Abstract
In mammals, global passive demethylation contributes to epigenetic reprogramming during early embryonic development. At this stage, the majority of DNA-methyltransferase 1 (DNMT1) protein is excluded from nucleus, which is considered the primary cause. However, whether the remaining nuclear activity of DNMT1 is regulated by additional mechanisms is unclear. Here, we report that nuclear DNMT1 abundance is finetuned through proteasomal degradation in mouse zygotes. We identify a maternal factor, Pramel15, which targets DNMT1 for degradation via Cullin-RING E3 ligases. Loss of Pramel15 elevates DNMT1 levels in the zygote pronuclei, impairs zygotic DNA demethylation, and causes a stochastic gain of DNA methylation in early embryos. Thus, Pramel15 can modulate the residual level of DNMT1 in the nucleus during zygotic DNA replication, thereby ensuring efficient DNA methylation reprogramming in early embryos.
Similar content being viewed by others
Introduction
Following fertilization, embryos undergo epigenetic reprogramming to reset epigenetic information inherited from the parental genome, which is a pre-requisite for the embryonic gene expression programs1,2,3. The mammalian genome contains a substantial level of methylation at CpG dinucleotides, specifically in the form of 5-methylcytidine (5mC). One of the most remarkable features of early preimplantation embryo development, particularly during the first cell cleavage, is the widespread loss of DNA methylation4,5,6,7. This process is conserved across mammals but exhibits variations in demethylation kinetics and extent8. The global demethylation process is considered crucial for the developmental competence of early embryos, as mouse embryos with hypermethylated genomes exhibited abnormalities9,10,11,12,13. In mammals, biological processes associated with a global decrease in DNA methylation are relatively unusual14. To achieve this, early embryos employ unique regulatory mechanisms that remain partially understood15.
The current consensus agrees that multiple mechanisms participate in global demethylation in early embryos. One such mechanism involves the dioxygenase enzyme TET3, which catalyzes the oxidation of the 5mC, leading to active demethylation16,17. In zygotes, both paternal and maternal pronuclei undergo active demethylation18,19,20. Additionally, in male pronuclei, TET-independent active demethylation for paternal DNA methylation has been reported, and the following de novo methylation by DNMT1 and DNMT3A provides the substrates to TET321. However, replication-dependent passive demethylation appears to be responsible for the majority of global demethylation, since zygotic DNA demethylation is severely abolished when DNA replication is inhibited18,19.
The maintenance of DNA methylation after DNA replication primarily relies on the DNA methyltransferase DNMT1 and its co-factor UHRF122,23,24,25,26. In contrast to somatic cells, DNMT1 and UHRF1 in mouse early embryos are predominantly localized to the cytoplasm27,28. Cytoplasmic retention of these factors is widely believed to be responsible for zygotic passive demethylation process15,29. Supporting this notion, in recent studies, mouse embryos lacking maternal factor NLRP14 and PADI6 were shown to exhibit impaired cytoplasmic retention of UHRF1 and/or DNMT1, and an increase of DNA methylation levels13,30,31,32. However, since NLRP14 loss resulted in pervasive proteomic dysregulation in oocyte31,33, the molecular basis of the nuclear exclusion of DNMT1 and UHRF1, as well as the extent to which demethylation depends on cytoplasmic exclusion, remains to be clarified.
Despite that the maintenance factors are predominantly localized in the cytoplasm, there is evidence that residual activity of maintenance methylation machinery functions in the early embryo nuclei. For example, gamete-inherited methylation patterns at specific genomic regions, such as imprinting control regions and retrotransposable elements, are maintained by DNMT1 and UHRF128,34,35,36,37. Additionally, studies investigating the genomic distribution of DNMT1 during early embryo development have demonstrated its preferential association with retroelement regions38. Besides, chromatin modulators, including Trim28, ZFP57, and ZFP44539,40,41,42, are involved in the recruitment of DNMT1 for the maintenance of methylation post-fertilization. Thus, the activity of the maintenance machinery is finely orchestrated in the nucleus during the global demethylation program, with the molecular details of this process have yet to be elucidated.
Therefore, we hypothesized that there are unknown regulatory mechanisms in early embryos that regulate the global DNA demethylation process. This study aims to investigate this hypothesis by searching for novel regulators of DNA methylation, whose expression is specific to early embryos. In pursuit of this objective, we have identified Pramel15, which functions through the ubiquitin-proteosome system to target DNMT1 for degradation. Furthermore, we have revealed that Pramel15 tunes the protein levels of DNMT1 within the embryonic nuclei. Notably, mouse embryos lacking Pramel15 display a stochastic increase in DNA methylation levels across the genome. These findings unveil a layer of regulation for embryonic DNA demethylation. We anticipate that these findings will contribute to our ongoing efforts in understanding the intricate mechanisms governing mammalian DNA methylation reprogramming.
Results
Screening for factors promoting DNA demethylation
To discover potential regulators of DNA demethylation, we established a luciferase reporter system using a Gal4-VP16 fusion protein and Gal4-binding site to drive luciferase expression (Supplementary Fig. 1a). The reporter was pre-methylated in vitro to prevent Gal4-VP16 binding, ensuring that the transiently transfected reporter could only be activated after demethylation. By mixing various ratios of unmethylated and methylated reporters, we observed that the presence of just 5% unmethylated reporter led to a significant increase in luciferase signal, indicating a high sensitivity of our system (Supplementary Fig. 1b, c). The reporter system was then co-transfected with a cDNA expression library derived from mouse oocytes, aiming to identify genes whose overexpression could activate the methylated reporter. We screened approximately 1.44 × 105 mouse oocyte cDNAs, which corresponds to approximately 4-fold of the number of mouse genes (Supplementary Fig. 1d). Consequently, we obtained 72 positive clones, covering a few candidate genes that are potentially involved in DNA demethylation regulation.
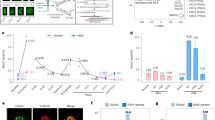
To verify our primary hits, we utilized another reporter cell line, B2-17, that we previously developed to monitor DNA demethylation events43. This reporter cell line contained an enhanced green fluorescence protein (EGFP) coding gene driven by a fully methylated CMV promoter, allowing us to determine demethylation events through an increase in fluorescence signal (Fig. 1a). By individually overexpressing the primary hits in B2-17 cells, we confirmed that two factors, Stella and Pramel15 (Preferentially expressed antigen of melanoma like 15), robustly activated the methylated EGFP gene (Fig. 1b and Supplementary Fig. 1e). We previously reported that Stella plays a role in sequestering UHRF1 from the nucleus and interfering with the maintenance of DNA methylation after replication13,44. In this study, we focus on exploring the potential role of Pramel15 in DNA methylation regulation.
a Scheme of B2-17, a DNA demethylation-sensitive cell line43. A CMV promoter-driven GFP fragment was methylated in vitro by HapII and then integrated into HEK293 cells. The clone with a low GFP background and high sensitivity to 5-aza-2′-deoxycytidine was selected as B2-17. b Left, fluorescence images show the GFP-activated cells in B2-17 transfected with Pramel15-expressing plasmids. Scale bars indicate 100 μm. Right, bar graph shows the ratio of GFP-positive cells in B2-17 transfected with Pramel15-expressing plasmids using fluorescence-activated cell sorting (FACS). Three biological replicates have been performed. c BS-seq shows the DNA methylation states of CMV promoter in B2-17 cells transfected with Pramel15-expressing plasmids. Genomic DNA was extracted 96 h later and followed by bisulfate sequence of CMV promoter. d Bar graphs show the relative level of 5-methylcytosine (mC) and 5-hydroxymethylcytosine (hmC) in genome of B2-17. Cells were harvested at 1–4 days following Pramel15 expression, and genomic DNA was extracted for nucleotide mass spectrum analysis. Normalization was conducted in reference to the 5mC level in Day 0 cells. e Genomic DNA of B2-17 was extracted for nucleotide mass spectrum analysis following transfection of plasmids expressing Pramel15-mCherry or mCherry, and cells were treated with 5 μM aphidicolin or vehicle for 2 days. For (d and e) each point indicates a biological replicate. Data are presented as mean value ± s.d.
Pramel15 overexpression leads to global DNA demethylation
To investigate the impact of Pramel15 on DNA methylation, we initially examined the methylation status of CMV promoter in B2-17 cells. Bisulfite sequencing analysis revealed a marked decrease in methylation level, from 92.1% to 29.8%, after the expression of Pramel15 for 4 days (Fig. 1c). This suggests that Pramel15 reactivates the methylated EGFP reporter through DNA demethylation. Then we performed mass spectrometry analysis to quantify the abundance of methylated cytosine (5mC) in genomic DNA. Consistent with our observations with the EGFP reporter, the expression of Pramel15 led to a progressive reduction in global 5mC level (Fig. 1d). Notably, there was a concurrent decrease in global 5hmC level (Fig. 1d), indicating that Pramel15 induces loss of DNA methylation independent of TET-mediated 5mC oxidation. We further explored whether Pramel15 impairs DNA methylation maintenance during DNA replication. The treatment of DNA synthesis inhibitor aphidicolin almost fully abolished Pramel15-induced global demethylation (Fig. 1e). These results suggest that Pramel15 likely interferes with the DNA methylation maintenance machinery, resulting in global passive demethylation.
Pramel15 interacts with DNMT1 RFTS domain and regulates DNMT1 protein stability
DNMT1 and UHRF1 are two indispensable factors for maintaining DNA methylation22,23,24. To investigate whether Pramel15 contributes to DNA methylation loss by affecting DNMT1 or UHRF1, we explored their potential interaction. Co-immunoprecipitation (Co-IP) analysis revealed interaction between Pramel15 and DNMT1 under a stringent washing condition (500 mM NaCl). In contrast, no interaction with UHRF1 was observed, even under a mild washing condition (150 mM NaCl) (Fig. 2a).
a Flag-immunoprecipitation from 293FT cells transfected with Pramel15-HA and DNMT1-Flag (upper) or UHRF1-Flag (lower) expressing plasmids. b High-content images show DNMT1-GFP level with or without Pramel15-mCherry induction by doxycycline (Dox) treatment for 48 h (left) and the boxplot graph shows the statistic results of DMSO group (n = 3973) and Dox group (n = 3109) (right). Boxplots show median (center line), 25th to 75th percentiles (hinges). The whiskers go down to the smallest value and up to the largest. Two-sided t-tests assuming unequal variances were performed. c Western blotting of DNMT1 protein levels in T-REx293 cells expressing Dox-inducible Pramel15-Flag with or without 5 μM MG132 treatment. d Schematic representation of DNMT1 and its truncations. e, f Flag-immunoprecipitation from 293FT cells transfected with DNMT1-Flag or its truncations and Pramel15-HA expressing plasmids. Asterisk: heavy chain of antibody. g Western blotting of Flag-DNMT1 and Flag-DNMT1(Δ350-609aa) protein levels in 293FT cells with or without Pramel15-HA. h Western blotting of Flag-DNMT1(350-609aa) protein levels in mES cells expressing Flag-DNMT1(350-609aa) and Dox-inducible Pramel15-HA. i Flag-immunoprecipitation in 293FT cells transfected with plasmids expressing His-ubiquitin, Flag-DNMT1(1-609aa), and Pramel15-HA or blank plasmids treated with 20 μM MG132. All immunoprecipitation and quantification experiments analyzed by Western blotting were repeated at least three times independently.
To further investigate the regulation between Pramel15 and DNMT1, we utilized an HEK293 cell line expressing DNMT1-GFP fusion protein and doxycycline-inducible Pramel15-mCherry fusion protein. Upon induction of Pramel15, we observed a significant decrease in DNMT1-GFP fluorescence signal (Fig. 2b). Moreover, when we inhibited proteasome activity using MG132, the DNMT1 signal was restored (Fig. 2c). Therefore, Pramel15 likely induce DNA demethylation by promoting proteasome-dependent degradation of DNMT1.
DNMT1 contains multiple domains, including DMAP1 binding domain, PCNA binding domain, nucleus localization signal (NLS) motif, RFTS domain, CXXC domain, BAH1 and BAH2 domain, and the C-terminal catalytic domain (Fig. 2d)45,46. We expressed several DNMT1 truncations in cell and performed Co-IP experiments to characterize its corresponding region responsible for the interaction with Pramel15. We first observed interaction between Pramel15 and the N-terminal region of DNMT1 (1–609 aa) (Fig. 2e), and then we identified the DNMT1 RFTS domain (350–609 aa) as the region that interacts with Pramel15 (Fig. 2f and Supplementary Fig. 2a).
To determine whether the DNMT1 RFTS domain is essential for Pramel15-induced DNMT1 degradation, we examined the degradation of a DNMT1 truncation lacking the RFTS domain (Δ350–609aa). In contrast to the full-length DNMT1 protein, DNMT1 lacking the RFTS domain was resistant to Pramel15-induced degradation (Fig. 2g). In addition, Pramel15 expression led to the degradation of a DNMT1 truncation that only contains the RFTS domain (Fig. 2h and Supplementary Fig. 2b). Taken together, our results suggest that Pramel15 associates to the RFTS domain of DNMT1, and that the RFTS domain is both sufficient and necessary for Pramel15-mediated proteasome degradation.
Cullin-RING ligases are involved in Pramel15-mediated DNMT1 degradation
To exam whether ubiquitination is involved in Pramel15-mediated DNMT1 degradation, we conducted an in vivo ubiquitination assay by co-transfecting constructs expressing His-tagged ubiquitin. Given that the full-length DNMT1 protein is large, and it is challenging to detect its ubiquitination, we used DNMT1(1–609aa) in this assay. The results showed an increase of ubiquitin levels on DNMT1(1-609aa) when Pramel15 was expressed (Fig. 2i).
To understand the mechanism by which Pramel15 facilitates ubiquitination and degradation of DNMT1, we performed anti-Flag affinity purification experiments using T-Rex-293 cells expressing a doxycycline-inducible Flag-Pramel15. We identified several proteins co-purified with Flag-Pramel15, including Cullin 5 (Cul5), Elongin-B (EloB) and Elongin-C (EloC) (Supplementary Fig. 3a), all of which are subunits of the Cul5-RING ubiquitin ligases (CRL5) complex47. Similarly, we performed immunoprecipitation experiments using mouse embryonic stem cells (mES cells) expressing Flag-Pramel15. Label-free quantitative mass spectrometry analysis revealed the enrichment of Cul5, EloB, EloC, and Rbx2, which is another subunit of the CRL5 complex (Fig. 3a). Other enriched factors are also highly relevant (Supplementary Fig. 3b). In addition to DNMT1, USP7 is a deubiquitinase of DNMT148,49, AMBRA1 targets EloC to modulate CRL5 complex50, ARIH2 is an atypical E3 ligase working together with CRL5 complex51,52, and PAF15 is involved in DNMT1 recruitment53.
a Label-free mass-spectrometry analysis of proteins co-immunoprecipitated with Pramel15-Flag in mES cells from two independent experiments. Pramel15, DNMT1, and the subunits of CRL5 complex (highlight in red) are among the top enrichment candidates. b Western blotting of DNMT1 protein levels in mES cells expressing Dox-inducible Pramel15-Flag with or without 2.5 μM MLN4924 treatment. c Western blotting of DNMT1 protein levels in mES cells and Cul5-KO mES cells expressing Dox-inducible Pramel15-Flag. d Flag-immunoprecipitation from 293FT cells transfected with plasmids expressing Pramel15-HA and different Flag-tagged Cullin proteins. e Model of Cullin-RING E3 ligase complex. Different Cullin complexes contain different adapters and Cul5 binds to Rbx2, while other Cullin proteins bind to Rbx1. f Western blotting of DNMT1 protein levels in mES cells and Cul5-KO mES cells expressing Dox-inducible Pramel15-Flag with or without Rbx1 siRNA transfected (left). Statistics of four biological replicates (right). Two-sided t-tests assuming unequal variances were performed. Data are presented as mean value ± s.d. All immunoprecipitation and quantification experiments analyzed by Western blotting were repeated at least three times independently.
Activation of CRL complex relies on the neddylation of Cullin proteins54. MLN4924 is a small molecule inhibitor of the Nedd8-activating enzyme E1 (NAE)55. MLN4924 treatment effectively blocked Pramel15-mediated DNMT1 degradation in mES cells (Fig. 3b), confirming the functional role of CRL complexes in mediating Pramel15-dependent DNMT1 degradation.
To examine whether the CRL5 complex is responsible for Pramel15-mediated DNMT1 degradation, we generated a Cullin5 knockout (Cul5-KO) mES cell line (Fig. 3c and Supplementary Fig. 3c, d). Cullin5 knockout attenuated, but did not fully abolish, Pramel15-induced DNMT1 degradation (Fig. 3c). This suggests that the other members of the CRLs may also participate in Pramel15-induced DNMT1 degradation. In agreement with this hypothesis, the remaining Pramel15-induced DNMT1 degradation activity in Cul5-KO cells was also inhibited by MLN4924 treatment (Supplementary Fig. 3e).
In mammals, there are 8 Cullin family proteins: Cul1, Cul2, Cul3, Cu4A, Cul4B, and Cul5, Cul7, and Cul956,57. To determine which Cullin proteins, in addition to Cul5, can mediate Pramel15-induced DNMT1 degradation, we then co-expressed Pramel15-HA and Flag-tagged individual Cullin proteins in HEK293 cells and performed anti-Flag Co-IP experiments. Considering that Cul7 and Cul9 are barely expressed in mES cells, we focused on the other Cullin proteins. In addition to Cul5, several other Cullin proteins, including Cul1, Cul2, Cul3, Cul4A, exhibited various degrees of interaction with Pramel15 (Fig. 3d). Since multiple Cullin proteins likely participate in Pramel15-induced DNMT1 degradation, we adopted an alternative approach to test their functional relevance. The RING component of CLRs consists of two family members: Rbx1 and Rbx2, responsible for the catalytic activity of CRL complexes. Rbx2 is a specific partner with Cul5, whereas all other Cullin proteins functionally cooperate with Rbx1 (Fig. 3e)58. Taking advantage of this feature, we knocked down Rbx1 in wild-type and Cul5-KO mES cells. The loss of both Rbx1 and Cul5 fully abolished Pramel15-induced DNMT1 degradation (Fig. 3f). The above results clearly support the involvement of multiple members of Cullin-RING ubiquitin ligase family in Pramel15-induced DNMT1 degradation.
The essential motifs of Pramel15 in mediating DNMT1 degradation
Through the identification of conserved amino acid motifs in Pramel15 that mediate its interaction with CRLs components, we discovered two interesting motifs at the N-terminal: Cul2-box, and Cul5-box, which mediate the interactions with scaffold proteins Cul2 and Cul5, respectively59 (Fig. 4a, b). This likely explains the versatility of Pramel15 in binding to various members of the Cullin family. We also identified a BC-box motif, which is responsible for anchoring substrate receptor proteins to Cullins (Fig. 4a, b). In addition, the C-terminal domain of Pramel15 contains 5 LRR motifs (Fig. 4c), which often function in protein-protein interactions60. The domain organization suggests that the N-terminus of Pramel15 is involved in Cullin association, while the C-terminus LRR motifs contribute to DNMT1 interaction. Supporting this notion, a Pramel15 truncation lacking the LRR domain (delta268-402) lost its interaction with DNMT1 but still bound to Cullin5 (Fig. 4d). Conversely, removing the N-terminus, including the CRL-binding motifs abolished its binding with Cullin5 (Fig. 4d). We note that Pramel15 lacking the N-terminus also lost its association with DNMT1, potentially due to compromised protein folding (Fig. 4d). Our analysis suggests that Pramel15 functions as a substrate receptor protein that delivers DNMT1 to CRLs.
a Schema representing the domain organization of Pramel15, containing predicted BC-box, Cul2-box, Cul5-box and LRR motifs. LRR, leucine-rich repeat. b Multiple sequence alignment of Pramel15 with proteins containing BC and Cul2 box or BC and Cul5 box motifs, highlighting conserved residues. Identical amino acids are in black, very similar ones are in dark grey, and similar amino acids are in light grey. Conserved Pramel15 residues are marked with asterisks. c Sequence of the predicted Pramel15 LRR motif, with the conserved leucine in LRR sequence patterns (LXXLXL) highlighted in yellow. X, any amino acid. d HA-immunoprecipitation from 293FT cells transfected with plasmids expressing Pramel15-HA and Flag-tagged Cul5. Western blotting shows the association of DNMT1 and Cul5 with Pramel15, Pramel15(Δ2-121aa), and Pramel15(Δ268-402aa). e Table listing the point mutants of Pramel15 at BC-box, Cul2-box, and Cul5-box. f Western blotting shows DNMT1 protein levels in 293FT cells expressing Dox-inducible HA-tagged mutants of Pramel15 as per (e). g Flag-immunoprecipitation from 293FT cells transfected with plasmids expressing HA-tagged Pramel15 or the N-terminal mutants and Flag-tagged Cul5. Western blotting shows the association of Cul5 with Pramel15 and the mutants. h Western blotting of DNMT1 protein levels in 293FT cells expressing Dox-inducible HA-tagged LRR motifs mutants (L/V/F to A, according to Fig. 4c) of Pramel15. i Flag-immunoprecipitation from 293FT cells transfected with plasmids expressing Flag-tagged DNMT1and HA-tagged Pramel15 or the LRRs mutants (L/V/F to A, according to (c)) and cultured with 2.5 μM MLN4924 to prevent DNMT1 degradation. Western blotting shows the association of DNMT1 with Pramel15 and the LRRs mutants. All immunoprecipitation and quantification experiments analyzed by Western blotting were repeated at least three times independently.
Next, we aimed to determine whether Pramel15 bridges DNMT1 to CRLs for degradation. According to this model, mutations in either the CRL-binding module or the DNMT1-recognition module of Pramel15 should impair DNMT1 degradation. Indeed, Pramel15 mutants with either a mutated BC-box or Cul2/5-box motif failed to induce DNMT1 degradation (Fig. 4e, f). Meanwhile, these mutants showed impaired association with Cul5 (Fig. 4g). Similarly, when the conserved residues in LRR motifs (L/V/F) were mutated to alanine, DNMT1 degradation was also impaired (Fig. 4h), and the association between DNMT1 and mutants was abolished (Fig. 4i). Collectively, our results suggest that Pramel15 serves as a substrate receptor module within the Cullin-RING E3 ligase complex to target DNMT1 for proteasomal degradation.
No apparent developmental and fertility defects in Pramel15 knockout mice
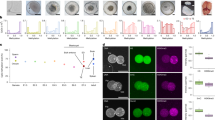
According to public data61, Pramel15 mRNA is specifically expressed and translated in oocytes, zygotes, and early two-cell embryos (Fig. 5a). To gain insight into the physiological function of Pramel15, we generated Pramel15 knockout mice using a gRNA targeting the first exon of Pramel15 to produce random nucleotide deletion, which resulted in a frameshift and the appearance of a premature stop codon (Supplementary Fig. 4a, b). The Pramel15 homozygotes knockout mice were born, and both male and female mutant mice were capable of producing offspring normally (Supplementary Fig. 4c). Furthermore, the development rate and dynamics of early embryos derived from Pramel15 KO oocytes showed no apparent defects (Supplementary Fig. 4d, e). In addition, the International Mouse Phenotyping Consortium (IMPC) has also analyzed various phenotypes and observed no significant difference (https://www.mousephenotype.org/data/genes/MGI:3712553). These analyses suggest that the loss of Pramel15 minimally impairs early embryonic development in mice.
a Line graph depicting the dynamics of Pramel15 mRNA and ribosome-protected fragments (RPF) during early development, along with a bar graph showing the translational efficiency (RFP/mRNA). Source data: GSE165782. b Representative fluorescence images illustrating Pramel15 localization in GV oocytes, zygotes, and 2 C embryos that were microinjected with Pramel15-mCherry mRNA. These results were repeated three times independently. c Schematic representation of 3D reconstruction from images acquired from a high-content imaging analysis system. d Bar graphs showing relative DNMT1 intensity in the nuclei of zygotes. Zygotes derived from WT (n = 118) and Pramel15-KO (n = 118) oocytes were collected 11–12 h post in vitro fertilization. e Bar graphs showing relative DNMT1 intensity of the male (WT, n = 58; KO, n = 56) and female (WT, n = 57; KO, n = 60) pronuclei, which were defined based on nucleus size. f Bar graphs showing the relative DNMT1 intensity in nuclei of zygotes (WT, n = 49; KO, n = 45), 2-cell embryos (2 C) (WT, n = 67; KO, n = 57), 4-cell embryos (4 C) (WT, n = 73; KO, n = 71), 8-cell embryos (8 C) (WT, n = 139; KO, n = 124), and morula embryos (WT, n = 337; KO, n = 185). DNMT1 intensity was normalized to the WT group at each stage. g, h Bar graphs showing relative DNMT1 intensity in the nuclei of zygotes treated with DMSO (WT, n = 107; KO, n = 145), MG132 (WT, n = 100; KO, n = 127), and MLN4924 (WT, n = 126; KO, n = 143). Zygotes were transferred to HTF containing DMSO, 10 μM MG132, and 5 μM MLN4924 respectively 2 h after fertilization and cultured further for an additional 9 h. For (d–h), each dot (n) in these figures stands for one analyzed nucleus. Unpaired two-sided t-tests assuming unequal variances were performed. Data are presented as mean value ± s.d. i Genome was divided into 1 Mb bins. Normalized counts of DNMT1 CUT&RUN represented DNMT1 level. CPM counts per million. j DNMT1 level represented by the normalized counts in each corresponding genomic bin. Genomic bins are ranked by read counts of H3K9me3 ChIP-seq in MII oocytes. For (i, j) Unpaired two-sided Wilcoxon tests were performed. Boxplots show median (center line), upper and lower quartiles (box limits), The upper whisker extends to the largest value no further than 1.5× IQR from the hinge. The lower whisker extends from the hinge to the smallest value at most 1.5× IQR of the hinge. H3K9me3 ChIP-seq data source: GSM2588560.
Pramel15 regulates DNMT1 protein level in early embryonic nucleus
Given that ectopic expression of Pramel15 induced significant degradation of DNMT1 in cultured cells, we investigated whether Pramel15 also regulates DNMT1 and, consequently, DNA methylation in vivo. In mouse oocytes and early embryos, DNMT1 protein is highly abundant but predominantly localizes in cytoplasm, with only a small fraction retained in the nuclei27,36,62 (Supplementary Fig. 5a, b), In contrast to our observations in cell lines, the loss of Pramel15 hardly affect the global level of DNMT1 in early mouse embryos, as determined by immunofluorescence quantification (Supplementary Fig. 5a, c). However, we observed that the fusion protein Pramel15-mCherry, but not mCherry alone, exhibited nuclear localization from GV oocytes to the 2-cell stage embryos (Fig. 5b). Consequently, we investigated whether Pramel15 is more crucial in controlling the nuclear DNMT1 level. To reliably quantify the DNMT1 level in the zygotic pronuclei, we acquired confocal images of whole embryos using a high-content imaging system and performed 3D-reconstitution (see details in “Methods”, Fig. 5c). In support with a role of Pramel15 in the regulation of DNMT1 level, our quantitative analysis revealed a significant increase in DNMT1 levels in both male and female pronuclei of Pramel15-deficient zygotes (Fig. 5d, e). Moreover, the elevated nuclear DNMT1 level in Pramel15-deficient embryos was consistently detected throughout preimplantation development until the morula stage (Fig. 5f).
To determine whether Pramel15 degrades nuclear DNMT1 through CRLs, we performed quantitative immunostaining analysis in zygotes treated with MG132 or MLN4924. Consistent with our observations in Pramel15 maternal KO (mKO) zygotes, the inhibition of proteasomes or CRLs in wild-type zygotes also led to an increase of nuclear DNMT1 levels (Fig. 5g). Importantly, this change did not occur in Pramel15 mKO zygotes (Fig. 5h). These results support a role of Pramel15 in regulating nuclear DNMT1 level in the zygotes via the CRL and proteasome pathway.
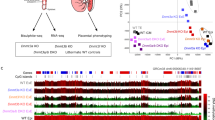
To assess the DNMT1 level on chromatin, we performed CUT&RUN of DNMT1 in parthenogenetic 1-cells. Consistent with our immunostaining quantification, the CUT&RUN signal for chromatin-bound DNMT1 increased in Pramel15-KO 1-cells in a statistically significant manner (Fig. 5i). Since DNMT1 has preference towards the H3K9me3-riched regions39,63,64, we analyzed the association between H3K9me3 and the genomic distribution of DNMT1. Indeed, we found that the increase of DNMT1 CUT&RUN signal preferentially occurred on chromatin domains enriched for H3K9me3 (Fig. 5j). Taken together, our results suggest that Pramel15 regulates nuclear level of DNMT1 in preimplantation embryos.
Increase of DNA methylation in Pramel15-deficient embryos
Since Pramel15 knockout led to an elevation of nuclear DNMT1 levels in zygotes, we further investigated whether this has any consequence on DNA methylation. We performed DNA methylome profiling using PBAT65 in MII oocytes, zygotes, and 2-cell embryos. While samples from distinct stages cluster together, Pramel15-KO samples are generally separated from Pramel15-Het samples, except in MII oocytes (Supplementary Fig. 6), suggesting that loss of Pramel15 altered the DNA methylome after fertilization. Indeed, we observed no significant change of global methylation in Pramel15 KO MII oocytes (Fig. 6a), but a statistically significant yet modest increase in methylation was observed in Pramel15-deficient zygotes and 2-cell embryos (Fig. 6b, c). We differentiated the maternal and paternal reads using SNPs and observed that such increase occurred in both the maternal and paternal genomes (Fig. 6d). SNP-based parent-of-origin analysis sacrificed a large number of sequencing reads. To acquire a higher coverage dataset for further analysis, we generated parthenogenetic embryos and profiled their DNA methylome. Consistently, we observed a significant methylation increase in parthenogenetic 1-cell embryos derived from Pramel15 KO oocytes (Fig. 6e). Thus, the loss of Pramel15 caused an increase in DNA methylation in zygotes but not in MII oocytes. DNA methylome of MII oocyte exhibits a bimodal distribution, in which highly methylated domains are separated by regions with very low levels of methylation6. We noticed that methylation gains in maternal genome of 1-cell embryo predominantly occurred on the methylated domains in MII oocyte (Fig. 6f), suggesting that Pramel15 regulates the zygotic demethylation process.
a–c Global DNA methylation levels in MII oocytes, zygotes, and 2-cell embryos derived from WT or Pramel15-Het and Pramel15-KO oocytes. d Bar graph shows zygotic global DNA methylation levels in paternal and maternal genomes of zygotes, splitting by single nucleotide polymorphisms (SNPs) of DBA/2J and C57BL/J, respectively. Two-sided t-tests assuming unequal variances were performed. Data are presented as mean value ± s.d. e Global DNA methylation levels in parthenogenetic 1-cell embryos derived from Pramel15-Het and Pramel15-KO oocytes. f Boxplots show differences in mean CpG methylation of 1-Mb bins between parthenogenetic 1-cells derived from Pramel15 Het and KO oocytes. Genomic bins are ranked by increase of DNMT1 CUT&RUN signal. For (a–c, e, f), Unpaired two-sided Wilcoxon tests were performed. Boxplots show median (center line), upper and lower quartiles (box limits), The upper whisker extends to the largest value no further than 1.5× IQR from the hinge. The lower whisker extends from the hinge to the smallest value at most 1.5× IQR of the hinge. g, Bar graph shows differences in mean CpG methylation of 1-Mb bins between parthenogenetic 1-cells derived from Pramel15 Het and KO oocytes. Genomic bins are ranked by read counts of H3K9me3 ChIP-seq in MII oocytes. h Genome browser view of the difference of CpG methylation and H3K9me3 density in parthenogenetic 1-cells derived from Pramel15 Het and KO oocytes. H3K9me3 ChIP-seq data source: GSM2588560. i Correlation of CpG methylation changes and CpG number of 1-kb bins grouped by methylation levels in MII oocytes j, The efficiency of DNA methylation maintenance during DNA replication in 1-kb bins with varying CpG density. Replication-coupled (0 min) and 5 min after replication are shown. Data source: GSE131098. k Bar graph shows differences in mean CpG methylation (mCG%KO − mCG%het) of 1-kb bins in parthenogenetic 1-cells, ranked by mean of total methylated CpG number for 1 kb bins in MII oocytes.
The gain in methylation occurred genome-wide, regardless of the chromosomes or genomic features (Supplementary Fig. 7). Notably, the gain of DNA methylation level also positively correlated with the increase of DNMT1 level in genome (Fig. 6f). The correlation between H3K9me3 and DNA methylation has been also reported in early embryonic development66,67. In line with the change in DNMT1 genomic distribution (Fig. 5j), we also noticed that the gain in DNA methylation level positively correlated with the levels of H3K9me3 in MII oocyte (Fig. 6g, h). To further determine the role of DNMT1 in the Pramel15 loss-induced increase in DNA methylation, we examined whether the observed increase contained signatures of DNMT1. We found that genomic regions that have higher CpG di-nucleotide density were preferentially affected by Pramel15 (Fig. 6i). This trend corresponds to the substrate preference of DNMT1, as previous measurement of maintenance methylation kinetics in replicating somatic cells revealed a similar pattern (Fig. 6j)68. Thus, both the initial methylation status, defined as the percentage of methylated CpG sites and the CpG density influence how a genomic region responds to Pramel15 deficiency. Consequently, the number of methylated CpG sites is clearly positively correlated with methylation gain in Pramel15 deficient embryo (Fig. 6k). As DNMT1-mediated maintenance methylation activity follows the same rule68,69, we conclude that the gain in methylation was driven by increased activity of DNMT1. Taken together, these results collectively support the functional role of DNMT1 in mediating the elevation of DNA methylation in embryos derived from Pramel15 KO oocytes.
Discussion
Global loss of DNA methylation is one of the most prominent and evolutionarily conserved epigenetic reprogramming events during mammalian preimplantation embryo development. However, uncovering the intricate molecular mechanisms governing this process has been hindered by challenges stemming from limited research materials and the need for unbiased investigative approaches. In our study, we employed a reverse genetic screen with an oocyte cDNA expression library to identify novel DNA methylation regulators. Through this approach, we discovered a proteasomal-dependent mechanism mediated by maternal factor Pramel15 in mouse early embryos. Pramel15 finely tunes the nuclear abundance of DNMT1 protein, thereby regulating the global demethylation process.
DNMT1 is primarily located in the cytoplasm of mouse oocytes and preimplantation embryos, making it initially unexpected to identify a nuclear regulatory mechanism. However, considering that mouse early embryos express significant levels of both DNMT1 and UHRF170,71,72, the presence of a residual fraction of these factors in the nucleus, if without additional layers of regulation, may lead to dysregulation in the embryo’s methylome. Supporting this notion, while the overall cellular abundance of DNMT1 remains relatively unchanged in response to Pramel15 loss, a slight increase in DNMT1 protein within the nucleus has been associated with higher DNA methylation levels. Therefore, the passive DNA demethylation process in mouse embryos appears to be sensitive to the nuclear level of DNMT1. Our findings suggest that mammalian embryos employ at least two independent strategies to control maintenance activity. These strategies involve the restriction of nuclear accumulation of maintenance factors and fine-tuning their abundance within the nucleus (Fig. 7).
To safeguard DNA methylation reprogramming in zygotes, DNMT1 and UHRF1 are regulated by multiple layers. NLRP14 has recently identified as a factor regulating UHRF1 cytoplasmic retention30,31, and DNMT1 is restricted in the cytoplasm by PADI632. Meanwhile, DNMT1 is actively exported from the nucleus62. Factors involving nucleus exportation and cytoplasmic retention of DNMT1 are still unknown. In nucleus, TET3 is involved in DNA reprogramming by actively demethylating some regions20. On the other hand, DNA methylation is retained at some regions, such as imprinted regions, during the reprogramming, which relies on DNMT1/UHRF1 in the nucleus28,35,36,37. To prevent excess DNMT1 from impairing DNA demethylation, Pramel15 target DNMT1 degradation via the ubiquitin-proteasome system (UPS) by Cullin-RING E3 ligases recruitment.
Although global loss of DNA methylation is a conserved feature during mammalian embryo development, there is a lack of evidence suggesting that DNMT1 and UHRF1 exclusively accumulate in the embryonic cytoplasm of other mammalian species, including humans73,74,75. We hypothesize that the nuclear regulation of maintenance factors may play a more important role in other mammalian species. Additionally, the intricate management of nuclear maintenance factor levels in mammalian embryos may determine inter-species variation in the kinetics and extent of global demethylation7,8.
The Pramel15-deficient embryos exhibited a slight yet significant gain of DNA methylation. We interpret this finding as each blastomere within the embryo randomly acquiring additional methylation at different CpG sites. This interpretation is supported by our observation that statistical changes in different genomic features largely recapitulated the global trend, along with the binary nature of CpG methylation status in each allele. Therefore, there is likely significant inter-blastomere variation in the impact of methylation gain within individual cleavage-stage embryos. Since not every blastomere is indispensable, a low-frequency random increase in methylation within individual blastomeres may not severely impede embryonic development. In line with our findings and previous analyses of Tet3 knockout embryos18,19,20, we propose that mouse embryos can tolerate a certain degree of inefficient demethylation. To gain a comprehensive understanding of how global DNA reprogramming regulates embryo development, further studies are warranted to decipher the underlying intricate regulatory mechanisms.
We have identified Pramel15 as a substrate-recognition module for the Cullin-family E3 ubiquitin ligase. Pramel15 belongs to the PRAME (Preferentially expressed antigen of melanoma) family, which is highly amplified in eutherian mammals76. More than thirty PRAME family members are specifically expressed during gametogenesis and development77. However, only a few proteins have been investigated to date, including Pramel778, Gm12794c (also called Pramel19)79, and Pramef12 (also called Pramel13)80,81,82. Notably, Prame and Pramel7 have also been characterized as substrate-recognition receptors of CRL2, which degrade p14/ARF and UHRF1, respectively78,83. Therefore, it is appealing to hypothesize that other PRAME family member(s) may also involve CRL complexes to function, and embryonic DNMT1 could be targeted by Pramel15 paralogs. The duplication of functionally redundant genes critical for early embryo development appears to be a common phenomenon, as exemplified by zygotic genome activation regulators such as the Dux and Obox family proteins84,85. Further studies are required to identify the protein targets for each Prame paralog and determine their functional redundancy during zygotic methylome reprogramming.
In conclusion, our discoveries elucidate a layer of regulation for DNA methylation reprogramming. While additional studies will be necessary to determine the significance of Pramel15 and address the issue of functional redundancy with other known and unknown regulatory mechanisms, our study sheds a light on our understanding of the regulation of global DNA methylation reprogramming that occurs at the beginning of life.
Methods
All experiments in this article comply with all relevant ethical regulations and are approved by the Institutional Committee of the Institute of Biophysics, Chinese Academy of Sciences. The animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the Institute of Biophysics, Chinese Academy of Sciences.
Cell lines and cell culture
HEK293-derived cell lines were cultured in DMEM (Life Technology) supplemented with 10% FBS (Hyclone) and 1× penicillin-streptomycin solution (BBI Life Sciences). TT2 mouse embryonic stem cells86-derived cell lines were cultured in DMEM (Gibco) with 10% KnockOut Serum Replacement (Life Technologies), 1% FBS (Hyclone), 1% non-essential amino acids (Sigma), 2 mM l-glutamine (BBI Life Sciences), 1 mM sodium pyruvate (Life Technologies), 0.1 mM β-mercaptoethanol, 1000 U/ml mLIF (Novoprotein) and 1% Penicillin/Streptomycin (BBI Life Sciences). Mouse ES cells used 0.1% Gelatin pre-coated the culture dishes. All cells were grown at 37 °C and 5% CO2.
Constitutive and inducible expression cell lines based on HEK293 cells were established using a lentiviral system87. Lentiviral-expressing vectors, together with packaging vectors pMDL, pRev, and pVSVG (Invitrogen), were transfected into 293FT cells using VigoFect (Vigorous). Virus-containing supernatant was collected, filtered, and used to incubate T-Rex-293 cells for doxycycline-inducible Pramel15, or HEK293 cells for constitutive expression of GFP-DNMT1. Stable transgenic cell lines were selected with puromycin, or fluorescence-activated cell sorting (FACS). Cell lines were further validated by Western blotting.
Constitutive and inducible expression cell lines based on TT2 mouse ES cells were established using a piggyBac (PB) transposon system88. For the inducible Pramel15 cell line, a Tet Response Element-driven Pramel15 and an rtTA gene, linked with a blasticidin-resistant gene using a P2A peptide, driven by a constitutive promoter, were inserted between PB elements. The vectors, along with plasmids expressing PB transposase (PBase), were transfected into TT2 cells using VigoFect. For the constitutive expression of Pramel15, a CAG-promoter-driven Pramel15 was inserted between PB elements. Polyclonal cells with stably integrated transgenes were selected through blasticidin treatment and validated by Western blotting.
Loci-specific DNA methylation analysis
For loci-specific DNA methylation analysis in cultured cells, purified genomic DNA was bisulfite-converted using EZ DNA Methylation-Lightning Kits (Zymo Research) following the manufacturer’s instructions. The loci of interest were PCR-amplified with TaKaRa Ex Taq (TaKaRa), and the PCR products were cloned into T-vector using pEASY-T5 Zero Cloning Kit (TransGen Biotech). Sanger sequencing results were analyzed using BiQ Analyzer89. Bisulfite sequencing analysis utilized the following primers: CMV-BS-F1, GGGTTATTAGTTTATAGTTTATATATGGA; CMV-BS-R1, ACCAAAATAAACACCACCCC.
UHPLC–MS/MS for 5mC and 5hmC
Genomic DNA was purified using Wizard Genomic DNA Purification Kit (Promega). The DNA was incubated with DNase I, calf intestinal phosphatase, and snake venom phosphodiesterase I at 37 °C for 24 h, and the resulting digest was filtered. A UHPLC-MS/MS method described previously was employed to measure 5mC and 5hmC90,91, using an Agilent 1290 Rapid Resolution LC system and a reverse-phase Zorbax SB-C18 2.1 × 100 mm column (1.8 μm particles). The digested DNA (5.0–10.0 μl) was injected onto the column, and nucleoside separation was achieved with a mobile phase consisting of 95% water (with 0.1% formic acid) and 5.0% methanol at a flow rate of 0.3 ml/min using isocratic elution. To quantify 5mC and 5hmC, the stable isotope internal standard was used. At least 2 biological replicates were performed.
Generation of Cul5 knockout (KO) cell lines
To generate Cul5-KO cell lines in TT2 mouse ES cells, we used a homology-independent intron targeting method based on CRISPR/Cas9 system with a minor modification92. Briefly, we constructed a donor plasmid by inserting a fragment containing a splice acceptor (SA) site, P2A linker, BleoR gene, and polyA terminator into the pUC19 vector (Supplementary Fig. 3c). The insertion sequence was flanked by two identical sequences that could be targeted by a single-guide RNA (sgRNA) (GTCGTATTCATCGTGGCCAT). We co-transfected a SpCas9 plasmid, a sgRNA targeting the first intron of Cullin5 (CCGGGAGCCGAGCGCATCGA), a sgRNA targeting the flanking sequence, and the donor plasmid into TT2 mouse ES cells. After 3 days, the population was screened with zeocin before single cells were sorted into 96-well plates by flow cytometry. The clonal cell lines were evaluated using genomic PCR, reverse transcriptional-quantitative PCR (RT-qPCR), and Western blotting to confirm Cul5 knockout.
Antibodies
Anti-Flag antibody (Abclonal, AE005), anti-HA antibody (CST, 3724), anti-Lamin B1 antibody (Protech, 12987-1-AP), anti-Cul5 antibody (Santa Cruz, sc-373822), anti-Rbx1 antibody (Santa Cruz, sc-393640), anti-H3 antibody (Abmart, MB9211), anti-GAPDH (Abclonal, AC002). Home-made anti-DNMT1 antibody(N118) and anti-DNMT1 antibody(M377) were described previously13.
Immunoprecipitation (IP)
Harvested cells were washed twice with cold PBS. When starting with cells that reached near confluency in a 3.5 cm cell culture dish, the pelleted cells were resuspended in 110 μl of IP buffer (20 mM HEPES-NaOH pH 7.9, 150 mM NaCl, 5 mM EDTA, and 5% glycerol) supplied with 1% NP-40 and 1x protease inhibitor cocktail. For IP experiments involving DNMT1 or UHRF1, the lysis buffer was supplemented with Cryonase Cold-Active Nuclease (Takara). Following 30 min incubation on ice, the lysates were cleared by centrifugation at 13,000 × g for 30 min at 4 °C. A portion of the supernatants were saved as input, while the remaining lysates were diluted to ten-fold volume using IP buffer. The lysates were then incubated with 10 μl of pre-balanced affinity agarose beads (anti-HA or anti-FLAG beads as indicated in figures) overnight at 4 °C. Subsequently, the beads were washed five times with Wash Buffer (20 mM HEPES-NaOH pH 7.9, 0.1% Triton X-100, 5 mM EDTA, and 150 mM or 500 mM NaCl as indicated in figures). After careful removal of the Wash Buffer, the beads were boiled with 1x Laemmli sample buffer for further analysis by Western blotting. For silver staining or label-free quantitative mass spectrum, elution was carried out by incubating the beads with 500 μg/ml FLAG peptide solution.
Western blotting
Harvested cells were lysed and boiled in WCL buffer (10 mM HEPES-NaOH pH 7.9, 2.5 mM EDTA, 1% SDS) for 30 min. After centrifugation, the supernatants containing the whole cell lysate were collected. The concentration of the lysate was determined with Detergent compatible Bradford kit (Beyotime). The proteins were then separated by electrophoresis on SDS polyacrylamide gradient gels and electroblotted onto PVDF membranes. Blocking was performed with 5% (w/v) fat-free milk in TBST (10 mM Tris-HCl pH 7.5, 500 mM NaCl, 0.1% Tween-20). The primary antibody was diluted in the blocking solution and incubation was performed overnight at 4 °C. The membranes were washed three times with TBST for 10 min each, followed by incubation with HRP-coupled secondary antibodies for 1 h at room temperature. After three washes with TBST, the membranes were developed using Immobilon Western HRP substrate (Millipore). The signal was captured by X-ray films. All Western blot results were repeated at least three times independently.
In vivo ubiquitin assay
HEK293 cells were transfected with plasmids expressing His-Ub and Flag-DNMT1(1-609aa), with or without Pramel15-HA as indicated. 40 h after transfection, MG132 (20 μM) was added into the culture medium. Cells were harvested after 8 h of MG132 treatment. The cells were washed twice with cold PBS and then lysed in denaturing lysis buffer (20 mM Tris-HCl 8.0, 150 mM NaCl, 5 mM EDTA, 0.1% Triton-X 100, 10% glycerol, and 1% SDS) by boiling at 100 °C for 30 min. Following centrifugation at 13,000 × g for 20 min, the supernatant was diluted with 10-fold volume of SDS-free denaturing lysis buffer. Immunoprecipitation was performed with anti-Flag M2-affinity Beads (Sigma) at 4 °C overnight. After extensive wash with Wash Buffer (20 mM Tris-HCl 8.0, 150 mM NaCl, 5 mM EDTA, 0.1% Triton-X 100), the immunoprecipitated proteins were eluted with 0.1 M glycine.
LC-MS/MS analysis for protein
After silver staining or Coomassie brilliant blue G-250 staining, the proteins were excised from the gel and subjected to in-gel digestion. The peptides were analyzed by nanoLC-MS/MS experiments on a Q Exactive (Thermo Scientific) equipped with an Easy n-LC 1000 HPLC system (Thermo Scientific).
The peptides were loaded onto a 100 μm id × 2 cm fused silica trap column packed in-house with reversed phase silica (Reprosil-Pur C18 AQ, 5 μm, Dr. Maisch GmbH) and then separated on a 75 μm id × 20 cm C18 column packed with reversed phase silica (Reprosil-Pur C18 AQ, 3 μm, Dr. Maisch GmbH). The peptides bound on the column were eluted with a 78 min linear gradient. The solvent A consisted of 0.1% formic acid in water solution and the solvent B consisted of 0.1% formic acid in acetonitrile solution. The segmented gradient was 4–8% B, 8 min; 8–22% B, 50 min; 22–32% B, 12 min; 32–90% B, 1 min; 90% B, 7 min at a flow rate of 300 nl/min.
The MS analysis was performed with Q Exactive mass spectrometer (Thermo Scientific). With the data-dependent acquisition mode, the MS data were acquired at a high resolution 70,000 (m/z 200) across the mass range of 300–1600 m/z. The target value was 3.00E + 06 with a maximum injection time of 60 ms. The top 20 precursor ions were selected from each MS full scan with isolation width of 2 m/z for fragmentation in the HCD collision cell with normalized collision energy of 27%. Subsequently, MS/MS spectra were acquired at resolution 17,500 at m/z 200. The target value was 5.00E + 04 with a maximum injection time of 80 ms. The dynamic exclusion time was 40 s. For nanoelectrospray ion source setting, the spray voltage was 2.0 kV; the heated capillary temperature was 320 °C.
The raw data from Q Exactive were analyzed with Proteome Discovery version 2.2.0.388 using Sequest HT search engine for protein identification and Percolator for FDR (false discovery rate) analysis.
Two biological replicates, containing one or two technical replicates, were performed.
RNA isolation and quantitative real-time PCR
Total RNA from cell lines and tissues was extracted using the Total RNA Extraction kit (Genstone Biotech) following the manufacturer’s instructions. Reverse transcription was performed using HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme). Real-time PCRs were performed on a 7500 Fast Real-Time PCR system (Thermo Fisher Scientific), using the Taq Pro Universal SYBR qPCR Master Mix (Vazyme). Primers of qPCR: GGAAGGTGAAGGTCGGAGT and GACAAGCTTCCCGTTCTCAG for Gapdh; CGCCAGGAATCTGTAACAAAACA and CAGTACACGAGCCTGTGCTT for Cullin5.
RNAi
siRNAs were synthesized by the Biological Resource Center of the National Institute of Biological Sciences, Beijing. To knockdown Rbx1, siRNAs (5′-UCCAUAAUGUGGUUCCUGCAGAUGG-3′) were transfected with RNAiMAX (Invitrogen) at a final concentration of 10 nM following the manufacturer’s instructions. The cells were harvested 72 h post-transfection for following experiments.
Mice
All animal experiments were performed in accordance with the institutional guides for the care and use of laboratory animals and were reviewed and approved by the institutional biomedical research ethics committee of the Institute of Biophysics of the Chinese Academy of Sciences. All mice used in this research were euthanized by CO2 asphyxiation.
To generate Pramel15 knockout mice, mRNA encoding CRISPR-Cas9 and gRNA targeting the first exon of Pramel15 (5′-CTGTTGAAGCAAGCCTACGA-3′), were injected into the zygotes of C57BL/6J mouse. At the F1 generation, mutant mouse strains carried premature stop codons were selected for further experiments. Genotyping was performed by using specific PCR primers: 5′-GATGGTTACCATCGTAGGC-3′ and 5′-ACCTGGAGTGGAATGAGCAG-3′ for distinguishing WT and KO alleles, and 5′-CTCCACCACATCCTTGTACCC-3′ and 5′-CAAGCCCTTACTCACACTCCAA-3′ for distinguishing wild-type, heterozygotes and homozygotes. For SNP analysis, sperms of DBA/2J male mice were used for in vitro fertilization.
WT and Ptamel15-KO mice were bred separately in the same facility in standard cages in a specific pathogen-free facility, maintained on a 12 h light/dark cycle, and given unrestricted access to food and water.
Oocyte collection
GV oocytes were collected from the ovaries of superovulated 3- to 8-week-old female mice. The mice were sacrificed 48 h after intraperitoneal injection with 5 IU of pregnant mare serum gonadotropin (PMSG) (San-Sheng Pharmaceutical). The ovaries were isolated and transferred to M2 medium containing 0.2 mM IBMX (Sigma). Inflated follicles were punctured with a 27-gauge needle to release the cumulus-oocyte complexes (COCs). The adhering cumulus cells were removed by gentle pipetting.
To collect MII oocytes, female mice (3- to 10-week-old) were superovulated by injecting 5 IU of PMSG, followed by injection of 5 IU human chorionic gonadotropin (hCG) (San-Sheng Pharmaceutical) 48 h later. MII oocytes were released from the cumulus cells by treating them with 0.1% hyaluronidase (Sigma) in M2 medium. Cumulus cells adhered to the zona pellucida were removed by gently pipetting.
In vitro fertilization, parthenogenesis, and development
The procedure of in vitro fertilization was carried out as previously described93. Briefly, the sperm was isolated from dissected epididymis and capacitated for 1 h in YTH medium supplemented with 0.75 mM methyl-β-cyclodextrin (MBCD). COCs were collected and incubated with capacitated sperm for 3–4 h, before being washed and transferred to HTF medium. Zygotes with two pronuclei were selected at 12 to 14 h post insemination (hpi) and cultured in KSOMAA medium (Millipore). Embryos at different development stages were collected at specific time points: early 2-cell embryos at 19–20 hpi, late 2-cell embryos at 34–36 hpi, 4-cell embryos at 42–44 hpi, 8-cell embryos at 56–58 hpi, early blastocysts at 74–76 hpi, and late blastocysts at 88–90 hpi.
Parthenogenesis was carried out as described previously94. Briefly, MII oocytes were collected 14 h post-hCG injection. Parthenogenesis activation was initiated in Ca2+-free M16 medium supplied with 10 mM SrCl2. Oocytes were transferred to HTF medium at 2.5–3 h post activation (hpa). At 12 hpa, oocytes with one pronucleus were collected as pseudo-zygotes.
Immunofluorescence
For immunofluorescence in cultured cells, cells were cultured on poly-D-lysine coated coverslips. Following washing twice with PBS, cells were fixed with 4% PFA for 15 min at room temperature. After three washes with PBS containing 0.1% Triton X-100 and 0.1 M glycine, cells were permeabilized with PBS containing 0.5% Triton X-100 for 15 min at room temperature. Following incubation in Blocking Buffer (PBS containing 3% BSA and 0.1% Triton X-100) for 1 h at room temperature, the samples were incubated with primary antibodies diluted in Blocking Buffer at room temperature for 2 h or at 4 °C overnight. After extensive wash with PBS containing 0.1% Triton X-100 (PBST) for 3 times, the samples were incubated with fluorophore-conjugated secondary antibodies diluted in Blocking Buffer for 1 h at room temperature. Cells were then washed three times with PBST, and stained with DAPI (Sigma).
For immunofluorescence in oocytes and whole embryos, the procedure was slightly modified: firstly, staining was performed in a U-bottom 96-well plate and the samples were transferred by mouth pipetting. Secondly, the blocking, primary antibody, and secondary antibody incubation steps were all performed at 4 °C overnight to ensure efficient antibody penetration and buffer exchange.
In vitro transcription
For microinjection, Pramel15-mCherry mRNA was produced using mMESSAGE mMACHINE™ T7 Transcription Kit (Invitrogen). The template DNA was prepared by PCR with the primer pairs containing T7 promoter at 5′ end (5′-TAATACGACTCACTATAGGGAGAATGAGCAGCAAGCCCTTACT-3′, 5′-TCACTTGTACAGCTCGTCCATGC-3′). Following instructions of the kit to perform in vitro transcription, the mRNA products were recovered by lithium chloride precipitation. The RNA pellet was resuspended in RNase-free H2O and determined concentration for further application or store at −80 °C.
Image acquisition and quantification
The fluorescent images were captured with Opera Phenix high throughput and high content imaging analysis system (PerkinElmer Life Sciences), and the raw data was processed and quantified with Harmony High-Content Imaging and Analysis Software. In some cases, fluorescent images were acquired with an LSM700 confocal microscope (Carl Zeiss), and the raw images were processed with ZEN software. Bright-field images of embryos were captured using an IX73 microscope (Olympus). The processed data were analyzed in GraphPad Prism 10 Software.
For confocal image acquisition of oocyte and embryo samples, the mid-section of a nucleus was manually determined based on maximal area. For oocytes stained with anti-DNMT1, the Z layer that was both closed to mid-section and contained DAPI-dense pericentric heterochromatin was chosen.
To quantify signal intensity in culture cells and embryos, multiple layers of images were captured with the Opera Phenix high throughput and high connotation imaging analysis system. To quantify the intensity of DNMT1 in nuclei of embryos, the images were captured at 1 μm intervals to reconstruct the 3D model of the whole embryos. The nuclear regions were defined by H3 immunofluorescent signal because DAPI or Hoechst tends to condense on heterochromatin, which makes it hard to define the entire nucleus accurately.
CUT&RUN
CUT&RUN of DNMT1 in parthenogenetic 1-cells was performed according to the protocol previously described95,96 with some modifications. Briefly, cumulus cell-free MII oocytes were activated in Ca2+-free M16 medium containing 10 mM SrCl2 and 5 μg/ml cytochalasin B. The parthenogenetic 1-cells were moved to KSOM medium 8 h post activation. The zona pellucida (ZP) was remove by 0.1% pronase E (Sigma, 107433). The ZP-free cells were incubated with Ab buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 2 mM EDTA, 0.02% Digitonin, and 1x protease inhibitor cocktail) containing DNMT1 antibody (M377) at 4 °C overnight. Following washing with Dig-wash buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 0.02% Digitonin, and 1x protease inhibitor cocktail) twice, the cells were incubated with Dig-wash buffer containing 500 ng/μl of pA-MNase (Vazyme, S701-01) at 4 °C for 3 h. Following washing with Dig-wash buffer twice, the 1-cells were moved into 1.5 ml tubes and incubated with activated Con-A beads at room temperature for 10 min. Removing the supernatant on magnetic stand, 21 μl of cold Reaction buffer (Dig-wash buffer with 2 mM CaCl2) was added and incubated on ice for 30 min. Then, 7 μl of cold 4x STOP buffer (80 mM EGTA, 0.02% Digitonin, 100 μg/ml RNase A) was added and mixed. The tubes were incubated at 37 °C for 30 min and then placed on magnetic stand. The supernatant was transferred into a 0.2 mL MAXYMum Recovery Thin Wall PCR tube (Axygen) and performed library construction by VAHTS Universal Pro DNA Library Prep Kit for Illumina (Vazyme, ND608) following the manufacturer’s instructions.
Whole-genome DNA methylation analysis
For whole-genome DNA methylation analysis in oocytes or embryos, PBAT libraries were generated according to the protocol previously described65 with some modifications. Briefly, 20–50 of oocytes or embryonic cells were collected into a 0.2 mL MAXYMum Recovery Thin Wall PCR tube. Cell was lysed in 20 μl of lysis buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.3% Triton X-100, 1 mg/ml Qiagen Protease), and incubated at 50 °C for 60 min, followed by 75 °C for 15 min. To evaluate the bisulfite conversion rate, 0.5 μg of unmethylated λDNA was also added to the lysate. Bisulfite conversion was carried out using EZ DNA Methylation-Lightning Kits. The converted DNA was eluted with 20 μl of ddH2O into a 1.5 ml DNA LoBind Tube (Eppendorf). A mixture containing 2.5 μl of 10x BLUE buffer, 1 μl of 10 mM dNTPs, and 1 μl of 10 μM Biotin-PBAT-F (5′-biotin-ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNN-3′) was added to the eluted DNA. The mixture was incubated at 75 °C for precisely 3 min and then cooled ice immediately. 1 μl of Klenow 3′−5′ exo- Fragments (50 U/μl, Tiangen) was added to the mixture on ice, and biotin-tagged strands were synthesized by following process: Incubate at 4 °C for 5 min then raise to 20 °C at a rate of 1 °C every 15 s, incubate at 20 °C for 5 min, then raise to 37 °C at a rate of 1 °C every 15 s, and incubate at 37 °C for 60 min. Three cycles of synthesis by replenishing the reaction components were recommended to improve coverage. Excess primers were digested by exonuclease I (NEB) and the DNA was purified using 0.8x VAHTS DNA Clean Beads (Vazyme). Biotin-tagged DNA strands were captured by Dynabeads M280 (Invitrogen, streptavidin-coupled). The Dynabeads were then washed with 0.1 M NaOH and 10 mM Tris-HCl pH 8.0 twice. Second DNA strands were synthesized directly on M280 beads using Klenow fragments with random primers (5′-AGACGTGTGCTCTTCCGATCTNNNN-3′). The beads were washed with 10 mM Tris-HCl pH 8.0 and directly subjected to PCR amplification. Final library was amplified with 2x KAPA HiFi HotStart ReadyMix for 15 PCR cycles. PCR products were purified with 0.8x VAHTS DNA Clean Beads. PBAT libraries were sequenced using Illumina NovaSeq 6000 platform.
Data processing
Each WGBS library was sequenced in 100-bp paired-end reads, with average data amount of 48G bp per sample. All samples were prepared with 2 or 3 biological replicates (Supplementary Table 1). Sequencing quality of raw reads was first assessed by FastQC (v0.12.1) and then filtered low-quality and adapter sequences with Trim_galore (v0.6.10) (trimming parameters: –clip_R2 3 –three_prime_clip_R1 4 –three_prime_clip_R2 4). Trimmed read pairs were aligned to GRCm38/mm10 assembly of the mouse genome using BitmapperBS (v1.0.2.3). Mapped reads were then deduplicated and sorted with Sambamba (1.0.0). Methylation information was then extracted from deduplicated reads using MethylDackel (v0.6.1). For allelic assignment of sequencing reads to their parental origin, SNPsplit (v0.6.0) was used based on the SNP information of DBA/2J (Supplementary Tables 2, 3). After consistency check among replicates, methylation information of replicates was merged for higher depth and clarity (C/T counts of each CpG site were added up).
For genomic segment analysis, the genome is divided into continuous intervals of 1 kb or 1 Mb. It is required that at least one CpG site is covered every 200 bp average. The methylation level of each bin is calculated as the ratio of C/(C + T) for all CpG sites within the interval. When comparing KO and Het samples, bins that are covered by both samples are selected for comparison. The total methylation level of each interval in MII oocytes is the sum of methylation levels for all CpG sites within the interval. The reads number of H3K9me3 within each interval are sorted from low to high, and then evenly divided into the desired number of groups according to their ranks. In the analysis related to the number of CpG sites, a limit is set so that the number of CpGs in a 1-kb interval does not exceed 30 (which includes over 90% of the sites), to avoid regions with low methylation in CpG islands. The above analysis was performed with custom R scripts, which are available upon request.
For the CUT&RUN data analysis, each library was sequenced with 150-base pair paired-end reads. The initial quality of these raw reads was evaluated using FastQC (version 0.12.1). After assessment, low-quality sequences and adapter sequences were filtered out using Trim Galore (version 0.6.10). Following trimming, the paired reads were aligned to the mouse reference genome (mm10) using Bowtie2 with its default parameters. Duplicate reads were identified and removed to ensure data accuracy. The mouse genome was segmented into 1-megabase (1-Mb) bins, and the reads aligning to each bin were counted. These counts were then normalized to a total sequencing depth of 1 million read pairs to standardize the signal across samples. The normalized counts represent the DNMT1 signal in each corresponding genomic region.
Statistics & reproducibility
The WGBS and CUT&RUN data were analyzed in R v.3.4.0. The other statistical analyses were performed with GraphPad Prism 10. No statistical method was used to predetermine sample size. At least two biological replicates were used in each experiment unless otherwise stated. In bar plot, data are presented as mean ± s.d. In box plots, the center line represents the median, the box limits show the upper and lower quartiles and the whiskers represent 1.5x the interquartile range. Unpaired two-sided t-tests or unpaired two-sided Wilcoxon tests were used to calculate the P values indicated in the figure legends, for assessment of the statistical significance of differences between groups. P values ≤ 0.05 were regarded as statistical significance. When analyzing DNMT1 levels in the nucleus by immunofluorescence, if the nuclear volume defined by the H3 signal was too large or too small compared to the population, the sample was omitted from subsequent analyses to avoid the effect of abnormal values (Fig. 5d–h, Supplementary Fig. 5c). The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequence data of WGBS and CUT&RUN generated in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences under GSA accession code CRA013324. The H3K9me3 ChIP-seq data was adopted from NCBI GEO database under the accession of GSM258856067. The Hammer-seq data was adopted from NCBI GEO database under the accession of GSE13109868. The mass spectrometry proteomics data generated in this study have been deposited in the ProteomeXchange Consortium via the iProX partner repository97,98 with the dataset identifier PXD052998. Source data are provided with this paper.
Code availability
Custom codes used for data analysis in this paper can be found on Figshare [https://doi.org/10.6084/m9.figshare.26493205].
References
Eckersley-Maslin, M. A., Alda-Catalinas, C. & Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018).
Xia, W. & Xie, W. Rebooting the epigenomes during mammalian early embryogenesis. Stem Cell Rep. 15, 1158–1175 (2020).
Ladstätter, S. & Tachibana, K. Genomic insights into chromatin reprogramming to totipotency in embryos. J. Cell Biol. 218, 70–82 (2019).
Guo, H. et al. The DNA methylation landscape of human early embryos. Nature 511, 606–610 (2014).
Smith, Z. D. et al. DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611–615 (2014).
Smith, Z. D. et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344 (2012).
Li, C. et al. DNA methylation reprogramming of functional elements during mammalian embryonic development. Cell Discov. 4, 41 (2018).
Ivanova, E. et al. DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin. Epigenetics 12, 64 (2020).
Zhang, J. et al. TET3 overexpression facilitates DNA reprogramming and early development of bovine SCNT embryos. Reproduction 160, 379–391 (2020).
Gao, R. et al. Inhibition of aberrant DNA re-methylation improves post-implantation development of somatic cell nuclear transfer embryos. Cell Stem Cell 23, 426–35.e5 (2018).
Peat, J. R. & Reik, W. Incomplete methylation reprogramming in SCNT embryos. Nat. Genet. 44, 965–966 (2012).
Biniszkiewicz, D. et al. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 22, 2124–2135 (2002).
Li, Y. et al. Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature 564, 136–140 (2018).
Messerschmidt, D. M., Knowles, B. B. & Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 28, 812–828 (2014).
Chen, Z. & Zhang, Y. Role of mammalian DNA methyltransferases in development. Annu. Rev. Biochem. 89, 135–158 (2020).
Wossidlo, M. et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2, 241 (2011).
Gu, T.-P. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011).
Guo, F. et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–459 (2014).
Shen, L. et al. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15, 459–471 (2014).
Peat, J. R. et al. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Rep. 9, 1990–2000 (2014).
Amouroux, R. et al. DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 18, 225–233 (2016).
Bestor, T. et al. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. J. Mol. Biol. 203, 971–983 (1988).
Bostick, M. et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 (2007).
Sharif, J. et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 (2007).
Ming, X., Zhu, B. & Li, Y. Mitotic inheritance of DNA methylation: more than just copy and paste. J. Genet. Genomics 48, 1–13 (2021).
Du, W. et al. Mechanisms of chromatin-based epigenetic inheritance. Sci. China Life Sci. 65, 2162–2190 (2022).
Carlson, L. L., Page, A. W. & Bestor, T. H. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 6, 2536–2541 (1992).
Cohen, P. E. et al. Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLOS Genet. 13, e1007042 (2017).
Zeng, Y. & Chen, T. DNA methylation reprogramming during mammalian development. Genes 10, 257 (2019).
Yan, R. et al. Dynamics of DNA hydroxymethylation and methylation during mouse embryonic and germline development. Nat. Genet. 55, 130–143 (2022).
Meng, T. G. et al. NLRP14 safeguards calcium homeostasis via regulating the K27 ubiquitination of Nclx in oocyte‐to‐embryo transition. Adv. Sci. 10, e2301940 (2023).
Giaccari, C. et al. A maternal-effect Padi6 variant causes nuclear and cytoplasmic abnormalities in oocytes, as well as failure of epigenetic reprogramming and zygotic genome activation in embryos. Genes Dev. 38, 131–150 (2024).
Uemura, S. et al. UHRF1 is essential for proper cytoplasmic architecture and function of mouse oocytes and derived embryos. Life Sci. Alliance 6, e202301904 (2023).
Kelsey, G. & Feil, R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. B: Biol. Sci. 368, 20110336 (2013).
Kurihara, Y. et al. Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Dev. Biol. 313, 335–346 (2008).
Hirasawa, R. et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 22, 1607–1616 (2008).
Cirio, M. C. et al. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev. Biol. 8, 9 (2008).
Min, B. et al. Dnmt1 binds and represses genomic retroelements via DNA methylation in mouse early embryos. Nucleic Acids Res. 48, 8431–8444 (2020).
Haggerty, C. et al. Dnmt1 has de novo activity targeted to transposable elements. Nat. Struct. Mol. Biol. 28, 594–603 (2021).
Alexander, K. A. et al. TRIM28 controls genomic imprinting through distinct mechanisms during and after early genome-wide reprogramming. Cell Rep. 13, 1194–1205 (2015).
Lorthongpanich, C. et al. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 341, 1110–1112 (2013).
Takahashi, N. et al. ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 33, 49–54 (2019).
Li, X. et al. Small molecules capable of activating DNA methylation–repressed genes targeted by the p38 mitogen-activated protein kinase pathway. J. Biol. Chem. 293, 7423–7436 (2018).
Du, W. et al. Stella protein facilitates DNA demethylation by disrupting the chromatin association of the RING finger–type E3 ubiquitin ligase UHRF1. J. Biol. Chem. 294, 8907–8917 (2019).
Takeshita, K. et al. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc. Natl Acad. Sci. 108, 9055–9059 (2011).
Song, J. et al. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331, 1036–1040 (2011).
Kamura, T. et al. MUF1, a novel elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276, 29748–29753 (2001).
Qin, W., Leonhardt, H. & Spada, F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J. Cell. Biochem. 112, 439–444 (2011).
Felle, M. et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 39, 8355–8365 (2011).
Chen, S. H. et al. CRL4 AMBRA1 targets Elongin C for ubiquitination and degradation to modulate CRL5 signaling. EMBO J. 37, e97508 (2018).
Kostrhon, S. et al. CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation. Nat. Chem. Biol. 17, 1075–1083 (2021).
Hüttenhain, R. et al. ARIH2 Is a Vif-Dependent Regulator of CUL5-Mediated APOBEC3G Degradation in HIV Infection. Cell Host Microbe 26, 86–99.e7 (2019).
Nishiyama, A. et al. Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation. Nat. Commun. 11, 1222 (2020).
Schwechheimer, C. NEDD8—its role in the regulation of Cullin-RING ligases. Curr. Opin. Plant Biol. 45, 112–119 (2018).
Soucy, T. A. et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009).
Skaar, J. R. et al. PARC and CUL7 form atypical cullin ring ligase complexes. Cancer Res. 67, 2006–2014 (2007).
Bosu, D. R. & Kipreos, E. T. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 3, 7 (2008).
Lydeard, J. R., Schulman, B. A. & Harper, J. W. Building and remodelling Cullin–RING E3 ubiquitin ligases. EMBO Rep. 14, 1050–1061 (2013).
Mahrour, N. et al. Characterization of cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 Modules to elongin BC-based ubiquitin ligases. J. Biol. Chem. 283, 8005–8013 (2008).
Kobe, B. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732 (2001).
Xiong, Z. et al. Ultrasensitive Ribo-seq reveals translational landscapes during mammalian oocyte-to-embryo transition and pre-implantation development. Nat. Cell Biol. 24, 968–980 (2022).
Cardoso, M. C. & Leonhardt, H. DNA methyltransferase is actively retained in the cytoplasm during early development. J. Cell Biol. 147, 25–32 (1999).
Liu, X. et al. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 4, 1563 (2013).
Ren, W. et al. Direct readout of heterochromatic H3K9me3 regulates DNMT1-mediated maintenance DNA methylation. Proc. Natl Acad. Sci. 117, 18439–18447 (2020).
Miura, F. et al. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 (2012).
Yang, H. et al. Allele-specific H3K9me3 and DNA methylation co-marked CpG-rich regions serve as potential imprinting control regions in pre-implantation embryo. Nat. Cell Biol. 24, 783–792 (2022).
Wang, C. et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20, 620–631 (2018).
Ming, X. et al. Kinetics and mechanisms of mitotic inheritance of DNA methylation and their roles in aging-associated methylome deterioration. Cell Res. 30, 980–996 (2020).
Wang, Q. et al. Imprecise DNMT1 activity coupled with neighbor-guided correction enables robust yet flexible epigenetic inheritance. Nat. Genet. 52, 828–839 (2020).
Zou, Z. N. et al. Translatome and transcriptome co-profiling reveals a role of TPRXs in human zygotic genome activation. Science 378, abo7923 (2022).
Zhang, H. M. et al. Stable maternal proteins underlie distinct transcriptome, translatome, and proteome reprogramming during mouse oocyte-to-embryo transition. Genome Biol. 24, 166 (2023).
Yan, R. et al. Decoding dynamic epigenetic landscapes in human oocytes using single-cell multi-omics sequencing. Cell Stem Cell 28, 1641–56.e7 (2021).
Lodde, V. et al. Localization of DNA methyltransferase-1 during oocyte differentiation, in vitro maturation and early embryonic development in cow. Eur. J. Histochem. 53, e24 (2009).
Shen, W. et al. A novel role for DNA methyltransferase 1 in regulating oocyte cytoplasmic maturation in pigs. Plos One 10, e0127512 (2015).
Virant-Klun, I. et al. Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol. Cell. Proteom. 15, 2616–2627 (2016).
Liberles, D. et al. The expansion of the PRAME gene family in eutheria. PLoS ONE 6, e16867 (2011).
Kern, C. H., Yang, M. & Liu, W.-S. The PRAME family of cancer testis antigens is essential for germline development and gametogenesis. Biol. Reprod. 105, 290–304 (2021).
Graf, U. et al. Pramel7 mediates ground-state pluripotency through proteasomal–epigenetic combined pathways. Nat. Cell Biol. 19, 763–773 (2017).
Napolitano, G. et al. A novel member of Prame family, Gm12794c, counteracts retinoic acid differentiation through the methyltransferase activity of PRC2. Cell Death Differ. 27, 345–362 (2019).
Li, N. et al. MAGE-B4, a binding partner of PRAMEF12, is dispensable for spermatogenesis and male fertility in mice. Biochem. Biophys. Res. Commun. 675, 46–53 (2023).
Haraguchi, D. & Nakamura, T. Pramef12 enhances reprogramming into naïve iPS cells. Biochem. Biophys. Rep. 30, 101267 (2022).
Wang, Z. et al. Sertoli cell-only phenotype and scRNA-seq define PRAMEF12 as a factor essential for spermatogenesis in mice. Nat. Commun. 10, 5196 (2019).
Zhang, W. et al. Tumor-associated antigen Prame targets tumor suppressor p14/ARF for degradation as the receptor protein of CRL2Prame complex. Cell Death Differ. 28, 1926–1940 (2021).
De Iaco, A. et al. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49, 941–945 (2017).
Ji, S. et al. OBOX regulates mouse zygotic genome activation and early development. Nature 620, 1047–1053 (2023).
Yagi, T. et al. A novel ES cell line, TT2, with high germline-differentiating potency. Anal. Biochem. 214, 70–76 (1993).
Tiscornia, G., Singer, O. & Verma, I. M. Production and purification of lentiviral vectors. Nat. Protoc. 1, 241–245 (2006).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005).
Bock, C. et al. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21, 4067–4068 (2005).
Zhao, B. et al. Redox-active quinones induces genome-wide DNA methylation changes by an iron-mediated and Tet-dependent mechanism. Nucleic Acids Res. 42, 1593–1605 (2014).
Yin, R. et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 135, 10396–10403 (2013).
Serebrenik, Y. V. et al. Efficient and flexible tagging of endogenous genes by homology-independent intron targeting. Genome Res 29, 1322–1328 (2019).
Takeo T., Nakagata N. In vitro fertilization in mice. Cold Spring Harb. Protoc. 2018, 415–421 (2018).
Ma, S. F. et al. Parthenogenetic activation of mouse oocytes by strontium chloride: a search for the best conditions. Theriogenology 64, 1142–1157 (2005).
Meers M. P. et al. Improved CUT&RUN chromatin profiling tools. Elife, 8, e46314 (2019).
Hayashi, R. & Inoue, A. Low-Input CUT&RUN for mouse oocytes and preimplantation embryos. Methods Mol. Biol. 2577, 83–92 (2023).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217 (2019).
Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 50, D1522–D1527 (2022).
Acknowledgements
We are grateful to Ya Wang (Protein Science Core Facility of the Institute of Biophysics) for her technical help in high-content screening determination, to Jifeng Wang and Mengmeng Zhang (Laboratory of Proteomics, Institute of Biophysics, Chinese Academy of Sciences) for assistance in mass spectrometry, to Junying Jia and Shu Meng for their technical assistance in FACS. This work was supported by grants from the Chinese Ministry of Science and Technology (2019YFA0801401), the China Natural Science Foundation (32288102, 32130021, and 92153302), the Chinese Academy of Sciences (XDB39010100, JZHKYPT-2021-05), the K. C. Wong educational foundation (GJTD-2020-06), and the New Cornerstone Science Laboratory. Z.Z. is supported by the Youth Innovation Promotion Association (2017133) of the Chinese Academy of Sciences. Y.L. is supported by the Youth Innovation Promotion Association (2022090) of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
J.T., Y.L. and B.Z. conceived and designed this project. J.T. and Y.L. performed the majority of experiments. Z.Z. performed bioinformatics analysis. X.L. and J.W. participated in immunoprecipitation experiments. L.L. performed the genetic screening under the supervision of Z.W. H.H. performed the UHPLC-MS/MS experiments under the supervision of H.W. X.Z. generated the Pramel15 knockout mice under the supervision of Y.T. J.T., Y.L., Z.Z., and B.Z. wrote the manuscript with comments and contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kathleen Stewart-Morgan, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tan, J., Li, Y., Li, X. et al. Pramel15 facilitates zygotic nuclear DNMT1 degradation and DNA demethylation. Nat Commun 15, 7310 (2024). https://doi.org/10.1038/s41467-024-51614-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51614-0