family, the ClC-3 is homologous to ClC-4 and ClC-5, which is widely expressed in the membrane of endosome. However, as the only available structure of this subbranch of ClC proteins is the intracellular cystathionine-beta-synthase (CBS) domains in ClC-54, whether ClC-3 employs similar mechanisms in Cl-/H+ exchange as other ClC exchangers like ClC-ec15,6 and ClC-77,8 remains to be investigated.
Moreover, the structural mechanisms underlying the physiological and pathological functions of ClC-3 are unexplored. Physiologically, ClC-3, together with ClC-4 and ClC-5, have been proposed as metabolic energy sensors as they are differentially modulated by ATP, ADP or AMP9. As ClC-3, ClC-4 and ClC-5 exchanger are mostly expressed on the membrane of endosome10,11 to assist acidification of endosome12,13,14 and shunt the electrical current of V-ATPase expressed there1, ATP modulation of this exchanger links the cellular metabolic energy state to the endosomal activities. However, whether adenine nucleotides bind to ClC-3 as in the CBS domains of ClC-54,15 and ClC-78, as well as how ClC-3 is differentially regulated are largely unknown.
Pathologically, point mutations in ClC-3 that cause global developmental delay (GDD), intellectual disability (ID) and neurodevelopmental disorders (ND) have been identified in 11 individuals16. This is supported by the systematically sick ClC-3 knockout mice, where the entire hippocampus of the mice was completely lost with severe retinal degeneration10,17 and abnormality in cellular protein degradation18. Moreover, as ClC-3 can form heterodimer with ClC-4, simultaneous introducing an “uncoupling” point mutation E224A (E282A in mClC-3b) in ClC-3 and knocking out of ClC-4 in mice lead to more severe neurodegeneration than that observed in ClC-3 knockout mice19. Furthermore, two of the human GDD/ID/ND causing mutations (I607T and T570I) in ClC-3 exhibited increased current amplitude at negative transmembrane voltages16, but how such mutations lead to gain-of-function behaviors in ClC-3 is unexplored.
To answer these questions, we first resolve the high-resolution structures of wildtype mouse ClC-3 in the apo state (mClC-3apo, 3.1 Å) and in complex with ATP (mClC-3ATP, 3.3 Å), ADP (mClC-3ADP, 4.0 Å) and AMP (mClC-3AMP, 3.1 Å), and the disease-causing I607T mutant in the apo (mClC-3I607T_apo, 4.0 Å) and ATP-bounded (mClC-3I607T_ATP, 3.0 Å) state by single-particle cryo-electron microscopy (Supplementary Table 1). We further perform patch-clamp recordings and molecular dynamic (MD) simulations to corroborate structural observations and elucidate mechanisms of physiological and pathological modulation in the ClC-3 exchanger.
Results
The overall structure of ClC-3
We expressed ClC-3 protein in HEK293 cells and then used cryo-EM to determine the structures of ClC-3 under various conditions. We observed that ClC-3 in apo state at a resolution of 3.1 Å (mClC-3apo, Fig. 1a–d and Supplementary Fig. 1) as a homo-dimer, with the overall architecture similar to other members in the ClC protein family1. Similar to previously resolved ClC protein structures6,7,8,20,21,22,23,24, in one subunit of the dimeric ClC-3, there are 17 α helices (Helix B to Helix R) in the transmembrane domains (Fig. 1e–h), where Helix H, I, P and Q form the dimer interface (Fig. 1h). We discovered such a dimer interface to be critical for the pathological function of ClC-3 as described later. The CBS domains for adenine nucleotides binding also located in the cytosol side as other ClC exchangers (Fig. 1a).
a–d 3D reconstruction and cartoon representation of ClC-3 in the apo state (mClC-3apo) with each subunit individually colored. e Topology and domain arrangement of ClC-3. f–h The structure of mClC-3apo subunit A. Helices located within the dimer interface are zoomed in. i The Cl- ions pathway identified by CAVER plugin in PyMOL and shown as blue shade.The region where pore radius less than 1.0 Å is displayed in orange. j Three Cl- ions observed in mClC-3apo are shown in green with their electron density mesh displayed. k Representative whole-cell recording of ClC-3 and mutant E282A (based on the mClC-3bS3/S2/S1 background). The voltage ramp protocol (from −120 mV to +180 mV for 450 ms) was used. l Statistical analysis of the ClC-3 and mutant E282A current-density in whole-cell recording at −120 mV. Data were represented as mean ± S.E.M. from biologically independent cells; n = 5. Two-sided t-test, the exact P values were labeled in the panels. m Statistical analysis of the ClC-3, mutants S453R and Y630A (based on the mClC-3bS3/S2/S1 background) current-density in whole-cell recording at +180 mV. Data were represented as mean ± S.E.M. from biologically independent cells; n = 5. Two-sided t-test, the exact P values were labeled in the panels. n The Cl- and H+ ions pathways identified by CAVER plugin in PyMOL and shown as shades in blue or yellow. The region where pore radius less than 1.0 Å is displayed in orange. o The electron density resembling one water molecule is shown as the mesh in red. Two helices next to the proton pathway are colored in blue. p The water molecule in mClC-3apo and the three interacting residues at the entrance of the proton pathway. q Mutations in key residues along the proton pathway significantly altered ClC-3 current-density measured by whole-cell recording at +180 mV. The mutants were based on the mClC-3bS3/S2/S1 background. Data were represented as mean ± SEM from biologically independent cells; n = 5. Two-sided t-test, the exact P values were labeled in the panels.
The Cl- and H+ conducting pathways
We have detected a chloride conducting pathway within the transmembrane domains of each subunit in mClC-3apo by the CAVER plugin in PyMOL25, which connect the endosomal lumen to the cytosol (Fig. 1i and Supplementary Fig. 4). Residues known critical for ion conducting ClC channels and exchangers, such as the “gating glutamate” E28219,20,26,27,28, also locate along this pathway with its sidechain pointed upward (Fig. 1i, j). We have also observed three electron densities likely representing three chloride ions right under E282 (Sext), near Y630 (Sin) and in between (Scen), respectively (Fig. 1j), which is similar to chloride ions observed in ClC-7 exchanger8 (Supplementary Fig. 2).
We then validated the functional roles of pathway-lining residues by site-directed mutagenesis and patch-clamp recording. To record current meditated by ClC-3, which is mostly expressed on endosomal membrane, we first introduced point mutations in the N terminus of ClC-3 to enable its plasma membrane localization as previously reported29,30. When this mutant (mClC-3bS3/S2/S1) was transiently expressed in HEK293T cells, it was indeed expressed on the cell membrane (Supplementary Fig. 3a–d).
On this background exchanger, we further mutated E282 to Ala. As expected, such a mutation led to reduction in outward rectification and increase in current amplitude under hyperpolarization (Fig. 1k, l) due to uncoupling of chloride and proton transport19,28. Moreover, when S453 near the Sext site was mutated to Arg, which was found in patients to cause neurodevelopmental disorders16, the extra positive charge introduced likely favored the enrichment of the negatively charge Cl- ions so that the outward current at positive membrane potential was increased (Fig. 1m). Y630 constitutes another conversed “gate” in ClC channels and exchangers31,32,33 (Supplementary Fig. 4), the Y630A mutation also caused enhancement in current at depolarization as observed before34,35 (Fig. 1m).
Furthermore, we observed that such a pathway for ions is bifurcated, starting near the Scen site behind Helix O and Helix P (Fig. 1n, o, pathways in blue and yellow, respectively). E339 is the conserved “proton glutamate” in ClC exchangers36,37,38, which located at the cytosolic entrance of this pathway as previously observed39,40,41 (Fig. 1o). We observed an electron density likely resembling a water molecule there, which has been observed in previous molecular dynamics studies42,43,44 and is also in close proximity to other charged residues like K245 and E631 (Figs. 1o and 1p). Mutating any of these residues caused significantly altered ClC-3 current (Fig. 1q).
However, as the steady-state current level is a function of three factors (open probability, conductance, and expression level), the changes in current ratio we observed in these mutants here and mutants later reported in this study may be caused by any or a combination of these three factors. We further performed surface biotinylation and confocal imaging to examine the expression of ClC-3 mutants on the cell surface. We observed that though the overall expression levels of the mutants K245A, E339A and E631A were reduced as compared to that of WT (Supplementary Fig. 3e), with the surface biotinylation assay, we observed that the expression levels of these mutants on cell surface were at least comparable to that of the WT ClC-3 (Supplementary Fig. 3e). The surface expression levels of E339A and E631A mutants, which exhibited much reduced current density (Fig. 1q), were even higher than that of WT (Supplementary Fig. 3e), indicating these point mutations disrupted the function of ClC-3. Moreover, all the point mutants, which were fused with GFP as in patch-clamp recordings, exhibited clear co-localization with the cell membrane labeled with the fluorescent membrane dye DiD, indicating that they were expressed on the cell membrane (Supplementary Fig. 3f). We believe that these mutants were expressed on the cell membrane with a level similar to or higher than the WT ClC-3. Therefore, we assigned this pathway for proton transport.
Adenine nucleotides binding in ClC-3
To investigate regulation of ClC-3 by adenine nucleotides, we first observed that this exchanger was robustly potentiated by 10 mM ATP added in the patch pipette solution under the whole-cell patch configuration (Fig. 2a and Supplementary Fig. 5a). We quantified the effect of ATP on ClC-3 by the parameter of current ratio, which was calculated by comparing the maximum current amplitude reached after ATP potentiation to the initial amplitude before ATP potentiation measured as the average current amplitude within the voltage range between -150 mV and +180 mV in the ramp protocol, and with the voltage step protocol as I-V curves. To further rule out the possibility that ATP indirectly potentiated ClC-3 after its hydrolyzation, we used the non-hydrolysable analog adenosine 5′-(γ-thio)triphosphate (ATPγS). ATPγS potentiated ClC-3 as ATP (Fig. 2b, e), while adding 10 mM ADP or AMP in the same way barely potentiated ClC-3 (Fig. 2c–e, Supplementary Fig. 5 and Supplementary Fig. 6). Therefore, we believe ATP is a potentiator of this exchanger.
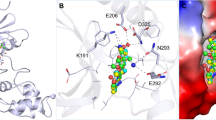
a–d The whole-cell current recordings of ClC-3 after about five minutes of break-in with 10 mM ATP, ATPγS, ADP, or AMP in pipette solution. The black represents the initial, the colored represents the current after five minutes. The voltage ramp protocol (from −120 to +180 mV in 450 ms) was used. e The current ratio stimulated by 10 mM ATP, ATPγS, ADP, and AMP to the initial current (mean ± SEM, n = 5 from biologically independent cells). Two-sided t-test, the exact P values were labeled. f–h The structure of mClC-3ATP. The electron density of ATP are shown in red. i The ATP binding pocket of ClC-3. j The current ratio after about five minutes of 10 mM ATP stimulation to the initial current of ClC-3 mutants (based on the mClC-3bS3/S2/S1 background).The corresponding n values are 5, 3, 5, 5, 5, 3, 6, 3, 3, 3, 3, 4, 5, 4, 4, 3, 5, 4, 3, respectively (mean ± S.E.M. from biologically independent cells). Two-sided t-test, the exact P values were labeled. k The backbone of the ATP binding site comparing the apo (brown) with the ATP (green) bound models. l The structure of mClC-3ADP. The electron density of ADP are shown in blue. m The structure of mClC-3AMP. The electron density of AMP are shown in orange. n The overlay of the ATP (red), ADP (blue) and AMP (orange) bound models. H89 and K798 close to gamma phosphate in ATP are shown. o The backbone of the ATP binding site comparing the ATP (green), ADP (grey), and AMP (pink) bound models. p A concentration-response for AMP or AMP-PNP with ATP showing that these nucleotides antagonise the current induced by ATP in a concentration-dependent manner. The ratio of the current after about five minutes of different concentration of AMP or AMP-PNP with 10 mM ATP to the initial. The corresponding n values are 5, 5, 6, 5, 5, 5, 5, 5, 5, respectively (mean ± S.E.M. from biologically independent cells). Two-sided t-test, the exact P values were labeled.
To understand how ATP binds to potentiate ClC-3, we determine the structures of ClC-3 in ATP binding state at a resolution of 3.3 Å (Supplementary Fig. 7). We first scrutinized our electron density map and structure model of mClC-3ATP and observed an extra density resembling ATP between the two intracellular CBS domains (Fig. 2f, g, density highlighted in red). We added ATP to the ClC-3 protein sample right before plunge freeze to minimize the hydrolysis of this molecule. The chemical structure of an ATP molecule nicely fitted within such a density, while no density was observed in the same region in the apo state (Fig. 2h, zoomed-in view and Supplementary Fig. 8a–e). The binding configuration of ATP and the sequence of ATP binding pocket in our ClC-3 were also very similar to that in ClC-54 and ClC-78 (Supplementary Fig. 9), where ATP binding was stabilized by residues from the N terminus (H89), CBS1 domain (N690 and Y689) and CBS2 domain (K798 and R659) (Fig. 2i). Indeed, when these residues were mutated (Supplementary Table 2), the mutants showed significantly reduced current potentiation by ATP (Fig. 2j).
We further performed all-atom MD simulation of mClC-3ATP, mClC-3ADP and mClC-3AMP. During the 300 ns simulation, the mClC-3 protein was stable with Root-mean-square-deviation (RMSD) plateaued after about 30 ns (Supplementary Fig. 10a–f). In the three independent trajectories, the ATP molecule stably bound within the binding pocket as shown in the ensemble plot of this molecule (Supplementary Fig. 10b).
We further observed that several residues within the ATP binding pocket exhibited conformational rearrangements as compared to those in the apo state, for instance, the sidechain in R659 flipped to accommodate the adenine group in ATP, while sidechains in H89 and K798 rotated and shifted, respectively, to accommodate the triphosphate group in ATP (Fig. 2k). ADP or AMP molecule can also stay bounded within the pocket with their phosphate groups pointing towards H89 and K798 (Supplementary Fig. 10c to 10f). Therefore, we have elucidated the binding configuration of ATP in the ClC-3 exchanger.
In contrast to the robust potentiation of ClC-3 by ATP, ADP or AMP barely enhance the current (Fig. 2c–e). Nevertheless, with the addition of saturating concentration of ADP or AMP (500 µM), we have observed the electron densities corresponding to these molecules in mClC-3ADP and mClC-3AMP structures (Fig. 2l, m, densities in blue and yellow, respectively; Supplementary Fig. 11 and Supplementary Fig. 12). ADP and AMP also bind to the ATP binding pocket in the CBS domains, with their binding configuration similar to that of ATP (Fig. 2n). However, we observed that while the gamma phosphate in ATP is close to H89 and K798, the beta phosphate in ADP or the alpha phosphate in AMP is away from these residues (Fig. 2o). Such distinctions in adenine nucleotides binding configuration prompted us to hypothesize that the electrostatic interactions between the negatively charged gamma phosphate in ATP and positively charged K798 and H89 residues are critical for ATP potentiation of ClC-3. If so, reducing the negative charge in the triphosphate group of ATP would decrease ATP potentiation. Indeed, as there are also three phosphate groups in the non-hydrolysable ATP analogue AMP-PNP but with less negative charge there as compared to ATP or ATPγS (Supplementary Fig. 13), AMP-PNP much less potentiated ClC-3 as compared to ATP or ATPγS (Fig. 2p). In fact, AMP or AMP-PNP antagonized ATP-potentiated current in a concentration-dependent manner (Fig. 2p). Therefore, removing the phosphate groups from ATP molecule reduced its binding affinity to ClC-3.
To corroborate these findings, we investigate the binding of ATP, ADP or AMP in MD simulations. The RMSD plot of ATP molecule exhibited smaller deviation as compared to that of ADP or AMP (Supplementary Fig. 10g to 10i), which was consistent with the ensemble plots of these ligands (Supplementary Fig. 10b, d, f), suggesting a tighter binding of ATP to ClC-3. We further performed MM/PBSA calculation in the MD simulation. We observed the calculated binding energy in ATP to ClC-3 was significantly larger than that of AMP (Supplementary Fig. 10j).
ATP potentiation of ClC-3
To investigate how ClC-3 current is enhanced upon ATP binding, we then examined the pathways for ion transport inside the transmembrane domains. While the overall structures of mClC-3ATP and mClC-3apo were similar (Fig. 3a) and there were three densities resembling Cl- ions in each of the two subunits (Fig. 3b–f, spheres in green with the electron density mesh), the location of Cl- ions are different in the presence of ATP. In the apo state, we found that Cl- ions occupied the Sext near E282, Scen near S239 and Sin below S239, respectively (Fig. 3f). Such a Cl- ion occupancy has also been observed in ClC-7 transporter (Supplementary Fig. 2). With the addition of ATP, we observed another Cl- ion near K281 and S453, which we referred to as the Sout, but the Sext was unoccupied (Fig. 3d, e, mesh in black). Occupancy of the Sout may push the Cl- ion in the Sext to move toward Scen and Sin to facilitate the ion transport. Expectedly, the nearby E282 and its corresponding residues in other ClC channels and transporters have been identified as the “gating glutamate” (Supplementary Fig. 14)19,20,26,27,28, the E282A mutant exhibited significantly increased inward current at negative membrane potential (Fig. 1l).
a–c Side view of mClC-3 in apo and ATP bound states with Cl- ions shown in green. d Sliced view of mClC-3ATP shows four Cl- ions binding sites in the transmembrane domains (TMD). The surface is colored by electrostatic potential (red, 10 kTe−1; blue, +10 kTe−1). e, f The four sites for Cl- ion binding in mClC-3 in apo and ATP bound states. Cl- ions are shown in green with their electron density mesh displayed. Residues close to these sites are shown. g The schematic diagram of the positions of the portals for the chloride ion and proton near CBS domains. h The portal outlined with green dashed line for the Cl- ion in the bottom view in mClC-3ATP, mClC-3AMP and mClC-3apo. The surface is colored by electrostatic potential. K521 residue was outlined in pink. i One subunit in mClC-3ATP, mClC-3AMP and mClC-3apo was compared in the bottom view, revealing specific conformational changes of the residues near the Sin Cl- ion. j The current ratio after about five minutes of 10 mM ATP stimulation to the initial current of ClC-3 mutant K521A and K521D (based on the mClC-3bS3/S2/S1 background). Mean ± S.E.M. from biologically independent cells, n = 5. Two-sided t-test, the exact P values were labeled. k The two sites of H2O of mClC-3 in apo, AMP bound and ATP bound states observed near the proton entrance. H2O molecules are shown in red with their electron density mesh displayed. Residues close to these sites are shown. l The specific conformational changes observed by structural alignment of mClC-3ATP, mClC-3AMP and mClC-3apo. Residues lining the H+ ion transport pathway showed altered sidechain orientations. m The water molecules in mClC-3AMP interacting with E339 and E631 simultaneously, suggesting the possibility of proton transport between the two residues.
In addition, we performed dynamic network analysis of our MD simulation of mClC-3ATP, where we observed a mutual information pathway starting from the ATP binding pocket in the CBS domains to the vicinity of Sout (Supplementary Fig. 10k, residues denoted as spheres in pink), which is in the vicinity of Cl- ions observed in cryo-EM (Fig. 3d, spheres in green). We have introduced point mutations (K245A, P640A, F641A, K777A and L778A, Supplementary Fig. 10k) to the mutual information pathway. We have measured the ATP potentiation in these mutants by patch-clamp recordings, where we observed that the majority of mutations (four out of five) significantly reduced ATP potentiation (Supplementary Fig. 10l). The local hydrogen bond network along this pathway was also altered (Supplementary Table 3). Therefore, we believe that ATP binding caused conformational changes in the critical residues in the transmembrane domains to enhance Cl- ion transport. Nevertheless, the changes in Cl- ion transport we observed in these mutants here and mutants later reported in this study may be caused by any or a combination of changes in open probability, conductance, and expression level of ClC-3.
ADP or AMP barely potentiates ClC-3
We further observed that ATP induced specific conformational changes as compared to the apo and AMP-bound states (Fig. 3h–i). For instance, upon ATP binding the sidechain of residues such as K521 pointed away from the Cl- ion in the Sin (Fig. 3h, i, Supplementary Fig. 8h–j), leading to the altered electrostatic distribution there and a wider exit portal for the Cl- ion. As expected, when we mutated K521, ATP potentiation of ClC-3 was significantly reduced (Fig. 3j).
Water molecules have previously been detected near this glutamate in molecular dynamics simulations of ClC exchangers and have been proposed to serve as a proton-conducting water-wire42,43,44. In mClC-3ATP, we found on electron density resembling water molecules near E339, which we referred as the H2O SE339 (Fig. 3k). In addition, there was another water resembling density close to E631 in Helix R, which we referred as the H2O SE631 (Fig. 3k–m). In either the apo or AMP-bound states, only one of the water binding sites was occupied, which is distinct to that in the ATP-bound state. Indeed, when these residues were mutated, the current of mutants was significantly reduced (Fig. 1q).
Structural changes in the disease-causing I607T mutant
Several point mutations in ClC-3 have been identified as the cause of GDD/ID/ND in 11 individuals, for instance, the individual with I607T mutation passed away at very early age16. The I607T mutation led to enhanced current with acidification in the endosomal lumen side to pH 5.016 (Fig. 4a–d). Such enhancements in I607T current were reversible (Supplementary Fig. 15). However, the structural basis of such a gain-of-function in I607T mutant remained unknown. Therefore, we resolved the structure of I607T mutant in the apo and ATP-bound states (Supplementary Figs. 16 and 17).
a, b Representative current recordings of ClC-3 expressed in H293T cells with endogenous proton sensitive PAC channel knocked out at extracellular pH 7.3 or pH 5.0. The voltage step protocol (ranging from −80 to +160 mV with a 20 mV step for 300 ms) was employed. c, d Representative current recordings of I607T mutant (based on the mClC-3bS3/S2/S1 background) expressed in H293T cells with endogenous proton sensitive PAC channel knocked out at extracellular pH 7.3 or pH 5.0. The voltage step protocol ranging from −80 to +160 mV (with a 20 mV step) for 300 ms was employed. e, f Overall cartoon structure of mClC-3I607T_apo or mClC-3apo was aligned to reveal clear conformational changes within the dimer interface. g Structural alignment of mClC-3I607T_ATP and mClC-3ATP revealed the substantially decreased size induced by the I607T mutation. h Packing of dimer interface in ClC-3 has been tightened by the I607T mutation. i Residues within the dimer interface of mClC-3I607T_ATP critical for dimer interface packing are shown in blue. j, k Representative current recordings of I607T-F598A double mutant (based on the mClC-3bS3/S2/S1 background) expressed in H293T cells with endogenous proton sensitive PAC channel knocked out at extracellular pH 7.3 or pH 5.0. The voltage step protocol was employed. l, m Representative current recordings of I607T-E599A double mutant (based on the mClC-3bS3/S2/S1 background) expressed in H293T cells with endogenous proton sensitive PAC channel knocked out at extracellular pH 7.3 or pH 5.0. The voltage step protocol was employed. n, o Average current-density voltage relationship of ClC-3 and mutants measured in H293T cells with endogenous proton sensitive PAC channel knocked out at extracellular pH 7.3 or pH 5.0. Mean ± SEM from biologically independent cells, n = 3. Two-way ANOVA, the exact P values were labeled. p Schematic model showing the proposed mechanism of ClC-3 potentiation by ATP. q Schematic model showing the proposed mechanism of I607T mutation induced functional changes in ClC-3.
We clearly identified the conformational changes in the I607T mutant in the apo state, where the Helix Q bearing the mutation tilted towards the dimer interface by 20° as compared to the wildtype mClC-3apo (Fig. 4e, f). Helix P also moved toward the dimer interface by about 2 Å (Fig. 4e, f). With the addition of ATP, the dimension of ClC-3 was substantially decreased from 106 Å in mClC-3ATP to 95 Å in mClC-3I607T_ATP (Fig. 4g), clearly indicating the packing of dimer interface in ClC-3 has been tightened by the I607T mutation (Fig. 4h). With ATP binding, the I607T mutation caused a counterclockwise rotation as compared to the WT ClC-3 (Supplementary Fig. 18a and 18b), which is similar to the recently reported ClC-7 conformation45. We also observed that the chloride ion occupancy was changed in the I607T mutant, as there were chloride ions occupying all the four Sout, Sext, Scen and Sin binding sites in the permeation pathway in I607T, but in the WT the Sext site was not occupied (Supplementary Fig. 18c to 18f). Such an increase in ion occupancy is in line with the gain-of-function phenotype of I607T when activated by ATP (Fig. 4b, d). We thus hypothesized that I607T mutation changes the dimer interface to alter the function of ClC-3.
To test this hypothesis, we reasoned that if the tighter packing of the dimer interface in I607T is weakened, the gain-of-function phenotype of this mutant may be rescued. Therefore, we introduced several point mutations to disrupt the dimer interface, including F598A, E599A and M611A (Fig. 4i), on the background of I607T mutant. Upon acidification to pH 5.0 in the endosomal lumen side, I607T mutant exhibited much increased current at negative membrane potential as compared to that in wildtype ClC-3 (Fig. 4a–d). However, double mutants I607T-F598A, I607T-E599A and I607T-M611A showed little increase in current, which is similar to the wildtype ClC-3 (Fig. 4j–o), indicating the gain-of-function phenotype of I607T was successfully rescued. Therefore, our hypothesis that the disease-causing I607T mutation alters the dimer interface to change ClC-3 function is supported by both cryo-EM structures and patch-clamp recordings.
Discussion
In this study, by determining high-resolution structures of the ClC-3 exchanger, we have found that similar Cl- and H+ ion transport mechanisms have been employed as compared to other ClC exchangers (Fig. 4p). For instance, electron densities resembling Cl- ions and water molecules have been observed near residue E282 and E339, respectively, in our ClC-3 structures. These residues have been well recognized as the “gating glutamate” and “proton glutamate” in ClC protein family1. Nevertheless, besides the Sext, Scent and Sin for Cl- ion binding in many ClC proteins, we observed another Cl- ion binding site (Sout) near K281 and S453 (Fig. 3e) in the ATP bound state, which has previously been detected in molecular dynamics simulations of ClC-ec146,47,48. Intriguingly, the S453R mutation in ClC-3 was found in patients to cause neurodevelopmental disorders16. Human mutation in the corresponding residue in ClC-4 (S395R) also lead to similar disorders49.
ClC-3 has been proposed as a sensor for metabolic energy state9. We have revealed the structural basis of the differential regulation of ClC-3 by adenine nucleotides. Though conformational changes, especially in the backbone of ClC-3 protein, between the apo, ATP, ADP, and AMP-bound state are relatively small, we observed that ATP binds to ClC-3 with a higher affinity and induced changes in ion occupancy between apo and ATP-bounded state, which may facilitate the transport of Cl- and H+ ions. However, the increase in Cl- ion transport by ATP may be caused by any or a combination of changes in open probability, conductance, and expression level of ClC-3. As ClC-3 is permissive for the release of insulin-containing secretory granules in pancreatic beta cells50,51 and essential in restricting proinflammatory signalling52,53 and regulating of reactive oxygen species (ROS) signaling54, the level of relative ATP/ADP/AMP concentration may control insulin production and contribute to the host defense function through their differential regulation of ClC-3.
Moreover, we have gained structural insights regarding the disease-causing I607T mutation in ClC-3 (Fig. 4q). Residue I607 locates on the dimer interface of ClC-3, so that a mutation there altered the dimer interface (Fig. 4e–g). Indeed, interactions there are known to be important in ClC proteins. For instance, in ClC-ec1 tryptophan mutation at residue I422, which corresponds to I607 in the ClC-3, completely disrupts the dimer assembly so that the ClC-ec1 mutant becomes a monomer55. In ClC-4, the asparagine mutation at residue I549 in patients with neurodevelopmental conditions, which corresponds to the I607 in ClC-3, also exhibited a gain-of-function phenotype49. Human mutations at or next to this residue have also been reported in ClC-1 (I553F)56 and ClC-6 (Y553C)57, which cause myotonia congenita and early-onset neurodegeneration, respectively. In ClC-5, some mutations in individuals with Dent disease are also clustered in the dimer interface58. On the I607T mutant of ClC-3, we have successfully rescued its gain-of-function phenotype by introducing extra mutations in the dimer interface (Figs. 4n, o), suggesting that the dimer interface could be targeted for drug discovery against GDD/ID/ND caused by this mutation or any other mutation with similar structural effects in ClC-3 or other ClC proteins in future.
Beyond the dimer interface, disease-causing point mutations of ClC-3 are also distributed in transmembrane domains, CBS domains and their interface (Supplementary Fig. 19). In fact, in ClC-4 or ClC-5 exchanger, mutations in individuals with neurodevelopmental conditions or Dent disease are also widely distributed49,58,59. How these mutations lead to abnormal function in ClC protein remains to be investigated in future.
Methods
Protein expression and purification
Mouse ClC-3apo, ClC-3I607T were cloned, expressed and purified using similar procedures. The full-length mouse ClC-3b gene (GenBankTM Accession Number: NM_007711.3) containing an N-terminal Flag and Strep tag II was cloned into a pEG-BM vector. The ClC-3 I607T mutation was introduced into the expression vector using a mutagenesis kit. All the ClC-3s were heterologously expressed in HEK293F GnTl− suspension cells (Life Technologies) using the BacMam system (Thermo Fisher Scientific). P3 baculovirus generated in Sf9 cells (Life Technologies) following the standard protocol was used to transfect HEK 293F GnTl− cells at a ratio of 1:10 (virus to HEK293F, v/v) when the cell density was around 4 × 106 cells per ml. Sodium butyrate (10 mM) was added 12 h post-transduction, and then cells were maintained at 30 °C to enhance protein expression. About 60 h post-transduction, cells were harvested by centrifugation at 4000 g, then flash-frozen in liquid nitrogen and stored at –80 °C.
The cell pellet was resuspended in lysis buffer A (20 mM Hepes pH 7.4, 150 mM NaCl) supplemented with a protease inhibitor cocktail (2 μg ml−1 DNase I, 0.5 μg ml−1 pepstatin, 2 μg ml−1 leupeptin, 1 μg ml−1 aprotinin and 100 μM phenylmethylsulfonyl fluoride) and homogenized by sonication on ice. ClC-3 protein was extracted with 1% (w/v) N-dodecyl-β-D-maltopyranoside (DDM, Anatrace) and 0.25% (w/v) cholesteryl hemisuccinate (CHS, Anatrace) by gentle agitation for 3 h at 4 °C. After extraction, the supernatant was collected following 1 h of ultracentrifugation at 48,000 g and then incubated gently with 0.5 ml of Strep-Tactin Sepharose resin (IBA) at 4 °C. After 1 h, the resin was collected on a gravity column (Bio-Rad) and washed in buffer B (buffer A + 0.05% lauryl maltose neopentyl glycol (LMNG) + 0.001% CHS) for 20 column volumes. The detergent was changed to 0.005% LMNG (Anatrace) and the protein sample was eluted with 10 mM desthiobiotin (Sigma) in buffer C (buffer A + 0.005% LMNG + 0.001% CHS). The protein sample was further purified by size-exclusion chromatography on a Superose 6 10/300 GL column (GE Healthcare) pre-equilibrated with buffer C. The peak fraction of ClC-3 was collected and concentrated in a 100 kDa concentrator (Millipore Sigma) to appropriate concentrations (5 mg ml−1) for cryo-EM analysis.
Cryo-electron microscopy sample preparation and data acquisition
The cryo-EM data of all ClC-3 structures were collected at the Center of Cryo-Electron Microscopy, Zhejiang University. For the ClC-3AMP, ClC-3ADP, ClC-3ATP and ClC-3I607T_ATP sample, AMP, ADP, ATP were added to the ClC-3 sample separately to a final concentration of 500 μM. Cryo-EM grids were prepared by applying 3 μl of ClC-3 protein sample to a glow-discharged holey carbon grid (Quantifoil Cu R1.2/1.3, 300 mesh) and blotting for 3.5 s under 100% humidity at 4 °C before plunging the grid into liquid ethane using a Mark IV Vitrobot (FEI).
Micrographs of all ClC-3 structures were acquired on a Titan Krios microscope (FEI) operated at 300 kV and equipped with the Selectris energy filter and Falcon4 detector. EPU software was used for automated data collection following standard procedures. A calibrated magnification of ×140,000 was used for imaging, yielding a pixel size of 0.93 Å on images. The defocus range was set from –0.8 to –1.5 μm. Each micrograph was dose-fractionated to 40 frames under a dose rate of 7.49 e− pixel−1 s−1, with a total exposure time of 6 s, resulting in a total dose of about 52 e− Å−2.
Image processing
Motion correction was performed using the MotionCorr2 program, and the CTF parameters of the micrographs were estimated using the Gctf program. Other image-processing steps were performed using cryoSPARC v.3.2.0.
For ClC-3apo, 1,462,270 particles were automatically picked and extracted from the full data set of 3099 micrographs via cryoSPARC. The particles were subjected to 2D classification. The particles extracted were subjected to one round of ab-initio reconstruction. One of the classes showed good secondary structural features and their particles were selected. After nonuniform refinement with C2 symmetry, the final 3D reconstruction of ClC-3apo from 116,977 particles yielded an EM map with a resolution of 3.06 Å.
For ClC-3ATP, 1,593,953 particles were automatically picked and extracted from the full data set of 3,152 micrographs via cryoSPARC. The particles were subjected to 2D classification. The particles extracted were subjected to one round of ab-initio reconstruction. One of the classes showed good secondary structural features and their particles were selected. After nonuniform refinement with C2 symmetry, the final 3D reconstruction of ClC-3ATP from 97,275 particles yielded an EM map with a resolution of 3.30 Å.
For ClC-3AMP, 1,293,826 particles were automatically picked and extracted from the full data set of 2506 micrographs via cryoSPARC. The particles were subjected to 2D classification. The particles extracted were subjected to one round of ab-initio reconstruction. One of the classes showed good secondary structural features and their particles were selected to one round of heterogeneous refinement. Then one of the classes showed good secondary structural features and their particles were selected. After nonuniform refinement with C2 symmetry, the final 3D reconstruction of ClC-3AMP from 84,022 particles yielded an EM map with a resolution of 3.14 Å.
For ClC-3ADP, 1,283,991 particles were automatically picked and extracted from the full data set of 2532 micrographs via cryoSPARC. The particles were subjected to 2D classification. The particles extracted were subjected to one round of ab-initio reconstruction. One of the classes showed good secondary structural features and their particles were selected to one round of heterogeneous refinement. Then one of the classes showed good secondary structural features and their particles were selected. After nonuniform refinement with C2 symmetry, the final 3D reconstruction of ClC-3ADP from 25,460 particles yielded an EM map with a resolution of 4.04 Å.
For ClC-3I607T_apo, 1,191,754 particles were automatically picked and extracted from the full data set of 2284 micrographs via cryoSPARC. The particles were subjected to 2D classification. The particles extracted were subjected to one round of ab-initio reconstruction. One of the classes showed good secondary structural features and their particles were selected to one round of heterogeneous refinement. Then one of the classes showed good secondary structural features and their particles were selected. After nonuniform refinement with C2 symmetry, the final 3D reconstruction of ClC-3I607T_apo from 122,975 particles yielded an EM map with a resolution of 3.96 Å.
For ClC-3I607T_ATP, 944,219 particles were automatically picked and extracted from the full data set of 1996 micrographs via cryoSPARC. The particles were subjected to 2D classification. The particles extracted were subjected to one round of ab-initio reconstruction. One of the classes showed good secondary structural features and their particles were selected. After nonuniform refinement with C2 symmetry, the final 3D reconstruction of ClC-3I607T_ATP from 57,969 particles yielded an EM map with a resolution of 2.98 Å.
The resolutions were estimated by applying a soft mask around the protein density and the gold-standard Fourier shell correlation =0.143 criterion. Local resolution maps were calculated via cryoSPARC.
Model building, refinement and validation
De novo atomic model building was performed in Coot based on the 3.1 Å resolution density map of ClC-3apo. The ClC-3apo atomic model was built using the ClC-3 model predicted by AlphaFold II as a starting template. The amino acid assignment was achieved based on the clearly defined density for bulky residues (Phe, Trp, Tyr and Arg). Models were refined against summed maps using real-space refinement in PHENIX50, with secondary structure restraints and non-crystallography symmetry applied. The initial EM density map enabled us to construct an ClC-3apo model containing residues 85–174, 188–368, 389–464, 497–559, 569–644, 651–661 and 753–805. The models of all other ClC-3 structures were built using the model of ClC-3apo as a template. The geometry statistics for models were generated using MolProbity. All of the figures were prepared in PyMoL, Chimera or Coot.
Cell culture for electrophysiology
HEK293T cells were cultured using DMEM medium containing 10% FBS in a cell incubator with 5% CO2 at 37 °C. Plasmids of mouse ClC-3b was labeled with green fluorescent protein as indicators for subsequent electrophysiological recordings.
When the cell density reaches 50–70%, the transfection reagent Lipofectamine 3000 (Invitrogen) and 3 μg plasmid DNA were used to transfect cells according to the instructions. Electrophysiological recordings were performed 16–24 h after transient transfection. Point mutations of mClC-3b were generated by Fast Mutagenesis Kit (Takara Bio). Primers used to generate point mutations are summarized in Table S2. All mutations were confirmed by sequencing.
Electrophysiology
Patch-clamp recordings were carried out with a HEKA EPC10 amplifier controlled by PatchMaster software (HEKA). Patch pipettes were prepared from borosilicate glass and fire-polished to a resistance of 3–6 MΩ using a P-97 puller for whole-cell recordings. Data from whole-cell recordings were analyzed in Igor Pro v.5.05 (WaveMatrix). The series resistance was about 25 MΩ under our whole-cell patch-clamp settings. We have compensated the series resistance by 60–70% in the PatchMaster software. We used consistent series resistance compensation in all whole-cell recordings.
Patch pipette solution contained 135 mM CsCl, 1 mM MgCl2, 5 mM EGTA, 10 mM HEPES (pH adjusted to 7.3 with CsOH). The extracellular solution: 150 mM NaCl, 2 mM KCl, 2 mM MgCl2, 1.5 mM CaCl2, 10 mM glucose, 10 mM HEPES (pH adjusted to 7.3 with NaOH). Adenine nucleotides (10 mM ATP, 10 mM ATPγS, 10 mM ADP, 10 mM AMP or 10 mM AMP-PMP) were added to the pipette solution shortly at 4 °C before the experiments. The acid extracellular solution with pH 5.0 was buffered with MES.
Two types of stimulation protocols were applied from a holding potential of -10 mV. The voltage ramp protocol (from −120 to +180 mV for 450 ms) was used to record the outward currents. The voltage step protocol consisted of steps ranging from −80 to +160 mV (in 20 mV steps) for 300 ms. The current was sampled at 10 kHz and filtered at 2.9 kHz. All recordings were performed at room temperature (25 °C) with the maximum variation of 1 °C.
The current ratio was calculated by comparing the maximum current amplitude reached after ATP potentiation (approximately five minutes after break-in to establish whole-cell patch-clamping) to the initial amplitude before ATP potentiation, where the current amplitude was measured as the average current between the range voltage of 150 mV to 180 mV.
A gravity-driven system (RSC-200, Bio-Logic) with freely rotating tubes was used for the ligand perfusion. The bath and ligand solutions were delivered through separate tubes to minimize mixing. Patch pipette holding cells were placed in front of the tube outlet for perfusion.
Western Blot
To quantify the whole cell expression and the cell surface expression level of ClC-3 mutants, the total cell lysates and the membrane protein of HEK293T cells of the same volume were respectively harvested. For extracting ClC-3 mutants of the cell surface, the Pierce™ Cell Surface Protein Biotinylation and Isolation Kit (Thermoscientific) was used, and the membrane protein was loaded according to the same volume and concentration. The total cell lysatesand and the membrane protein were resolved on 4–20% ExpressPlus SDS-PAGE gels with MOPs (3-(N-morpholino) propanesulfonic acid) running buffer and transferred to 0.2 µm PVDF membranes. The membranes were incubated with anti-GFP antibody (proteintech) and GAPDH antibody (Abcam) (1:1000), and then reprobed with the goat anti-rabbit IgG H&L Alexa Fluor 680 (Abcam) (1:5000) secondary antibodies.
All-atom molecular dynamic simulation
With the cryo-EM structures of mClC-3apo and mClC-3ATP, we used the Membrane Builder function60 of the CHARMM-GUI webserver61,62 to embed mClC-3ATP in a bilayer of 1-palmitoyl-2-oleoyl phosphatidylcholine (POPC) lipids, surrounded by a box of water and ions with a buffer distance of 15 Å with an overall system size of 170 Å × 170 Å × 165 Å. To ensure 0.15 M ionic concentration and zero net charge, 224 K+ and 224 Cl- ions were added. Equilibrations of the system were performed with 6 steps of gradually decreasing position restraints on the protein and lipids. Temperature coupling was done with Berendsen thermostat at a reference temperature of 303 K during the entire equilibration phase63. Berendsen barostat with a reference pressure of 1 bat and a compressibility of 4.5 × 10 − 5/bar was applied for pressure control during NPT equilibration. For production runs, Nose-Hoover thermostat64,65 and Parrinello-Rahman barostat66 were used for temperature and pressure coupling respectively. Covalent bonds are constrained to their equilibrium length by the LINCS algorithm67 and the long-range electrostatics (<1 nm) were modelled using the Particle Mesh Ewald (PME) method68. Three repeats of 300 ns unrestrained atomistic simulations were performed using GROMACS69 version 2021.3 with the CHARMM3670 force field and TIP3P71 water model for each system. VMD72 was used for visualization and analysis.
To identify hydrogen bond altered by the change of sidechain orientations, we use VMD hydrogen bonds plugin to calculate the average number of hydrogen bonds formed per frame between different residues for both apo and ATP-bound states. For each frame, hydrogen bonds are formed between donor and acceptor atoms provided that their distance less than 3.0 Å and the angle donor-hydrogen-acceptor less than 30 degrees. Going through each frame in the trajectory and we can calculate the average number of hydrogen bonds formed per frame between different residues.
MM/PBSA calculation
We used the MM/PBSA (molecular mechanics with Poisson-Boltzmann and surface area solvation) method to estimate the free energy of the binding of the ligands to ClC-3. In MM/PBSA, the free energies of the protein (P), the ligand (L), and the protein-ligand complex (PL) are estimated from the following sum73,74:
The first three terms are energy terms from bonded (bond, angle and dihedral), electrostatic and van der Waals interactions which can be directly obtained from forcefields used for simulation, Gpol and Gnp are the polar and non-polar contributions to the solvation free energies. Gpol is typically obtained by solving the Poisson-Boltzmann equation. Gnp is estimated from a linear relation to the solvent accessible surface area (SASA). The binding free energy is estimated as:
where \( < \ldots { > }_{{PL}}\) indicates ensemble average estimated from molecular dynamics simulation of the protein-ligand complex, GP, GL and GPL are free energies of the protein, the ligand and the protein-ligand complex respectively. The initial 50 ns trajectories where the ligands still stayed bound to ClC-3 were taken from 300 ns simulations for MM/PBSA calculations using gmx_MMPBSA75,76.
Dynamic network analysis
To obtain the mutual information pathway between ATP binding pocket and the ion transport channel, we follow the dynamic network analysis method77. First, we represent all residues in our system by nodes. Amino acid residues are represented by a single node located in their alpha-carbons, and ions are trivially represented by one node. Second, we determine which nodes are in contact. For each frame, the shortest distance between heavy atoms (all atoms excluding hydrogen atoms) represented by two nodes is calculated. If the distance is shorter than 4.5 Å, the pair of nodes is said to be in contact within in that frame. If a pair of nodes is in contact in more than 75% of simulation, they are considered to be in contact for the purposes of network analysis. Third, we calculate the generalized correlation coefficient between all pairs of contacting nodes by the formula \({{{{\rm{r}}}}}_{{MI}}={\left(1-{e}^{-\frac{2I}{3}}\right)}^{\frac{1}{2}}\), where \({{{\rm{I}}}}\) is the mutual information between the position of the two nodes. Next, we determine two target nodes by finding the amino acid residue which has the most hydrogen bond interactions with ATP and the amino acid residue in the channel that is in the vicinity of Cl- ions. Finally, we determine the shortest path between two target nodes using Floyd-Warshall algorithm by defining d = −log(rMI) as the distance between two contacting nodes.
Chemicals
All ligands such as ATP, ADP, AMP, AMP-PNP, and ATPγS were purchased from Sigma-Aldrich. The sodium salt of ATP, ADP and AMP were used.
Statistical analyses
All statistical data are represented as mean ± s.e.m. An unpaired two-tailed t-test or one-way analysis of variance was used to analyze the statistical significance in GraphPad Prism v.7.0 software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding authors upon request. Structure coordinates have been deposited in the Protein Data Bank under accession numbers 8JEV for mClC-3apo, 8JGJ mClC-3ATP, 8JGK for mClC-3ADP, 8JGL for mClC-3AMP, 8JGS for mClC-3I607T_apo, 8JGV for mClC-3I607T_ATP. Cryo-EM density maps have been deposited under accession numbers EMD-36200 for mClC-3apo, EMD-36236 for mClC-3ATP, EMD-36237 for mClC-3ADP, EMD-36238 for mClC-3AMP, EMD-36245 for mClC-3I607T_apo, EMD-36246 for mClC-3I607T_ATP. The following previously published structural models were referenced in this work: 2J9L, 6COY, 7JM7. The source data underlying Figs. 1l-m, 1q, 2e, 2j, 2p, 3l, 4n-o and Supplementary Figs. 1, 3, 6, 10, 16, are provided as a Source Data file. Source data are provided with this paper.
References
Jentsch, T. J. & Pusch, M. CLC chloride channels and transporters: structure, function, physiology, and disease. Physiol. Rev. 98, 1493–1590 (2018).
Stauber, T., Weinert, S. & Jentsch, T. J. Cell biology and physiology of CLC chloride channels and transporters. Compr. Physiol. 2, 1701–1744 (2012).
Bose, S., He, H. & Stauber, T. Neurodegeneration upon dysfunction of endosomal/lysosomal CLC chloride transporters. Front. Cell Dev. Biol. 9, 639231 (2021).
Meyer, S., Savaresi, S., Forster, I. C. & Dutzler, R. Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5. Nat. Struct. Mol. Biol. 14, 60–67 (2007).
Accardi, A. & Miller, C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl- channels. Nature 427, 803–807 (2004).
Lobet, S. & Dutzler, R. Ion-binding properties of the ClC chloride selectivity filter. EMBO J. 25, 24–33 (2006).
Zhang, S. et al. Molecular insights into the human CLC-7/Ostm1 transporter. Sci. Adv. 6, eabb4747 (2020).
Schrecker, M., Korobenko, J. & Hite, R. K. Cryo-EM structure of the lysosomal chloride-proton exchanger CLC-7 in complex with OSTM1. eLife 9, https://doi.org/10.7554/eLife.59555 (2020).
Grieschat, M., Guzman, R. E., Langschwager, K., Fahlke, C. & Alekov, A. K. Metabolic energy sensing by mammalian CLC anion/proton exchangers. EMBO Rep. 21, e47872 (2020).
Stobrawa, S. M. et al. Disruption of ClC-3, a chloride channel expressed on synaptic vesicles, leads to a loss of the hippocampus. Neuron 29, 185–196 (2001).
Scheel, O., Zdebik, A. A., Lourdel, S. & Jentsch, T. J. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature 436, 424–427 (2005).
Hara-Chikuma, M. et al. ClC-3 chloride channels facilitate endosomal acidification and chloride accumulation. J. Biol. Chem. 280, 1241–1247 (2005).
Mohammad-Panah, R. et al. The chloride channel ClC-4 contributes to endosomal acidification and trafficking. J. Biol. Chem. 278, 29267–29277 (2003).
Novarino, G., Weinert, S., Rickheit, G. & Jentsch, T. J. Endosomal chloride-proton exchange rather than chloride conductance is crucial for renal endocytosis. Sci. (N. Y., N. Y.) 328, 1398–1401 (2010).
Zifarelli, G. & Pusch, M. Intracellular regulation of human ClC-5 by adenine nucleotides. EMBO Rep. 10, 1111–1116 (2009).
Duncan, A. R. et al. Unique variants in CLCN3, encoding an endosomal anion/proton exchanger, underlie a spectrum of neurodevelopmental disorders. Am. J. Hum. Genet. 108, 1450–1465 (2021).
Riazanski, V. et al. Presynaptic CLC-3 determines quantal size of inhibitory transmission in the hippocampus. Nat. Neurosci. 14, 487–494 (2011).
Yoshikawa, M. et al. CLC-3 deficiency leads to phenotypes similar to human neuronal ceroid lipofuscinosis. Genes Cells 7, 597–605 (2002).
Weinert, S. et al. Uncoupling endosomal CLC chloride/proton exchange causes severe neurodegeneration. EMBO J. 39, e103358 (2020).
Dutzler, R., Campbell, E. B., Cadene, M., Chait, B. T. & MacKinnon, R. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature 415, 287–294 (2002).
Feng, L., Campbell, E. B., Hsiung, Y. & MacKinnon, R. Structure of a eukaryotic CLC transporter defines an intermediate state in the transport cycle. Science 330, 635–641 (2010).
Park, E., Campbell, E. B. & MacKinnon, R. Structure of a CLC chloride ion channel by cryo-electron microscopy. Nature 541, 500–505 (2017).
Park, E. & MacKinnon, R. Structure of the CLC-1 chloride channel from Homo sapiens. eLife 7, https://doi.org/10.7554/eLife.36629 (2018).
Wang, K. et al. Structure of the human ClC-1 chloride channel. PLoS Biol. 17, e3000218 (2019).
Schrodinger, L. L. C. The PyMOL Molecular Graphics System, Version 1.8 (2015).
Dutzler, R., Campbell, E. B. & MacKinnon, R. Gating the selectivity filter in ClC chloride channels. Science 300, 108–112 (2003).
Miller, C. ClC chloride channels viewed through a transporter lens. Nature 440, 484–489 (2006).
Rohrbough, J., Nguyen, H. N. & Lamb, F. S. Modulation of ClC-3 gating and proton/anion exchange by internal and external protons and the anion selectivity filter. J. Physiol. 596, 4091–4119 (2018).
Stauber, T. & Jentsch, T. J. Sorting motifs of the endosomal/lysosomal CLC chloride transporters. J. Biol. Chem. 285, 34537–34548 (2010).
Guzman, R. E., Miranda-Laferte, E., Franzen, A. & Fahlke, C. Neuronal ClC-3 splice variants differ in subcellular localizations, but mediate identical transport functions. J. Biol. Chem. 290, 25851–25862 (2015).
Jayaram, H., Accardi, A., Wu, F., Williams, C. & Miller, C. Ion permeation through a Cl–selective channel designed from a CLC Cl-/H+ exchanger. Proc. Natl Acad. Sci. USA 105, 11194–11199 (2008).
De Jesús-Pérez, J. J. et al. Electro-steric opening of the CLC-2 chloride channel gate. Sci. Rep. 11, 13127 (2021).
Basilio, D., Noack, K., Picollo, A. & Accardi, A. Conformational changes required for H(+)/Cl(-) exchange mediated by a CLC transporter. Nat. Struct. Mol. Biol. 21, 456–463 (2014).
Bennetts, B. & Parker, M. W. Molecular determinants of common gating of a ClC chloride channel. Nat. Commun. 4, 2507 (2013).
Walden, M. et al. Uncoupling and turnover in a Cl-/H+ exchange transporter. J. Gen. Physiol. 129, 317–329 (2007).
Zdebik, A. A. et al. Determinants of anion-proton coupling in mammalian endosomal CLC proteins. J. Biol. Chem. 283, 4219–4227 (2008).
Grieschat, M. & Alekov, A. K. Glutamate 268 regulates transport probability of the anion/proton exchanger ClC-5. J. Biol. Chem. 287, 8101–8109 (2012).
Kieseritzky, G. & Knapp, E. W. Charge transport in the ClC-type chloride-proton anti-porter from Escherichia coli. J. Biol. Chem. 286, 2976–2986 (2011).
Lim, H. H., Shane, T. & Miller, C. Intracellular proton access in a Cl(-)/H(+) antiporter. PLoS Biol. 10, e1001441 (2012).
Lim, H. H. & Miller, C. Intracellular proton-transfer mutants in a CLC Cl-/H+ exchanger. J. Gen. Physiol. 133, 131–138 (2009).
Accardi, A. et al. Separate ion pathways in a Cl-/H+ exchanger. J. Gen. Physiol. 126, 563–570 (2005).
Han, W., Cheng, R. C., Maduke, M. C. & Tajkhorshid, E. Water access points and hydration pathways in CLC H+/Cl- transporters. Proc. Natl Acad. Sci. USA 111, 1819–1824 (2014).
Chavan, T. S. et al. A CLC-ec1 mutant reveals global conformational change and suggests a unifying mechanism for the CLC Cl(-)/H(+) transport cycle. eLife 9, https://doi.org/10.7554/eLife.53479 (2020).
Li, C. & Voth, G. A. A quantitative paradigm for water-assisted proton transport through proteins and other confined spaces. Proc. Natl Acad. Sci. USA 118, https://doi.org/10.1073/pnas.2113141118 (2021).
Zhixuan, Z., Long, C., Jin, H. & Ji, S. Structure of the human CLC-7/Ostm1 complex reveals a novel state. JUSTC 53, 0306-0301–0306-0307 (2023).
Mayes, H. B., Lee, S., White, A. D., Voth, G. A. & Swanson, J. M. J. Multiscale kinetic modeling reveals an ensemble of Cl(-)/H(+) exchange pathways in ClC-ec1 antiporter. J. Am. Chem. Soc. 140, 1793–1804 (2018).
Faraldo-Gómez, J. D. & Roux, B. Electrostatics of ion stabilization in a ClC chloride channel homologue from Escherichia coli. J. Mol. Biol. 339, 981–1000 (2004).
Leisle, L. et al. Divergent Cl(-) and H(+) pathways underlie transport coupling and gating in CLC exchangers and channels. eLife 9, https://doi.org/10.7554/eLife.51224 (2020).
Palmer, E. E. et al. Functional and clinical studies reveal pathophysiological complexity of CLCN4-related neurodevelopmental condition. Mol. Psychiatry 28, 668–697 (2023).
Deriy, L. V. et al. The granular chloride channel ClC-3 is permissive for insulin secretion. Cell Metab. 10, 316–323 (2009).
Li, D. Q. et al. Suppression of sulfonylurea- and glucose-induced insulin secretion in vitro and in vivo in mice lacking the chloride transport protein ClC-3. Cell Metab. 10, 309–315 (2009).
Huang, L. Y. et al. ClC-3 chloride channel/antiporter defect contributes to inflammatory bowel disease in humans and mice. Gut 63, 1587–1595 (2014).
Dai, Y. P., Bongalon, S., Hatton, W. J., Hume, J. R. & Yamboliev, I. A. ClC-3 chloride channel is upregulated by hypertrophy and inflammation in rat and canine pulmonary artery. Br. J. Pharmacol. 145, 5–14 (2005).
Miller, F. J. Jr. et al. Cytokine activation of nuclear factor kappa B in vascular smooth muscle cells requires signaling endosomes containing Nox1 and ClC-3. Circulation Res. 101, 663–671 (2007).
Robertson, J. L., Kolmakova-Partensky, L. & Miller, C. Design, function and structure of a monomeric ClC transporter. Nature 468, 844–847 (2010).
Burgunder, J. M. et al. Novel chloride channel mutations leading to mild myotonia among Chinese. Neuromuscul. Disord. 18, 633–640 (2008).
Polovitskaya, M. M. et al. A recurrent gain-of-function mutation in CLCN6, encoding the ClC-6 Cl(-)/H(+)-exchanger, causes early-onset neurodegeneration. Am. J. Hum. Genet. 107, 1062–1077 (2020).
Mansour-Hendili, L. et al. Mutation update of the CLCN5 gene responsible for dent Disease 1. Hum. Mutat. 36, 743–752 (2015).
Palmer, E. E. et al. De novo and inherited mutations in the X-linked gene CLCN4 are associated with syndromic intellectual disability and behavior and seizure disorders in males and females. Mol. Psychiatry 23, 222–230 (2018).
Jo, S., Kim, T. & Im, W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE 2, e880 (2007).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Berendsen, H. J. C., Postma, J. P. M., Gunsteren, W. F. V., DiNola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81, 511–519 (1984).
Hoover, W. G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 31, 1695–1697 (1985).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput 8, 3257–3273 (2012).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Humphrey, W., Dalke, A. & Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Kollman, P. A. et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models.
Genheden, S. & Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities.
Valdés-Tresanco, M. S., Valdés-Tresanco, M. E., Valiente, P. A.-O. & Moreno, E. A.-O. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS.
Miller, B. R., 3rd et al. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations.
Melo, M. C. R., Bernardi, R. C., de la Fuente-Nunez, C. & Luthey-Schulten, Z. Generalized correlation-based dynamical network analysis: a new high-performance approach for identifying allosteric communications in molecular dynamics trajectories. J. Chem. Phys. 153, 134104 (2020).
Acknowledgements
Single-particle cryo-EM data were collected at the Center of Cryo-Electron Microscopy at Zhejiang University. We thank Dr. Xing Zhang and Dr. Cheng Ma for their support in facility access and data acquisition. We thank Drs. Haoxin Xu, Wei Yang, Yan Zhang and Jiangtao Guo for constructive suggestions. This work was supported by the National Natural Science Foundation of China (32122040 and 31971040 to F.Y.; 82200784 to L.H.), the Ministry of Science and Technology of China (2022YFF1201901 to H.W.), STI2030-Major Project (2022ZD0212700 to H.W.) and Zhejiang Provincial Natural Science Foundation (LR20C050002 to F.Y.). F.Y. was supported by MOE Frontier Science Center for Brain Science & Brain-Machine Integration, Zhejiang University. This work was supported by the Fundamental Research Funds for the Central Universities. This work was also supported by the Core Facilities in Zhejiang University School of Medicine, including the Zeiss LSM800 confocal fluorescence imaging microscope.
Author information
Authors and Affiliations
Contributions
F.Y. conceived and supervised the project. Y.W., L.X. and X.C. did sample preparation, data acquisition, and structure determination. S.G. and W.Z. did functional studies. F.L., Y.S., and S.L. did confocal imaging. X.W., F.Y., Q.W. and H.W. did the MD simulation. F.Y., L.H. and H.W. performed data analysis. All authors participated in the data analysis and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wan, Y., Guo, S., Zhen, W. et al. Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger. Nat Commun 15, 6654 (2024). https://doi.org/10.1038/s41467-024-50975-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-50975-w
This article is cited by
-
Endosomal chloride/proton exchangers need inhibitory TMEM9 β-subunits for regulation and prevention of disease-causing overactivity
Nature Communications (2025)
-
Structural basis of ClC-3 transporter inhibition by TMEM9 and PtdIns(3,5)P2
Nature Structural & Molecular Biology (2025)