Abstract
Lujo virus (LUJV) is a human pathogen that was the cause of a deadly hemorrhagic fever outbreak in Africa. LUJV is a divergent member of the Arenaviridae with some similarities to both the “Old World” and “New World” serogroups, but it uses a cell-entry receptor, neuropilin-2 (NRP2), that is distinct from the receptors of OW and NW viruses. Though the receptor binding domain of LUJV has been characterized structurally, the overall organization of the trimeric spike complex and how NRP2 is recognized in this context were unknown. Here, we present the structure of the membrane-embedded LUJV spike complex determined by cryo-electron microscopy. Analysis of the structure suggested that a single NRP2 molecule is bound at the apex of the trimeric spike and that multiple subunits of the trimer contact the receptor. The binding of NRP2 involves an intriguing arginine-methionine interaction, which we analyzed using quantum mechanical modeling methods. We compare the LUJV spike structure with the only other available structure of a complete arenaviral spike, which is the Lassa virus. The similarities and differences between them shed light on Arenavirus evolution, inform vaccine design, and provide information that will be useful in combating future Arenavirus outbreaks.
Similar content being viewed by others
Introduction
Zoonotic viruses, which circulate in animal hosts and can be transmitted to humans, pose a major risk to human health. An important family of zoonotic viruses is the Arenaviridae. The mammarenavirus genus, which are arenaviruses that infect mammals, is geographically and genetically classified into the Old World (OW) and the New World (NW) groups, which are mostly endemic to Africa and the Americas, respectively. Both these groups contain human pathogens, including the OW Lassa (LASV) and lymphocytic choriomeningitis (LCMV) viruses and the NW Junín, Guanarito, Sabiá, and Machupo (MACV) viruses.
Lujo virus (LUJV) is a contagious and highly fatal mammarenavirus that emerged in a limited but deadly outbreak in South Africa1. Interestingly, LUJV is genetically distinct from both the OW and NW groups of mammarenaviruses1. The LUJV also differs in the cell surface receptor that it utilizes for cell entry. To attach to and enter their target cells, the OW and some NW viruses use a special glycan modification, termed matriglycan, that specifically modifies the cell-surface protein α-dystroglycan (α-DG). Most NW viruses use transferrin receptor 1 (TfR1)2,3. LUJV is the only arenavirus known to use neuropilin-2 (NRP2) to attach to and enter its target cell4.
Regardless of the receptor used for cell entry, all mammarenaviruses display homotrimeric class-I glycoprotein spikes that mediate the attachment of the viruses to their cells and fuse the viral and host cell membranes5,6. The spikes are translated as glycoprotein precursor proteins (GPCs), which are processed by a signal peptidase (SPase) and a subtilisin kexin isozyme-1/site-1 (SKI-1) protease7 to yield a complex of a unique structured signal peptide (SSP), glycoprotein-1 (GP1), and glycoprotein-2 (GP2). Three copies of this complex constitute the spike. The SSP, which is unique to arenaviruses, is unusually long for a signal peptide (i.e., 58 amino acids) and is critical for spike trafficking and maturation by proteolysis8,9,10,11,12,13,14. GP1 serves as the receptor-binding domain, and GP2, which has a transmembrane segment, serves as the fusogenic module of the spike6,15,16,17.
The mechanisms by which diverse arenaviruses engage different receptors and activate fusion are poorly understood, as structural information for receptor recognition by an intact, functional arenaviral spike complex is currently available only for LASV17. The structure of the LASV spike complex showed how the three GP1 domains cooperate to engage a single matriglycan17. Quaternary-structure-dependent binding of NRP2 was also suggested in the case of LUJV but was not directly demonstrated16. More generally, it is not known how the trimeric organization of spike complexes differs among distinct mammarenaviruses.
Here, we present the cryo-EM structure of the full-length, membrane-embedded spike complex of LUJV. This structure reveals both the organization of the spike in the membrane and the architecture of its ectodomain. A comparison of the LUJV and LASV spike structures elucidates elements common to arenaviruses as well as virus-specific. This information is important for combating LUJV.
Results
The structure of the LUJV spike complex
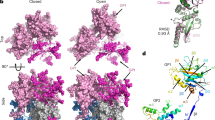
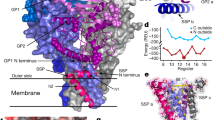
To determine the structure of the LUJV spike complex, we expressed the full-length GPC in mammalian cells with a Flag-tag fused to its C-terminus. We solubilized the cell membranes in detergents, purified the spike, and determined a three-fold symmetric, 3.0 Å resolution structure using single-particle cryo-EM (Supplementary Fig. 1 and Supplementary Table 1). All three domains that result from proteolytic cleavage of GPC (Fig. 1a) are visible in the density map and are part of the final model (Fig. 1b), which consists of the spike transmembrane and ectodomain but not the cytoplasmic portion. The overall architecture of the spike resembles that of the LASV spike complex17. In the membrane, the spike forms a six-helical bundle made of three central helices of GP2 surrounded by three helices of SSP (Fig. 1b). The central three GP2 helices are wrapped around each other such that each helix penetrates the membrane underneath a neighboring GP2 protomer (Supplementary Fig. 2a). This is achieved by a long helix (α4, Fig. 1b and Supplementary Fig. 2a) that extends to a neighboring GP2 and closely interacts at a kink that forms between the neighbor’s α4 and α5 GP2 helices (Supplementary Fig. 2b). The interaction is facilitated by sandwiching Glu386 and Tyr381 from each of the α4 helices between Thr374/Lys378 and Gln385/Ile390 on the neighboring α4 helices, respectively (Supplementary Fig. 2b, c). Above the surface of the membrane, the GP2-GP2 interaction is mainly driven by hydrophobic interactions that helix α3 makes with a groove between helix α2 and a preceding loop of an adjacent GP2, as well as electrostatic interactions between two oppositely charged patches (Supplementary Fig. 3a). Arenaviruses have a short fusion peptide and a fusion loop at the N-terminus of GP2, which are both assumed to be the membrane interacting elements and hence required for membrane fusion18. This, however, may or may not be correct as recent structural data for the SARS-CoV-2 spike in a post-fusion state reveals that only the fusion loop and not the fusion peptide is actually inserted into the membrane19. Nevertheless, in the LUJV spike, the GP2 N-terminus points toward the core of the complex (Supplementary Fig. 3b), sequestering the fusion peptide between two GP2 subunits. The fusion loop is sandwiched between N-linked glycans and interacts with the N-terminus of GP1 (Supplementary Fig. 3c).
a Schematic diagram showing the domain partitioning of the spike. Shaded regions indicate parts that are missing from the final model. The residue numbering of the different subunits, as well as the location of the transmembrane helices, are indicated. b The LUJV spike complex is presented using a ribbon diagram of a ‘side’ view. The three GPC protomers are colored pink, blue, and green; for each protomer, the SSP, GP1, and GP2 domains are indicated using different tones. The secondary structure organization is indicated for representative GP1 and GP2 domains using two ribbon representations, which are rainbow-colored from their N- to the C- termini. The naming of secondary structure elements in GP1 preserves and extends the naming in PDB ID: 6GH8. N-linked glycans are shown as spheres. The estimated location of the membrane is indicated. The inset shows the organization of the transmembrane helices, presented along the three-fold symmetry axis of the spike (indicated with a triangle) and shown from the ‘bottom’ of the spike. c The interaction between the GP1 domains. A top view shows only the GP1 domains, using the same coloring scheme as in (b). The main GP1 elements that mediate the interaction are indicated. d A close-up view of the ‘RKLM’ latch. One GP1 domain is shown using a surface representation colored by its electrostatic potential (red -5 kT/e; blue 5 kT/e). The side chains of the ‘RKLM’ latch are shown as sticks and indicated with arrows. e The hydrophobic pocket of the ‘RKLM’ latch. The side chains of the residues, which form the hydrophobic pocket that accommodates the ‘RKLM’ latch (shown using a transparent surface representation), are shown as sticks and indicated by arrows.
The SSP starts with a short helical segment that is parallel to, and partially embedded in, the membrane, followed by a trans-membrane helix (residues 15 to 34) (Fig. 1b). This configuration indicates a topological rearrangement of the C-terminal part of SSP from the outer/luminal side to the inner/cytoplasmic side following cleavage by the signal peptidase (SPase) (Supplementary Fig. 4). The previously observed17 structural role of SSP in stabilizing the spike complex seems to be conserved as the SSP interacts in the membrane with two adjacent GP2 helices, apparently preventing them from unwinding. The SSP itself is held fixed with respect to the trimer by interactions between its short helical N’-segment and GP2 (Fig. 1b).
The apex of the spike is stabilized by mutual interactions between the GP1 domains (Fig. 1c). The C-terminus of GP1, resulting from cleavage by the cellular protease SKI-1, together with the preceding helix α4, forms a latch that extends toward and interacts with the neighboring GP1 near the three-fold symmetry axis of the spike (Fig. 1c). These latch-mediated interactions create deep groves between the GP1 domains (Fig. 1c). The tip of the latch is made from the two hydrophobic residues of the ‘RKLM’-SKI-1 recognition site (Fig. 1a, d). These hydrophobic residues fit into a hydrophobic pocket on the neighboring GP1 made by Phe113, Phe124, Phe127, Leu143, and the aliphatic portions of Arg128 and Lys110 (Fig. 1e). The rest of the interacting surfaces are very polar and include Arg218 and Lys219 from the ‘RKLM’ motif, as well as several histidine residues: His131, His132, and His214 (Fig. 1d). Glu212 is also at the interface (Supplementary Fig. 5a). This glutamic acid points toward the symmetry axis of the trimer where it chelates a metal ion (Supplementary Fig. 5b). Although this interaction likely stabilizes the spike, is not required, as mutating Glu212 (i.e., E212A or E212Q) results in spikes that are as efficient as the WT in promoting cell entry (Supplementary Fig. 5c).
Shared and unique properties of the LUJV spike complex
Besides the LUJV spike complex, structural information for a complete arenaviral spike complex is available so far only for LASV17. Despite sharing a similar overall organization, superimposing the two structures (Fig. 2a) yields a root-mean-square deviation (RMSD) value of 22.5 Å for 1095 Cα atoms. This high RMSD value reflects different relative orientations of the GP1, GP2, and SSP domains. Indeed, superimposing the individual domains yields RMSD values of 8.2 Å, 3.0 Å, and 1.0 Å for the GP1 (155 Cα atoms), GP2 (176 Cα atoms), and SSP (33 Cα atoms), respectively (Fig. 2b). While the transmembrane region and the juxtaposed GP2 region are structurally similar, the GP1 domains themselves and their relative orientation in the spikes differ greatly between LUJV and LASV (Fig. 2a, b). The packing of the GP1 domains in LUJV seems to be looser compared with the packing in LASV (Fig. 2c). Narrow cavities are seen between the three LUJV GP1 subunits, and the interacting surfaces are limited to the vicinity of the symmetry axis (Fig. 2c). No cavities are observed in LASV, and the packing of the GP1 domains spans a larger interface (Fig. 2c). Nonetheless, in both spike complexes, the C-terminal ends of the GP1s that contain the SKI-1 recognition sequences are critical elements for trimerization.
a Superimposition of the complete LUJV and LASV (PDB ID: 7PUY) spikes, calculated based on the helix α4 regions of the GP2s. The LUJV spike is colored using the same coloring scheme as in Fig. 1, and the LASV spike is shown in gray. b Superimpositions of the individual domains of LUJV and LASV. Root Mean Square Deviation (RMSD) values for matching mutual Cα atoms are indicated. c Top views of the GP1 domains from the LUJV (left) and LASV (right) spikes, using surface representations and a distinct color for each GP1 domain. Arrows point to gaps between the GP1 domains of the LUJV spike complex. d Glycosylation of the spike complexes. N-linked glycans are shown as spheres on the LUJV (left) and the LASV (right) spikes. The glycan sites that are closest to the spikes’ three-fold symmetry axes are indicated. Two dashed lines highlight a region of the LUJV spike that has no glycans.
Arenavirus spikes have evolved to display glycans that aid in evading neutralizing antibodies and reducing viral clearance20. Comparing the distribution of N-linked glycans on the LUJV and LASV spike complexes provides a few interesting observations. The apex of the LUJV spike complex is significantly more exposed compared to the apex of the LASV spike (Fig. 2d). The closest glycan to the symmetry axis in LUJV is attached to Asn112, the Cα of which is 17.1 Å from the axis (Fig. 2d). The LASV spike, on the other hand, has a glycan attached to Asn119, which is only 13.8 Å from the axis and thus forms a cluster with its symmetry mates near the spike apex (Fig. 2d). Also, while the rest of the N-linked glycans of LASV seem to be evenly distributed on the surface of the spike, there is an evident belt on the surface of the LUJV spike that is lacking N-linked glycans (Fig. 2d). While viral glycan shields are never completely sealed, and will always leave patches that are free of glycans, as in the case of LASV (Fig. 2d) or other viral class-I spike complexes (Supplementary Fig. 9), having a complete belt without glycans seems to be unusual. Having such a belt free of glycans could be a pure coincidence, or it may confer some beneficial properties to the spike, like creating an antibodies-accessible immunodominant region to subvert immune response.
Recognition of NRP2 by the LUJV spike complex
We previously determined a crystal structure of an isolated, partially truncated LUJV GP1 domain in a complex with its NRP2 receptor16. Superimposing this isolated GP1 domain on the GP1 domains from the trimeric spike reveals almost identical structures with RMSD of 1.2 Å, without any apparent conformation changes (Fig. 3a). This observation paves the way to use the information from the GP1/NRP2 crystal structure to understand how the trimeric spike recognizes NRP2. The most noticeable aspect of this interaction is that NRP2 is bound at the apex of the spike (Fig. 3a), at the region that lacks N-linked glycans (Fig. 2d), with an orientation that overlaps the spike’s three-fold symmetry axis (Fig. 3b). This observation reveals that each spike complex if it maintains the closed configuration as seen for the unbound spike, could interact only with a single NRP2 molecule at a time.
a NRP2 binds at the apex of the spike. The inset shows the superimposition of two GP1 domains from the structure of the spike (pink) and the structure of the NRP2/GP1 complex (blue, PDB ID: 6GH8). The Root Mean Square Deviation (RMSD) value based on a common set of 110 Cα atoms is indicated. The spike complex is shown in a ‘side’ view using surface representations for all chains except a single GP1 domain (pink) that uses a ribbon diagram. The coloring scheme is the same as in Fig. 1. The NRP2/GP1–spike superimposition places the NRP2 (purple ribbon diagram) at the apex. b A ‘top’ view of the spike. Bound NRP2 overlaps with the spike’s three-fold symmetry axis, which a triangle indicates. c Lys219 from the same GP1 domain that NRP2 binds is positioned such that it can form a salt bridge with Asp48 of NRP2. d Arg130 of NRP2 serves as a lid that closes on the side-chain of Met221 from a nearby GP1. e Methionine’s sulfur atom/guanidinium interaction. The orange chart shows the preferred angle between the guanidinium and the Cγ-Sδ-Cε planes, as defined in the lower image, for arginine/methionine interactions based on statistical data derived from the PDB (n = 663). The red dots show calculated energy values (kcal/mol) for the bond strength between methylguanidinium and dimethyl sulfide as a function of the angle between the planes. A linear fit for the data points (R2 = 0.33) is shown. f NRP2 fits between two N-linked glycans on Asn112 residues from two separate GP1 domains. The inset shows a magnified view of the glycan from the cognate GP1 domain that binds the NRP2. In the NRP2/GP1 crystal structure, the first N-Acetylglucosamine is tilted with respect to the same sugar in the apo-spike structure. g Normalized infectivity of pseudotyped viruses bearing either the WT or a T114A mutant of the LUJV spike complex. Dots represent technical replicates (n = 27). The p-value (two-tailed Student’s t test) for the difference is p = 5.03427E-08. Whiskers indicate the minimum and maximum values. Central lines indicate means, and boxes show the interquartile ranges. Results for a representative experiment out of two independent repeats are shown. Below the graph, IC50 values for neutralization by NRP2-Fc, and NRP2-His are indicated. Source data are provided as a Source Data file.
Besides the various Ca2+-dependent interactions that a single LUJV GP1 domain forms with NRP216, superimposing the GP1/NRP2 crystal structure on the spike’s structure suggests additional interactions enabled by the spike quaternary structure. Specifically, Lys219 from the C-terminal end of one GP1 is positioned to make an electrostatic interaction with Asp48 of NRP2 (Fig. 3c). The position of this lysine in the spike is determined by the interaction of the hydrophobic latch with the neighboring GP1. In addition, Lys219 of a second GP1 may interact with the main-chain carbonyl of NRP2’s Arg130 by adopting a different rotamer (Supplementary Fig. 6).
Arg130 of NRP2 is involved in an interaction that is quaternary-enabled. This arginine residue makes important polar interactions with GP1 in the context of the crystal structure complex16. In the context of the trimeric spike, the side-chain of Arg130 further forms a lid that covers the hydrophobic cavity to which Met221 from a neighboring GP1 is inserted (Fig. 3d). In this configuration, the aliphatic tail of Arg130 makes van der Waals interactions with the side chain of Met221. Interestingly, this configuration also positions the sulfur atom of Met221 (Sδ) below the plane of the Arg130 guanidino group (Fig. 3d). The contribution of a sulfur-guanidino interaction to the stability of the complex is not clear as this kind of interaction has not been analyzed for its energetic contribution before. Bioinformatic analysis of unique 34,376 structures at a resolution better than 2.2 Å from the PDB reveals n = 771 instances of methionine sulfur atoms at a distance of 3.2 Å to 3.8 Å from the Cζ atom of arginine (Fig. 3e). This set was split into interactions in polar or hydrophobic environments (i.e., nearest water molecule is at a distance smaller, or greater than 3.5 Å from the Cζ atom of arginine and the Sδ atom of methionine). For n = 663 interactions that are in a less polar environment, there is a preference for orientations that minimize the Sδ - guanidino group distance while maximizing the distances of the methionine Cγ and Cε atoms (i.e., the angle between the normal of the guanidino group’s plane and the normal of the Cγ-Sδ-Cε plane approach 90°) (Fig. 3e). To determine the interaction energy, we employed Double-Hybrid Density Functional Theory (DH-DFT) at the quantum mechanical level. We selected representative arginine/methionine pairs from hydrophobic (n = 34) (Fig. 3e) and polar (n = 48) (Supplementary Fig. 7a, b) environments, and for each pair, we performed geometry optimization to converge to its closest local minimal-energy conformation prior to the energy calculations. For the DH-DFT calculations, we used methyl-guanidinium and dimethyl-sulfide to represent arginine and methionine, respectively (Fig. 3e). The methionine Sδ engages with the arginine guanidinium in a van der Waals-like interaction that can contribute up to − 3.3 kcal/mol in a low dielectric environment (Fig. 3e) as in the hydrophobic pocket of the LUJV spike (Fig. 3d) and up to −2.5 kcal/mol in a high dielectric environment (Supplementary Fig. 7a). Pairs with the more abundant angle between the arginine and methionine planes near 90°, as seen in the PDB-derived statistical data, also show somewhat stronger interactions (Fig. 3e). Taking into account the sulfur-guanidino interaction and the additional contribution of the aliphatic tails, the Arg130-Met221 interaction is a significant quaternary-enabled interaction for the NRP2/LUJV spike complex.
Receptor-binding sites are known vulnerability spots on viral spike complexes. Viruses must balance the need to create a dense glycan shield to avoid humoral immune responses while not interfering with receptor binding. In the case of the LUJV, the apex of the spike is decorated by several N-linked glycans that are visible in the density maps (Supplementary Fig. 1). Two Asn112-linked glycans (Fig. 2d) are particularly close to NRP2: a glycan attached to Asn112 on the GP1 domain that makes the main association site with NRP2, and an Asn112-linked glycan on the neighboring (clockwise, looking from the top) GP1 domain (Fig. 3f). NRP2 fits very tightly between these two glycans. The Asn112-linked glycan on the main GP1 domain, as modeled in the EM structure, would clash with NRP2 unless the glycan tilts to adopt a different conformation with respect to GP1 (Fig. 3f). Indeed, the first GlcNAc of this glycan is tilted away from NRP2 in the NRP2/GP1 crystal structure but not in the EM structure, which was determined in the absence of NRP2 (Fig. 3f). Interestingly, while the Asn112-linked glycan clearly has conformational flexibility that allows binding to NRP2, it also has a preferred ground state that gives rise to the observed EM density, which was sufficient for modeling several consecutive sugar monomers of this glycan tree (Supplementary Fig. 1). Glycans in proximity to the NRP2 binding site may affect complex formation by interfering sterically with NRP2 binding or by contributing favorable molecular interactions with NRP2. Interestingly, pseudo-viruses decorated with LUJV spikes lacking glycans at position 112 (due to a T114A mutation that abrogates the N-X-T/S glycosylation motif) can infect cells as efficiently, and perhaps even more efficiently, compared with pseudo-viruses bearing WT spikes (Fig. 3g). This observation suggests that the Asn112-linked glycans may restrict binding by steric interference. Testing the capacity of NRP2-based competitors to prevent cell entry of pseudo-viruses strengthens this notion. A bulky NRP2-Fc immunoadhesin, made of the first CUB domain of NRP2 fused to the Fc portion of IgG1, neutralizes LUJV better when the Asn112-linked glycan is absent (Fig. 3g and Supplementary Fig. 8), and a smaller reagent that comprises only the first CUB domain of NRP2 (NRP2-His) achieves better IC50 values for neutralization compared with NRP2-Fc (Fig. 3g). Hence, the Asn112-linked glycans impose steric constraints for interacting with the NRP2 receptor.
Evaluating the potential of an anti-LUJV immunoadhesin
Previously, we demonstrated that host-derived receptors could be utilized to make powerful immunoadhesins against zoonotic viruses21,22. In the case of LUJV, however, the exact host is unknown, and the NRP2 interface that GP1 recognizes is highly conserved in mammals16, complicating the construction of high-affinity immunoadhesin. Potentially, NRP2-Fc may be used as an immunoadhesin (Fig. 3g), but such a reagent is unlikely to have sufficient potency for clinical use. As we noted above, the N-linked glycan on Asn112 restricts the binding of the LUJV spike to NRP2 (Fig. 3f, g). A close inspection of the LUJV spike structure with NRP2 superimposed revealed that the N-linked glycan on Asn112 clashes with Tyr128 of NRP2 (Fig. 4a). Eliminating the bulky side-chain of Tyr128 in NRP2 with a Y128G mutation increases the potency of NRP2-Fc nearly tenfold (Fig. 4b), as measured using MLV-based pseudotyped viruses bearing the LUJV spike complex. Moreover, the structure suggests that Gln47 of NRP2 is forced to assume an uncommon rotamer to avoid clashing with a neighboring GP1 domain (Fig. 4a). Restricting the available rotamers for Gln47 confers an entropic cost for binding and hence reduces affinity. Indeed, eliminating the Gln47 side chain with a Q47G mutation improves the neutralization of NRP2-Fc (Fig. 4b). Lastly, the structure further suggests that Ile108 will be in very close proximity, perhaps will even clash, with Asn112-linked glycan from the neighboring GP1 monomer (Fig. 4b). Introducing an I108S mutation to NRP2-Fc also results in more potent neutralization of LUJV (Fig. 4b).
a Closeup views of the three residues that are predicted to interfere with the binding of NRP2 by the LUJV spike. Tyr128 of NRP2 is on the cognate GP1 (pink), and Ile108 and Gln47 are on adjacent GP1 (light blue). b Neutralization of LUJV pseudoviruses by NRP2-Fc WT and the indicated mutants. The graph is from a representative experiment (each data point is the average value from four technical repeats, and error bars show standard deviations), and the IC50 values are averaged values from several independent experiments. c Fold improvement of neutralization IC50 values of the NRP2-Fc mutants compared with WT NRP2-Fc. Each point represents an independent experiment (n = 2 for Q47G, Y128G, and Q47G-I108S; n = 3 for I108S, and I108S-Y128G; and n = 4 for Q47G-I108S-Y128G). Averaged values are indicated, and error bars show standard deviations. Statistical significance for the differences between the mutants and the WT NRP2-Fc are indicated (ns not significant, **− p = 0.009524 two-sided Wilcoxon-Mann-Whitney test, no adjustments were made). The Source Data File lists the exact p values for all other pairs. d Conservation of the mutated sites. Multiple sequence alignments of 447 NRP2 sequences from mammals are summarized as WebLogos around the Gln47, Ile108, and Tyr128 sites (top, middle, and bottom, respectively). In each WebLogo, the middle residue (noted with an asterisk) is the mutation site. Source data are provided as a Source Data file.
Next, we tried to combine two or three mutations in a single NRP2-Fc reagent. Combining two mutations did not have any added value compared with the single mutations, reaching 5- to 10-fold improvements in IC50 values compared with WT NRP2-Fc (Fig. 4c and Supplementary Fig. 10). Combining all three alterations resulted in a more potent reagent that shows up to 15-fold improvement compared to WT (Fig. 4c). NRP2(Q47G, I108S, Y128G)-Fc achieved a significantly improved (p < 0.01, Wilcoxon-Mann-Whitney test) IC50 value of 1.6 nM (i.e., 0.12 μg/ml) compared with NRP2-Fc. Interestingly, this NRP2-Fc variant achieved fairly high neutralizing capacity despite the fact that its binding mechanism clearly lacks avidity (Supplementary Fig. 10). The absence of avidity is further reflected in the shallow slopes (Hill slopes smaller than 1) of the neutralization curves (Fig. 4b, Supplementary Figs. 10, 8). As noted above, only a single NRP2 can bind to the LUJV spike at a time (Fig. 3b). Hence, avidity could be enabled only if the NRP2-Fc were to contain a hinge sufficiently long to allow the two NRP2 moieties in the reagent to bind two adjacent spikes.
Discussion
Comparison of the cryo-EM structures of the LUJV and LASV spike trimers highlights common structural elements of arenaviral spikes. The LUJV and the LASV spikes both organize as six-helix bundles in the membrane, in which the SSP functions as a stabilizing element. The observed topology of SSP in the membrane is the same in both spikes, indicating that topology rearrangement is a general requirement for this segment in arenaviral spikes. Also, the C’-end of GP1, which results from the cleavage by SKI-1, is an important trimerization factor at the apex of both spikes. Albeit the actual structures differ, the critical function of the SKI-1 recognition motif in mediating trimerization is preserved. This special structural role adds to the primary function of this motif in cleaving the GPC polypeptide chain for the spikes’ maturation and indicates that spike maturation involves conformational rearrangements following cleavage by SKI-I. This structural role further rationalizes why these arenaviruses adhere to SKI-1 as their cellular protease for maturation rather than evolving to utilize other cellular proteases like furin, for example, which other viruses use to enhance their virulence23.
Another important observation that emerges from this study is the predicted stabilization of the spikes due to the binding of the receptor. The quaternary-enabled interactions that NRP2 forms with the LUJV spike (Figs. 2d, 3c and Supplementary Fig. 6) inevitably favor the trimeric closed form of the spike by locking the C’-ends of the GP1 subunits at their bound conformation. These quaternary-enabled interactions include the identified interaction between Met221 and Arg130. Such interaction between sulfur atoms of methionine and the guanidino groups of arginine has a significant energetic contribution and hence can promote protein’s structural stability. Receptor binding also stabilizes the spike in the case of LASV. Similarly to LUJV, the C’-ends of the GP1 subunits in the spike of LASV are also critical elements in the formation of the matriglycan binding site17. Likewise, binding to the matriglycan receptor favors the trimeric form of the LASV spike. In addition to stabilizing the trimeric form, in the case of the LASV spike, matriglycan also serves as an N’-cap for exposed helices in the GP1 subunits. Therefore, the binding of matriglycan further stabilizes the folded state of the GP1 domains. Interestingly, both LUJV and LASV are known to dissociate from their primary cell-host receptors in a pH-driven process and utilize a secondary receptor for triggering4,24,25. Taken together, the pH-driven dissociation from NRP2 in the case of the LUJV or matriglycan in the case of the LASV also destabilizes the trimeric organization of the spikes. This may be a required step on the path to the induction of membrane fusion.
The fact that the binding of the LUJV spike to the human NRP2 is sterically restricted, as exemplified by the identification of three different binding-enhancing alterations to NRP2, is intriguing. NRP2 is a conserved protein, and analysis of 447 NRP2 sequences from mammals (Supplementary Note 1) indicates that Gln47, Ile108, and Tyr128 are completely conserved (Fig. 4d). Viruses typically adapt to their natural host receptor26, and the apparent steric constraints on binding to the human NRP2 could be explained in two different ways: First, it might be that the natural host of LUJV is not a mammal, and has a cell surface receptor with a more compatible sequence than the human NRP2. Second, it might be that LUJV sacrifices some binding affinity to its receptor in order to sterically conceal its receptor-binding site from the immune system. Regardless of the exact reason for this interesting binding mode, its mere existence provides an opportunity to construct an NRP2-based immunoadhesin that binds the LUJV spike complex better than the human NRP2. While in the absence of avidity, the potency of the NRP2-Fc immunoadhesin could be insufficient for clinical use, the NRP2(Q47G-I108S-Y128G) variant itself is a promising reagent, warranting further research since having an off-the-shelf remedy for a future LUJV outbreak is highly desired.
Methods
LUJV GPC production and purification
Expression of the full-length LUJV GPC was carried out in HEK293F cells (Invitrogen) using FreeStyle Medium (Life Technologies). Cells were grown to a density of approximately 1.0 × 106 cells per ml before transfection. HEK 293F cells were transfected using 40 kDa polyethylenamine (PEI-MAX) (Polysciences) at 1 mg ml−1, pH 7 with DNA at a ratio of 1:2 (DNA:PEI solution). Codon-optimized LUJV GPC was chemically synthesized (Genescript) and then subcloned with a C-terminal Flag tag into pcDNA3.1 using BamHI–NotI restriction sites. The LUJV GPC-expressing cells were collected at 48 h post-transfection by centrifugation at 700 × g, 4 °C for 5 min. Membranes were then resuspended in a cold lysis buffer (10 mM Tris, 150 mM NaCl, 100 μM MgCl2, 1 mM EDTA, 100 μM phenylmethyl sulfonyl fluoride (PMSF), 15% glycerol) and homogenized for 5 min on ice. The lysis mixture was then incubated while rotating for 1 h at 4 °C. A second homogenization was carried out and the lysis mixture was centrifuged at 33,300 × g for 25 min, 4 °C. The supernatant was discarded, and pellets were dissolved in solubilization buffer (20 mM Tris, 150 NaCl, 15% glycerol, 3% (% w/v) n-dodecyl-β-D-maltoside (DDM; Anatrace)). The solubilization mixture was then homogenized and incubated for 4 h; after which it was centrifuged at 370,000 × g for 25 min, 4 °C. The supernatant of this solubilization step was then incubated overnight with 50 μl of EZview red anti-Flag beads (Sigma Aldrich). The insoluble material from the solubilization step was discarded. The anti-Flag beads were then spun down (800 × g, 2 min) and washed by subsequently decreasing amounts of glycerol to 0.75% and DDM to 0.03%. The protein was eluted after the last washing step with 100 μl of 0.40 mg/ml of 1 × Flag peptide (Genescript) in a buffer containing 0.03% DDM, 20 mM Tris-HCl, 150 mM NaCl, from a 1-h incubation on ice.
Cryo-EM image acquisition, data analysis, and 3D reconstruction
A purified LUJV spike sample (3.5 μl at ~ 1 μg ml−1) was applied on glow-discharged (6 s, 12 mA; Pelco easiGlow, Ted Pella) graphene oxide Quantifoil copper grids, R1.2/1.3, (Electron Microscopy Sciences) using a Vitrobot system (Thermo Fischer/FEI) (4.5 s blotting time, 4 °C, 100% humidity). Samples were incubated on the grid for 1 minute before blotting was carried out. cryo-EM data was then collected on the Titan Krios microscope (FEI) operated at 300 kV, using a Gatan K3 direct detection camera. The beam size was 705 nm diameter (fringeless illumination), the exposure rate was 18 e− s−1 pixel−1, and movies were obtained at 105,000 × magnification with a pixel size of 0.824 Å. The nominal defocus range was − 0.6 to − 1.8 μm. A total of 11,381 movies were automatically collected using EPU 2.14. Data processing was carried out with the cryoSPARC v4.1.2 suite27. Patch motion correction and patch CTF estimation were carried out using cryoSPARC Live. Blob picking was used to pick 5,877,899 particles. Particles were extracted using a 256-pixel box, and the data set was cleaned from junk particles by carrying iterative 2D classifications, followed by 3D classifications. The final map was obtained from non-uniform refinement, imposing C3 symmetry from 87,868 particles. This map was then filtered using DeepEMhancer28 to obtain the initial working map that was used for model building. For the final refinement of the model, we used a local-resolution-based filtered map.
Model building, refinement, and analysis
An initial model was generated by docking the GP1 structure (6GH8) and the GP2 structure (7PUY) into the density map using ChimeraX 1.729. Then, using Coot 0.9.430 and real-space refinement in Phenix 1.21.131, we manually completed and refined the model of the spike. We used PyMol 2.4.2 for structural analysis and representation32.
Pseudoviral Particle Production
MLV virus-like particles (VLPs) pseudotyped with LUJV GPC were produced by transfecting retroviral transfer vector pLXIN-Luc encoding luciferase as a reporter gene together with LUJV-flag or mutated LUJV-flag in pcDNA3.1 into the GP2-293 retroviral packaging cell line (Clontech). GP2-293 cells were seeded at 5 × 106 on 10- cm plates and transfected 24 h later with 5 µg of LUJV-flag/LUJV-flag-mut and 5 µg Luciferase using Lipofectamine 2000 (Invitrogen). Cells’ media were replaced 5 h later to full medium, i.e., DMEM (Biological Industries) supplemented with 1% Pen-Strep (v/v), 1% Glutamine (v/v), and 1% sodium pyruvate (v/v). At 48 h post-transfection, media containing pseudoviruses were harvested, and VLPs were concentrated 10 times by the addition of PBS/8 % (w/v) PEG 6000 (Sigma), incubation at 4 °C for 24 h, centrifugation at 10,000 × g for 20 min and resuspension in full medium. The concentrated VLPs were stored at − 80 °C until use. Mutated genes of LUJV-flag were generated by PCR mutagenesis.
Infectivity assays
For infectivity assays, HEK293T cells were seeded on a poly-L-lysine-precoated white, chimney 96-well plate (Greiner Bio-One). Cells were left to adhere for 3 h, followed by the addition of LUJV VLPs. Cells were washed from the viruses at 18 h post-infection, and luminescence from the activity of luciferase was measured at 48 h post-infection using a Tecan Infinite M200 Pro plate reader after applying Bright-Glo reagent (Promega) to the cells.
NRP2-his and NRP2-Fc purification
Expression constructs of the first CUB domain of Neuropilin-2 (NRP2) fused to a 6 x His tag or Fc were previously constructed16. Expressions of the proteins were carried out in HEK 293 F cells (Invitrogen) using FreeStyle Medium (Life Technologies). Cells were grown to a density of approximately 1.0 × 106 cells per ml before transfection. HEK 293 F cells were transfected using 40 kDa polyethylenamine (PEI-MAX) (Polysciences) at 1 mg ml−1, with DNA at a ratio of 1:3 (DNA:PEI solution). Media containing the NRP2 constructs were collected at 6 d post-transfection. The NRP2-his was purified using a HiTrap IMAC FF Ni2+ (GE Healthcare) affinity column and Superdex 75 10/300 size exclusion chromatography (GE Healthcare) and then concentrated to an optical density at 280 nm (OD280) of 4, in 20 mM Tris-HCl pH 8.0, 150 mM sodium chloride, 10 µM CaCl2, 0.02% (wt/vol) sodium azide using an Amicon concentrator (Millipore). The NRP2-Fc was purified using a Protein-A affinity column (GE Healthcare). NRP2-Fc point mutations were introduced by PCR mutagenesis using Kapa HiFi DNA polymerase (Kapa Biosystems), according to the QuikChange site-directed mutagenesis manual. Mutated variants were expressed in HEK 293 F cells. Media were collected 6 d post-transfection. The mutated proteins were purified from the media using a Protein-A affinity column (GE Healthcare).
Bioinformatic structural analysis
A total of 34,376 structures from the PDB that were determined at 2.2 Å resolution or better and are unique (sequence similarity cutoff of 90%) were used for our analysis. We employed a custom-made Python script (see Supplementary Data 1) to identify interacting arginine and methionine residues and further used this script to perform geometry analysis. Raw output from this analysis is included as supplementary data. Plots were generated using R.
Calculating binding energies
The binding energies, which represent the energy difference between the optimized pairwise conformation and the energies of the optimized separated residues, were computed using the ORCA 5.0.3 software33. These energies were calculated at the quantum mechanics (QM) level, employing the double-hybrid revDSD-PBEP86-D4 functional34, with def2-qzvppd’ basis set, following a brief optimization at the revDSD-PBEP86-D4/QZ QM level. The choice of the DH-DFT functional revDSD-PBEP86-D434 was influenced by its significantly accurate performance with both π-π and cation-π datasets35,36. To save computational cost, arginine was represented as methylguanidinium, while methionine was represented as dimethyl sulfide. To account for structures located in hydrophobic regions, an implicit solvent model Conductor-like Polarizable Continuum Model (CPCM), was utilized with a dielectric constant of 4.0. In contrast, conformations in well-dissolved regions were calculated using CPCM (water) with a dielectric constant of 80.4. The partial charge was calculated using the CHELPG calculation scheme for the global minimum energy37.
Neutralization data and statistical analysis
IC50 values (signify the concentration at which 50% neutralization of LUJV by the NRP2 variant was achieved) were derived by fitting neutralization curves to Four Parameter Logistic Regression using Prism (GraphPad) software version 8.0.2. Wilcoxon-Mann-Whitney test for comparing groups of IC50 values was calculated using R.
Bioinformatic sequence analysis
447 mammalian NRP2 amino acid sequences were aligned using the EMBL-EBI Clustal-Omega multiple sequence alignment tool38. The Uniprot accession codes corresponding to these sequences can be found in Supplementary Note 1. Sequence logos were generated by using WebLogo 3.1 (http://weblogo.threeplusone.com/).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Coordinates file and experimental density maps were deposited at the PDB and EMDB under accession codes 8P4T and EMD-17428, respectively. Source data are provided with this paper in the Source Data File. All data are freely available. Source data are provided in this paper.
Code availability
The code that was used in this research is included in the Supplementary Data 1.
References
Briese, T. et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 5, e1000455 (2009).
Radoshitzky, S. R. et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446, 92–96 (2007).
Flanagan, M. L. et al. New world clade B arenaviruses can use transferrin receptor 1 (TfR1)-dependent and -independent entry pathways, and glycoproteins from human pathogenic strains are associated with the use of TfR1. J. Virol. 82, 938–948 (2008).
Raaben, M. et al. NRP2 and CD63 are host factors for lujo virus cell entry. Cell Host Microbe 22, 688–696.e685 (2017).
Eschli, B. et al. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J. Virol. 80, 5897–5907 (2006).
Harrison, S. C. Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008).
Pasquato, A. et al. Arenavirus envelope glycoproteins mimic autoprocessing sites of the cellular proprotein convertase subtilisin kexin isozyme-1/site-1 protease. Virology 417, 18–26 (2011).
York, J., Romanowski, V., Lu, M. & Nunberg, J. H. The signal peptide of the Junin arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J. Virol. 78, 10783–10792 (2004).
York, J. & Nunberg, J. H. Role of the stable signal peptide of Junin arenavirus envelope glycoprotein in pH-dependent membrane fusion. J. Virol. 80, 7775–7780 (2006).
Nunberg, J. H. & York, J. The curious case of arenavirus entry, and its inhibition. Viruses 4, 83–101 (2012).
York, J. & Nunberg, J. H. Intersubunit interactions modulate pH-induced activation of membrane fusion by the Junin virus envelope glycoprotein GPC. J. Virol. 83, 4121–4126 (2009).
Eichler, R. et al. Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep. 4, 1084–1088 (2003).
Eichler, R., Lenz, O., Strecker, T. & Garten, W. Signal peptide of Lassa virus glycoprotein GP-C exhibits an unusual length. FEBS Lett. 538, 203–206 (2003).
Bederka, L. H., Bonhomme, C. J., Ling, E. L. & Buchmeier, M. J. Arenavirus stable signal peptide is the keystone subunit for glycoprotein complex organization. mBio 5, e02063 (2014).
Abraham, J., Corbett, K. D., Farzan, M., Choe, H. & Harrison, S. C. Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nat. Struct. Mol. Biol. 17, 438–444, (2010).
Cohen-Dvashi, H., Kilimnik, I. & Diskin, R. Structural basis for receptor recognition by Lujo virus. Nat. Microbiol. 3, 1153–1160 (2018).
Katz, M. et al. Structure and receptor recognition by the Lassa virus spike complex. Nature 603, 174–179 (2022).
Klewitz, C., Klenk, H. D. & Ter Meulen, J. Amino acids from both N-terminal hydrophobic regions of the Lassa virus envelope glycoprotein GP-2 are critical for pH-dependent membrane fusion and infectivity. J. Gen. Virol. 88, 2320–2328 (2007).
Shi, W. et al. Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane. Nature 619, 403–409 (2023).
Sommerstein, R. et al. Arenavirus glycan shield promotes neutralizing antibody evasion and protracted infection. PLoS Pathog. 11, e1005276 (2015).
Cohen-Dvashi, H. et al. Rational design of universal immunotherapy for TfR1-tropic arenaviruses. Nat. Commun. 11, 67 (2020).
Cohen-Dvashi, H. et al. Anti-SARS-CoV-2 immunoadhesin remains effective against Omicron and other emerging variants of concern. iScience 25, 105193 (2022).
Cantuti-Castelvetri, L. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860 (2020).
Jae, L. T. et al. Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science 344, 1506–1510 (2014).
Cohen-Dvashi, H., Israeli, H., Shani, O., Katz, A. & Diskin, R. Role of LAMP1 binding and pH sensing by the spike complex of Lassa virus. J. Virol. 90, 10329–10338 (2016).
Diskin, R. A structural perspective on the evolution of viral/cellular macromolecular complexes within the arenaviridae family of viruses. Curr. Opin. Struct. Biol. 79, 102561 (2023).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 (2010).
The PyMOL Molecular Graphics System. Version 2.4.2 Schrödinger, LLC.
Neese, F. The ORCA program system. WIREs Comput. Mol. Sci. 2, 73–78 (2012).
Santra, G., Sylvetsky, N. & Martin, J. M. L. Minimally empirical double-hybrid functionals trained against the GMTKN55 database: revDSD-PBEP86-D4, revDOD-PBE-D4, and DOD-SCAN-D4. J. Phys. Chem. A 123, 5129–5143 (2019).
Santra, G., Semidalas, E., Mehta, N., Karton, A. & Martin, J. M. L. S66x8 noncovalent interactions revisited: new benchmark and performance of composite localized coupled-cluster methods. Phys. Chem. Chem. Phys. 24, 25555–25570 (2022).
Spicher, S., Caldeweyher, E., Hansen, A. & Grimme, S. Benchmarking London dispersion corrected density functional theory for noncovalent ion-pi interactions. Phys. Chem. Chem. Phys. 23, 11635–11648 (2021).
Breneman, C. M. & Wiberg, K. B. Determining atom-centered monopoles from molecular electrostatic potentials. The need for high sampling density in formamide conformational analysis. J. Comput. Chem. 11, 361–373 (1990).
Madeira, F. et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic acids Res. 50, W276–W279 (2022).
Acknowledgements
The Diskin lab is supported by research grants from the Ernst I Ascher foundation, Ben B. and Joyce E. Eisenberg Foundation, Estate of Emile Mimran, Jeanne and Joseph Nissim Center for Life Sciences Research, Dov and Ziva Rabinovich Endowed Fund for Structural Biology, Donald Rivin, Stanley and Tanya Rossby Endowment Fund, Natan Sharansky, Dr. Barry Sherman Institute for Medicinal Chemistry, as well as from the Israel Science Foundation (grant No. 209/20). We thank Deborah Fass for her critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
R.D. conceived and oversaw this research. M.E.-A. produced and purified proteins. M.E.-A. and R.D. performed EM analysis and solved the structure. M.E.-A. and S.B. performed infectivity and neutralization assays. R.S. performed a statistical analysis of structural information from the PDB. R.C. and Y.L. performed DH-DFT calculations and analyzed data. H.C.-D. provided reagents. All authors contributed to the data analysis. R.D. and M.E.-A. prepared the manuscript with the help of all the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Ilona Rissanen and the other anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Eilon-Ashkenazy, M., Cohen-Dvashi, H., Borni, S. et al. The structure of the Lujo virus spike complex. Nat Commun 15, 7175 (2024). https://doi.org/10.1038/s41467-024-51606-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51606-0
This article is cited by
-
Molecular organization of the New World arenavirus spike glycoprotein complex
Nature Microbiology (2025)
-
Metal-induced conformational changes in the Sabiá virus spike complex
Nature Microbiology (2025)