Abstract
During the epithelial-mesenchymal transition driving mouse embryo gastrulation, cells divide more frequently at the primitive streak, and half of those divisions happen away from the apical pole. These observations suggest that non-apical mitoses might play a role in cell delamination. We aim to uncover and challenge the molecular determinants of mitosis position in different regions of the epiblast through computational modeling and pharmacological treatments of embryos and stem cell-based epiblast spheroids. Blocking basement membrane degradation at the streak has no impact on the asymmetry in mitosis frequency and position. By contrast, disturbance of the actomyosin cytoskeleton or cell cycle dynamics elicits ectopic non-apical mitosis and shows that the streak region is characterized by local relaxation of the actomyosin cytoskeleton and less stringent regulation of cell division. These factors are essential for normal dynamics at the streak and favor cell delamination from the epiblast.
Similar content being viewed by others
Introduction
Epithelial-mesenchymal transition (EMT) is a reversible process by which a static epithelial cell becomes motile through the acquisition of a mesenchymal phenotype. It involves a loss or change in intercellular junctions, reorganization of the cytoskeleton, a switch from apical-basal to front-rear polarity, and remodeling of the extracellular matrix1,2. EMT is a fluid non-linear spectrum in which cells can adopt intermediate phenotypes with various levels of epithelial and mesenchymal features2.
Gastrulation, an evolutionary conserved developmental event through which multiple germ layers arise from a single epithelium, occurs through EMT-mediated cell delamination. In the mouse embryo, gastrulation takes place in the primitive streak (PS), a structure specified in the posterior region of the epiblast at embryonic day (E) 63.
One of the earliest steps of gastrulation EMT is the degradation of the epiblast basement membrane4,5. Perforations are already present globally before the onset of gastrulation, then the mesh size progressively increases and displays an anterior to posterior gradient when gastrulation begins, so that the basement membrane in the streak region becomes a highly fenestrated sheet5. Cells retain markers of apical-basal polarity6, possibly thanks to an interaction with the fragments of the basement membrane. PS cells maintain tight and adherens junctions until they exit the epiblast3, then their E-Cadherin levels diminish as they travel in the mesodermal wings. Snail is a major EMT master transcription factor implicated in E-Cadherin down-regulation; it is critical for the adequate establishment of the three germ layers3. Interestingly, PS cells can delaminate in Snail knockout embryos, but nascent mesoderm maintains an intermediate level of E-cadherin and fails to migrate away7. PS cells asynchronously delaminate through apical constriction followed by retraction of the apical process and exit on the basal side4,8. 3D time-lapse imaging of gastrulating mouse embryos revealed that apical constriction occurs in a pulsed ratchet-like fashion, through asynchronous shrinkage of apical junctions6. Apical polarity proteins play an important role during delamination6,8. Analysis of Crumbs2-deficient embryos suggested that it regulates the anisotropic accumulation of actomyosin at junctions of ingressing cells after E7.5. Moreover, cells with high levels of aPKC undergo apical constriction and preferentially ingress into the streak9. However, the mechanism for cell ingression during the early stages of gastrulation remains unclear.
In mouse, cell number is tightly regulated at the gastrulation stage. Indeed, experiments involving reduction or increase of embryo size highlighted a remarkable ability to compensate through change in proliferation rate10,11. Consistent with what was first observed by Snow in 197712, immunostaining on sections and live imaging of mosaically labeled mouse embryos at the gastrulation stage showed that the frequency of mitoses is two times higher at the streak, compared to the rest of the epiblast13. Accordingly, single-cell RNA sequencing revealed that cells at the PS have a higher G2/M phase score than other epiblast cells14. Quantification of cell cycle length in rat embryos also indicated that cells at the PS are cycling faster than in the rest of the epiblast15.
The epiblast is a pseudostratified epithelium, in which nuclei undergo an Interkinetic Nuclei Migration (INM); in short pseudostratified epithelia (2-3 pseudolayers), INM is usually mostly dependent on the actomyosin cytoskeleton16. In S phase, nuclei are basal. They are rapidly translocated toward the apical side during G2 so that mitosis occurs apically16. Live imaging confirmed the existence of such nuclear movements in the mouse epiblast17. Staining for Phospho-Histone H3, a marker for M phase, indicated that mitosis is indeed apical in the epiblast, except in the PS where about 40% of mitoses happen away from the apical pole12,13. Live imaging of embryos in which a small number of epiblast cells were labeled with membrane targeted GFP (thereby allowing tracking of individual cell shape and behavior) showed that most cells undergoing non-apical mitotic rounding retained a basal attachment until cytokinesis, yet preferentially gave rise to one or two cells that delaminated to become mesoderm13. Non-apical mitoses have been described in other pseudostratified epithelia. For example, in the dorsal neuroepithelium of chick embryos, in which neural crest cells arise by EMT, non-apical mitoses are found in similar proportions than in the mouse PS. Importantly, abrogation of non-apical mitosis leads to a severe delay in neural crest cell delamination18,19.
All these observations indicate that PS cells have specific cycling properties and a high rate of non-apical mitoses that might be linked to their ability to delaminate. In this work, we use experimental and in silico approaches to assess the functional relationship between proliferation rate, INM, cell contractility, and the occurrence of non-apical mitoses in the epiblast and whether some of these parameters actively contribute to cell delamination at the streak. Here, we find that non-apical mitoses can be favored by multiple stimuli such as cytoplasmic actomyosin relaxation, a higher proliferation rate, a shorter G2, or detachment from the basement membrane. All these inputs act in concert at the PS. Importantly, non-apical mitoses promote but are not sufficient to provoke cell delamination, which can only occur if epithelial tissue integrity is impaired.
Results
Gastrulation pharmacological screen and in silico modeling
We previously established that non-apical mitoses at the PS primarily give rise to basal delamination of nascent mesoderm cells13. Therefore, we set out to identify the factors controlling the occurrence of non-apical divisions and assess whether they are necessary and/or sufficient for cell delamination. Variations in the position within the epithelium at which a cell divides could come from differences in apicobasal polarity, actomyosin contractility or cell cycle parameters (such as total duration of the cycle or duration of the G2/M phase). Thus, we performed a pharmacological screen targeting these cellular characteristics. While genetic manipulation allows for spatial control, it lacks precision in intensity and duration, which can complicate the interpretation of a phenotype, particularly in a highly dynamic context. The pharmacological approach addresses these limitations as it allows establishing optimal posology and recording the consequences of short-term alterations. One caveat is that all cells (epiblast and PS) are simultaneously targeted. To circumvent this, we used a computational model of proliferating pseudostratified epithelium in which modifications of cell cycle parameters, attachment to the basement membrane, and fine regulation of apical and medial actomyosin can be adjusted at the single cell level.
For biological experiments, we established a systematic protocol of measurements on embryos sectioned along the proximal-distal axis (Fig. 1a, b). Sections were positioned along the anterior-posterior axis based on anatomical landmarks such as the long axis of the oval embryo shape and the morphology of visceral endoderm cells, as well as molecular markers including collagen IV for the basement membrane. The epiblast was divided into three regions: anterior and posterior halves of the epiblast, and PS, defined as the subregion of the posterior epiblast where the basement membrane is discontinuous. Staining for mitosis (Phospho-Histone H3) and nuclei (DAPI) allowed calculating the mitotic and non-apical mitotic indexes (Fig. 1b). We quantified the percentage of basement membrane (collagen IV) degradation as a readout for EMT initiation and progression (Fig. 1a).
a Graphic representation of a transverse section from a E6.5 embryo. The epiblast is divided in three regions: anterior (pink), posterior (green) and the primitive streak (PS, blue). Anterior and posterior regions are defined by dividing the epiblast in halves. The PS region is the subsection of the posterior region where the basement membrane (BM) is discontinuous (Collagen IV, yellow). Mesoderm cells that exited the epiblast are in purple. The black double head arrow represents the distance in µm between the apical and the basal surface. b Graphic representation of a transverse section from a E6.5 embryo. Nuclei are in purple, apical mitosis in pink and non-apical mitosis in green. A mitosis is considered non-apical if it occurs at least 10 µm away from the apical surface (lumen), in a cell that retains apical and basal contact. c Diagram representing simulated cells controlled by various springs: apical cell adhesions (apical-to-apical spring), and the cytoplasmic cytoskeleton with basal cell-matrix adhesion (nucleus-to-basal spring and apical-to-nucleus spring). Cell-matrix adhesions are modeled by the attachment of the nucleus-to-basal spring to the basal line. Nuclei are modeled with two layers. The soft layer can adjust to the environment to model pseudo-stratified morphology, the hard core cannot be deformed and increases during mitosis. d Representation of a simulation setup (t = 0 h) for the in silico model adapted to the epiblast dynamics, with 19 PS (purple nuclei) cells and 40 epiblast cells (green nuclei). The red line represents the apical surface, and the black line the basal surface.
For the modeling of pseudo-stratified epithelium, we relied on a model originally developed on the chick neuroepithelium20. We used an extended version that allows the control of each parameter in time and space at the single cell level, described in ref. 21. Briefly, each cell is approximated by a nucleus, an apical point, and a basal point. The two points are connected to the nucleus through dynamically adjustable springs representing the viscoelastic properties of the cytoplasm (Fig. 1c). The nucleus is composed of an outer soft core and an inner hard core. Hard cores of neighboring cells cannot overlap while soft cores can but are subjected to a repulsion force. This allows representing packing and compression occurring between cells in a dense tissue. Apical cell-cell adhesions are modeled by springs between the apical points of neighboring cells (aa). On the basal side, basal points are attached to a fixed basal line representing the basement membrane. Basal points cannot overlap or swap positions. When a cell detaches its basal point in the model, it is equivalent to a local fenestration of the matrix as it locally increases the mean distance between the basal points of the nearest neighbors. The cell cycle and INM movements are divided into three phases. The first phase corresponds to G1, S, and early G2, where movements are controlled by environmental constraints generated by neighboring cells. The second phase corresponds to late G2, where the pre-mitotic rapid apical movement (or PRAM) is simulated by contraction of the apical-nuclei string (aN) to bring nuclei to the apical pole, and extension of the basal-nuclei spring (bN) to accompany the movement. These settings are maintained in the third phase, the M phase, which results in the formation of two daughter cells. As the model represents a 2D view of a tissue in 3D, only a few daughter cells would actually divide exactly within the simulated 2D plane. As a dimensional correction, we neglect the second daughter cell in 95% of the cases. To adapt the model to the epiblast size and configuration, a maximum height was set to allow no more than three pseudo-layers of nuclei (Fig. 1d). A user-friendly online version of this model is available as a web-based stand-alone EMT simulator (sEMTor) at https://semtor.github.io/gastrulation/.
Basement membrane degradation triggers non-apical mitoses
In the mouse embryo, perforations in the basement membrane separating epiblast and visceral endoderm are distributed uniformly prior to migration of the Anterior Visceral Endoderm (AVE), the organizer that determines the anterior-posterior axis and therefore the position of the streak. Upon axis specification, perforations become more abundant in the posterior half of the embryo. These holes in the basement membrane are generated by a Nodal-dependent increase in the expression of Matrix Metalloproteinases (MMP) 2 and 14, and are necessary for embryo growth, PS morphogenesis, and gastrulation5. In EMT, loss of basal attachment due to basement membrane degradation is frequently associated with a disruption in apical-basal polarity. However, during mouse gastrulation, polarity markers are present in all PS cells4,6,22. We wondered whether basement membrane degradation might nonetheless be required for the occurrence of non-apical mitosis, possibly through modification of mechanical constraints in the epiblast.
To test this, we cultured pre-gastrulation E6 embryos for 6 hours with a combination of 20 µM Pan-MMPs inhibitor prinomastat hydrochloride and 100 µM (NSC405020) MMP14 inhibitor, as previously described5. After incubation, embryos were fixed, and transverse cryosections were immunostained for Phospho-Histone H3, Collagen IV, nuclei (DAPI), and F-actin (phalloidin). Additionally, adjacent sections were stained for Cerberus 1 (Cer1) to identify the location of the AVE. We confirmed the abrogation of basement membrane degradation in embryos treated with MMPs inhibitors (Fig. 2a–c). However, there was no significant difference in mitotic and non-apical mitotic indexes (Fig. 2d, e), nor in the non-apical mitosis ratio (Supplementary Fig. 1a) throughout the epiblast relative to untreated embryos.
Z-projection of transverse sections from representative E6 embryos cultured for 6 hours with vehicle or MMPs inhibitors, stained for (a) nuclei (DAPI, cyan), a mitosis (Phospho-histone H3 (Phh3), magenta), a, b basement membrane (collagen IV, yellow), and (a, b) F-actin (phalloidin, grey). Arrowheads indicate non-apical mitoses. Scale bars in main panels: 25 µm. Zooms in b show the basement membrane at the PS. Scale bars: 10 µm. c Violin plots representing the percentage of basement membrane degradation in embryos cultured with control vehicle or MMPs inhibitors. Inner doted lines indicate third quartile, median, and first quartile. Normality was assessed using a Shapiro–Wilk test followed by a two-tailed Mann Whitney test. Split violin plots representing the mitotic index (d) and non-apical mitotic index (e), in anterior, posterior and PS regions embryos cultured with control vehicle or MMPs inhibitors. Normality was assessed using a Shapiro–Wilk test followed by a two-tailed Mann-Whitney or unpaired t-test. Controls: n = 14 slides from 8 embryos; MMPs inhibitors: n = 21 slides from 7 embryos. f Graph from in silico simulations representing the percentage of non-apical mitoses depending on aN strength in epiblast cells (6 h cell cycle) and PS cells (3 h cell cycle), 100 simulations with 30 proliferating cells. g Table presenting the simulated parameters set for epiblast and PS cells in control conditions in the in silico epiblast model. Event B designates the detachment of cells’ basal point from the basal line. h Scatterplot representing the ratio of non-apical mitosis (%), in the region of the epiblast and PS in simulations ran with control settings or in situations where PS cells do not undergo event B. For each condition, 100 simulations with 19 PS cells surrounded by 40 control epiblast cells. Error bars: SEM. (i) Table recapitulating the settings of epiblast and PS cells in control and test simulations. For the simulation, only the modified parameters are indicated. A-P Anterior – Posterior, Ap-Bas Apical – Basal, PS Primitive Streak, CC cell cycle, aN apical-Nuclei spring, bN basal-Nuclei spring, aa apical adhesion spring.
Before modelling the impact of detachment from the basement membrane in silico, we first had to calibrate the parameters of cell cycle and INM allowing to recapitulate the percentage of non-apical mitoses observed in vivo. As the velocity of PRAM is controlled by the contraction of the apical-to-Nucleus (aN) spring in the model, we tested a range of strength values for aN (Fig. 2f). We simulated 200 repetitions of 30 proliferating cells with a cell cycle of 6 ± 0.5 h or 3 ± 0.5 h, corresponding to measured values for epiblast and PS cells, respectively12,15. The percentages of non-apical mitoses observed at the PS (circa 40%) could only be reached at very low values of aN, suggesting that an almost complete loss of PRAM is necessary. In addition, we noted that a faster cell cycle favors non-apical mitoses even at higher PRAM intensity. Indeed, the frequency of non-apical mitoses for medium values of aN reached a plateau at 7% with a cell cycle length of 3 h, and only 1% with a length of 6 h. Based on these simulations, we used the following as default parameters: 6 ± 0.5 h of cell cycle and 3 of aN strength for epiblast cells, 3 ± 0.5 h of cell cycle and 0.25 of aN strength for PS cells (Fig. 2g, Supplementary Movie 1). We counted the number of PS cells on cross sections of stage E6.5 mouse embryos and found a mean population size of 19 PS cells per section13. Thus, we set the size of the PS population at 19 cells, surrounded by 20 epiblast cells on each side. Simulations start with a period of 6 hours during which all cells are attached basally. Then there is an 18 h long window of opportunity for PS cells to detach from the basal line, which is referred as event B21 (Fig. 2g) and represents basement membrane degradation21.
We then ran simulations with or without detachment of PS cells from the basal line and plotted the rate of non-apical mitoses (Fig. 2h, i; Supplementary Movie 1). As observed in embryos treated for 6 h with MMPs inhibitors, impairing detachment from the basal line did not suppress the occurrence of non-apical mitoses at the PS (Fig. 2h, i).
Collectively, these data indicate that preventing basement membrane degradation does not influence the rate of mitoses and non-apical mitoses at the PS.
To determine if basement membrane degradation was sufficient to trigger the appearance of non-apical mitoses, we examined the position and frequency of mitoses in embryo deficient for FLRT3 (Fibronectin leucin-rich transmembrane protein 3). Knockout of FLRT3 in mouse leads to disorganization of the basement membrane between visceral endoderm and epiblast and the emergence of ectopic sites of epiblast delamination23. Since basement membrane degradation could no longer be used to define the region of the streak in mutant embryos, we approximated the position of the PS as the posterior 1/3rd of the epiblast, opposite the AVE. The rest of the epiblast, referred as anterior 2/3rd, approximates the anterior and posterior epiblast (Supplementary Fig. 2a, b). We detected no difference regarding the frequency of mitosis in WT vs FLRT3KO embryos (Supplementary Fig. 2c). However, there was a significant increase in the proportion of non-apical mitosis frequency in the anterior 2/3rd epiblast in FLRT3KO embryos, compared to wild-type embryos (Supplementary Fig. 2d). In ectopic sites of delamination in FLRT3KO embryos, 45% of mitoses happened apically, 30% non-apically, and 25% in delaminated cells (Supplementary Fig. 2e, f).
These data indicate that ectopic basement membrane degradation is sufficient to trigger non-apical mitosis, associated with cell delamination, in regions of the epiblast that are presumably not exposed to the signaling pattern characteristic of the PS region.
Actomyosin relaxation drives non-apical mitoses
As INM movements are usually driven by actomyosin in short pseudo-stratified epithelia16, we tested whether a regional modification in actomyosin contractility could be responsible for the increased rate of non-apical mitosis at the PS.
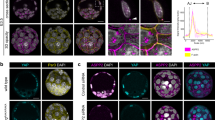
The shape of nuclei in interphase can be used as a readout of the mechanical constraints exerted on cells24,25, which are believed to be particularly high in pseudo-stratified epithelia26,27. We marked nuclear membrane via staining for Lamin B1, the first Lamin subtype expressed during early development in mouse28,29, and calculated the nuclei aspect ratio by dividing the short axis length by the long axis length (Fig. 3a). A ratio close to 0 corresponds to an elongated nucleus, indicating that the cytoskeleton is contracted. A ratio close to 1 indicates a round nucleus, reflecting relaxation of the cytoskeleton network. Nuclei aspect ratios were recorded in the anterior, posterior, and PS regions of E6.25, E6.5, and E7 embryos (Fig. 3b-c”). Nuclei in the PS were rounder, compared to the rest of the epiblast, from E6.25 onwards, with a peak at E6.5 and maintenance until E7 (Fig. 3c-c”). This is compatible with a local relaxation of the cytoskeleton at the streak, leading to nuclei shape changes, upon the onset of gastrulation.
a Nucleus representation. b-b’ Z-projection of a transverse section from a E7 embryo, stained for Lamin B1 (green) (b), with zooms on nuclei from the anterior, posterior and PS regions (b’). Scale bars: 25 µm (b) and 10 µm. c-c” Violin plots representing nuclei aspect ratio at E6.25 (c), E6.5 (c’) and E7 (c”). Inner doted lines indicate third quartile, median, and first quartile. Normality was assessed using a Shapiro–Wilk test followed by a two-tailed Kruskall-Wallis test with Dunnet post hoc. E6.25: n = 4 embryos, E6.5: n = 4 embryos, E7: n = 4 embryos, with 20 nuclei per region. d Z-projection of transverse sections from E6.5 embryos cultured for 2 hours with vehicle or ROCK inhibitor at 40 µM, stained for nuclei (DAPI, cyan), basement membrane (collagen IV, yellow), mitosis (Phh3, magenta) and F-actin (phalloidin, grey). Arrowheads indicate non-apical mitoses. Scale bars: 25 µm. Split violin plots representing the mitotic index (e) and non-apical mitotic index (f), in embryos cultured with control vehicle or 40 µM ROCK inhibitor. Normality was assessed using a Shapiro–Wilk test followed by a two-tailed Mann-Whitney. Control: n = 26 slides from 12 embryos; 40 µM ROCK inhibitor: n = 14 slides from 7 embryos. g Violin plots representing nuclei aspect ratio in embryos cultured with vehicle or 40 µM ROCK inhibitor. Normality was assessed using a Shapiro–Wilk test followed by a two-tailed Mann-Whitney or unpaired t-test. Control: n = 11 embryos; 40 µM ROCK inhibitor: n = 5 embryos. h Scatterplot representing the percentage of non-apical mitosis in the region of the epiblast and PS in various simulations: control, relaxation of INM (aN/bN) springs, relaxation of all springs and relaxation of apical adhesion springs. For each condition, 100 simulations with 19 PS cells surrounded by 40 control cells. Error bars: SEM. i Table recapitulating the settings of epiblast and PS cells in control and test simulations. For the simulations, only the modified parameters are indicated. A-P Anterior - Posterior, Ap-Bas Apical – Basal, PS primitive streak, INM interkinetic nuclei migration, CC cell cycle, B detachment of basal adhesions from the basal line, aN apical-Nuclei spring, bN basal-Nuclei spring, aa apical adhesion spring.
The small Rho-GTPase RhoA regulates both actomyosin contractility and cell cycle progression. In addition, during EMT, RhoA participates in the cellular changes required to acquire a mesenchymal-like phenotype and migrate30. Rho-associated kinase (ROCK), a downstream effector of RhoA, regulates actomyosin contractility via myosin light chain phosphorylation31,32. In the pseudostratified epithelium of the Drosophila imaginal discs, pharmacological treatment with ROCK inhibitor Y27632 leads to basal accumulation of nuclei and the appearance of non-apical mitoses33. To investigate the link between mitosis location and actomyosin dynamics in the mouse epiblast, we cultured E6.5 embryos for 2 h with 40 µM Y27632. Embryos were then fixed, and transverse cryosections were immunostained for Phospho-Histone H3, Collagen IV, nuclei (DAPI), and F-actin (phalloidin) (Fig. 3d). There was no change in the mitotic index (Fig. 3e). However, a 2-fold increase in the non-apical mitotic index (and a 10-fold increase of the non-apical mitosis ratio) were recorded in the anterior and posterior epiblast (Fig. 3f, Supplementary Fig. 1b). This is compatible with an actomyosin-driven INM in the epiblast: indeed, it is likely that disturbing actomyosin contractility affects the ability of cells to drive their nuclei to the apical surface in G2, which results in mitosis occurring in ectopic location. Interestingly, non-apical mitotic index remained unchanged at the PS (Fig. 3f), strongly suggesting that actomyosin contractility is intrinsically low in PS cells. Upon ROCK inhibition, nuclei shape was rounder in the anterior and posterior epiblast confirming it induced a relaxation of the actomyosin cytoskeleton, while the shape of nuclei in the PS did not change (Fig. 3g). The effect of the inhibitor was reversible, as embryos treated for 2 h with 40 µM of ROCK inhibitor then washed were able to adjust rapidly and survive overnight in culture (Supplementary Fig. 1e).
To further explore the influence of cytoskeleton contractility on the occurrence of non-apical mitoses, we performed simulations with a reduced strength for all springs: aN and bN for the cell body/cytoplasm and aa for apical cell-cell adhesion (Fig. 3h, i; light purple). Interestingly, such global relaxation increased the frequency of non-apical mitoses in the epiblast and reduced it in the PS (Fig. 3h, Supplementary Movie 2). Then, we set out to assess the relative contributions of the cytoplasmic springs (aN, bN) that control INM/PRAM and of the apical adhesion spring (aa) by relaxing them independently (Fig. 3h, i; pink and orange; Supplementary Movie 2). Relaxing aN/bN strongly promoted non-apical mitoses in the epiblast but did not affect PS cells. Relaxing aa mildly promoted non-apical mitoses in the epiblast and lowered their frequency in the PS. It is interesting to note that in all simulations with aa relaxation the tissue experienced severe deformations (Fig. 3h, i; Supplementary Movie 2). The changes obtained in the scenario where all springs were relaxed are compatible with what we observed in embryos treated with a high dose of ROCK inhibitor (400 µM): those displayed severe morphological defects (Supplementary Fig. 3a, b); the mitotic index was decreased in all regions of the epiblast (Supplementary Fig. 3c) while the non-apical mitotic index was increased in the anterior and posterior epiblast but decreased in the PS (Supplementary Fig. 3d). We interpret that the effects observed at 40 µM of ROCK inhibitor correspond to a mild inhibition of the cell cytoskeleton contractility that can be modeled by the relaxation of only the INM springs, while the buckling effect seen at 400 µM requires stronger perturbation including that of the apical actomyosin.
Collectively, these experimental and simulated data indicate that non-apical mitoses in the epiblast can be elicited by affecting cytoplasmic actomyosin activity rather than apical actomyosin. This suggests that INM is governed by actomyosin in the epiblast and that, comparatively to the rest of the epiblast, actomyosin contractility is weaker in PS cells, leading to the appearance of non-apical mitoses.
Cell cycle regulation is less stringent in the PS
The complex formed by Cyclin-dependent Kinase 1 (CDK1) and CyclinD1 is critical for the control of cell entry into mitosis. In the avian neuroepithelium, overexpression of CyclinD1 leads to the appearance of non-apical mitoses34. In zebrafish retina, CDK1 is required for the rapid basal to apical movement of nuclei during G235. Therefore, we chose to pharmacologically target CDK1, negatively using a CDK1 inhibitor (R03366), and positively via an inhibitor (PD166285) of Wee1, a nuclear kinase that regulates mitosis by phosphorylating CDK1. Wee1 inhibition thereby enables to bypass the G2/M checkpoint36.
A 2 h incubation with 175 µM of CDK1 inhibitor abrogated all mitosis, confirming that targeting CDK1 regulation can rapidly affect the cell cycle in the epiblast (Fig. 4a–c). Embryos were able to recover from the treatment and developed normally (Supplementary Fig. 1e). With the goal to increase the frequency of mitoses without major morphological anomaly, we performed a dose-response analysis for the Wee1 inhibitor (5, 10, 20, 40, and 80 µM for 2 hours). Embryos were fixed directly after incubation. We measured the percentage of basement membrane degradation as well as the mitotic and non-apical mitotic indexes on transverse cryosections immunostained for Phospho-Histone H3, Collagen IV, nuclei, and F-actin (Fig. 4d). In the anterior and posterior epiblast, we observed a dose-dependent increase of the mitotic index, indicating an acceleration of the cell cycle, as well as an increase of the non-apical mitotic index and non-apical mitosis ratio (Fig. 4e, f; Supplementary Fig. 1c). We previously showed that an acceleration of the cell cycle is sufficient to trigger non-apical mitoses (Fig. 2f), therefore some of the increase of non-apical mitoses in the epiblast upon treatment with Wee1 inhibitor might be linked to the global increase in mitotic index. However, as PRAM takes place in G2, Wee1 inhibition might also contribute to the higher rate of non-apical mitoses by shortening the window of opportunity for PRAM to occur. By contrast, the frequency and location of mitoses in the PS stayed constant except at high doses of drug (40–80 µM) at which most cells were entering mitosis (Fig. 4e, f). These data indicate that cells at the PS were less sensitive to Wee1 inhibition compared to the rest of the epiblast, strongly suggesting an intrinsically shorter cell cycle and G2 duration in PS cells.
a Z-projections of transverse sections from E6.5 embryos cultured for 2 hours with control vehicle or 175 µM of CDK1 inhibitor (R03366), stained for nuclei (DAPI, cyan), basement membrane (collagen IV, yellow), mitosis (phh3, magenta) and F-actin (phalloidin, grey). Scale bars: 25 µm. Violin plots representing the mitotic index (b) and non-apical mitotic index (c) in anterior, posterior, and PS regions in embryos cultured with control vehicle or 175 µM CDK1 inhibitor. Inner doted lines indicate third quartile, median, and first quartile. Normality was assessed using a Shapiro–Wilk test followed by a two-tailed Mann-Whitney. Control: n = 24 slides from 8 embryos; 175 µM CDK1 inhibitor: n = 11 slides from 7 embryos. d Z-projection of transverse sections from E6.5 embryos cultured for 2 hours in vehicle or with 20 µM, 40 µM or 80 µM of Wee1 inhibitor, stained for nuclei (DAPI, cyan), basement membrane (collagen IV, yellow), mitosis (phh3, magenta) and F-actin (phalloidin, grey). Arrow heads indicate non-apical mitoses. Scale bars: 25 µm. Violin plots representing the mitotic index (e) and non-apical mitotic index (f), in anterior, posterior, and PS regions of embryos cultured with control vehicle or 5 µM, 10 µM, 20 µM, 40 µM or 80 µM of Wee1 inhibitor for 2 hours. Ns nonsignificant, *P-value ≤ 0.05, **P-value ≤ 0.01 and ***P-value ≤ 0.001. Normality was assessed using a Shapiro–Wilk test followed by a two-tailed Mann-Whitney or unpaired t-test. Control: n = 27 slides from 13 embryos; 5 µM Wee1 inhibitor: n = 12 slides from 5 embryos; 10 µM Wee1 inhibitor: n = 16 slides from 10 embryos; 20 µM Wee1 inhibitor: n = 21 slides from 9 embryos; 40 µM Wee1 inhibitor: n = 9 slides from 4 embryos; 80 µM Wee1 inhibitor: n = 12 slides from 6 embryos. g Scatterplot representing the percentage of non-apical mitosis in the region of the epiblast and PS in various simulations: control, reduction of the total cell cycle duration (3 h), reduction of G2 and M phase duration (15 minutes) and reduction of both the total duration of the cell cycle (3 h) and of G2 and M duration (15 minutes). For each condition, 100 simulations with 19 PS cells surrounded by 40 control cells. Error bars: SEM. h Table recapitulating the settings of epiblast and PS cells in control and test simulations. For the simulation, only the modified parameters are indicated. A-P Anterior – Posterior, Ap-Bas Apical – Basal, PS primitive streak, CC cell cycle, B detachment of basal adhesions from the basal line, aN apical-Nuclei spring, bN basal-Nuclei spring, aa apical adhesion spring.
To assess the relative impact of G2 and total cell cycle duration, we performed simulations in which we modulated their duration independently or simultaneously. The model has a simplified version of the cell cycle in which PRAM forces are applied through G2 and M. Thus, to mimic the effect of the Wee1 inhibitor in shortening biological G2 and the associated duration of PRAM forces, we reduced the total duration of simulated G2 + M by half. In the control situation, epiblast cells cycle every 6 ± 0.5 h and PS cells every 3 ± 0.5 h. All cells have a G2/M of 30 minutes each (Fig. 4g, h; dark purple). If all cells were forced to cycle every 3 ± 0.5 h at constant G2/M duration, it only generated a marginal increase of non-apical mitoses in the epiblast (Fig. 4g, h; light purple). If both cell types kept their respective cell cycle length but were forced to have a G2/M of 15 minutes each, there was a massive increase of non-apical mitoses in the epiblast (Fig. 4g, h; pink, Supplementary Movie 3). If both changes were combined (cell cycle 3 ± 0.5 h, G2/M 15 min in all cells), it did not generate a further increase of non-apical mitoses in the epiblast (Fig. 4g, h; orange; Supplementary Movie 3). By contrast, all these changes had negligible effects on the PS. These results indicate that specifically affecting G2/M duration has more impact on the rate of non-apical mitoses than shortening the whole cell cycle without affecting G2/M duration. Overall, these data indicate that an acceleration of the cell cycle and a shortened G2 duration contribute to the increased incidence of mitoses and non-apical mitoses observed at the PS.
Non-apical mitoses occur independently of EMT progression
There is evidence that mitosis can influence gene regulation and EMT progression, notably through the process of gene bookmarking37,38. Therefore, we asked whether mitosis frequency and location could favor EMT.
In control conditions, non-apical mitoses are followed by basal delamination of mesodermal precursors13. Thus, we wondered if any of the pharmacological treatments that led to ectopic non-apical mitoses might be sufficient to promote ectopic EMT in the epiblast. We monitored markers of EMT progression, notably the expression of the EMT marker Snail, the pattern of E-cadherin and N-cadherin, and the percentage of basement membrane degradation, upon pharmacological inhibition of ROCK, Wee1, and CDK1.
In embryos treated with 40 µM of ROCK inhibitor, immunostaining for Snail showed that cells at the PS were still able to transition, and no ectopic EMT site was detected (Fig. 5a). This was confirmed by staining for E-cadherin and N-cadherin, as there was no alteration in the cadherin switch (Fig. 5b). Similarly, we observed no significant change regarding the percentage, pattern, or localization of basement membrane degradation (Figs. 3d and 5c).
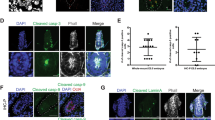
Z-projections of transverse sections from E6.5 embryos cultured for 2 hours with control vehicle, 40 µM ROCK inhibitor, 20 µM Wee1 inhibitor or 175 µM CDK1 inhibitor, stained for (a) EMT marker Snail (yellow) and F-actin (phalloidin, grey), or (b) E-cadherin (cyan) and N-cadherin (magenta). Scale bars: 25 µm. Representative images from Control n = 9 embryos; 40 µM ROCK inhibitor n = 3 embryos; 20 µM Wee1 inhibitor: n = 5 embryos, 175 µM CDK1 inhibitor: n = 4 embryos. c-c” Violin plots representing the percentage of basement membrane degradation in embryos cultured with control vehicle, 40 µM ROCK inhibitor (c), 20 µM Wee1 inhibitor (c’), 175 µM CDK1 inhibitor (c”). Inner doted lines indicate third quartile, median, and first quartile. Normality was assessed using a Shapiro–Wilk test followed by a two-tailed unpaired t-test. Control: n = 27 slides from 13 embryos; 40 µM ROCK inhibitor: n = 21 slides from 9 embryos; 175 µM CDK1 inhibitor: n = 11 slides from 7 embryos. d In situ hybridization for Brachyury (T) (top), and Eomesodermin (bottom) on E7.5 WT, RhoAΔepiblast, Ect2Δepiblast embryos. Scale bars: 100 µm. Brachyury (T): RhoAΔepiblast n = 2; Ect2Δepiblast n = 4. Eomesodermin: RhoAΔepiblast n = 5; Ect2Δepiblast n = 5. e Z-selection from E7.5 WT (n = 7), RhoAΔepiblast(n = 4), Cdk1Δepiblast(n = 5), Ect2Δepiblast(n = 6) embryos stained for F-actin (Phalloidin, grey) and Vimentin (green). Scale bar: 50 µm.
For Wee1, we focused on embryos treated with 20 µM of the inhibitor for 2 hours. Immunostaining for Snail, E-cadherin, and N-cadherin indicated that EMT was initiated normally at the PS (Fig. 5a, b). While these conditions led to a global increase of non-apical mitoses in all the epiblast (Fig. 4f; Supplementary Fig. 1c), there were no ectopic site of EMT nor significant change regarding the percentage of basement membrane degradation (Figs. 4d and 5a, b, c’). In addition, preventing entry in mitosis by culturing embryos for 2 hours with 175 µM of CDK1 inhibitor did not affect basement membrane degradation, the expression of Snail or the cadherin switch (Figs. 4a and 5a, b, c”), indicating that cell division per se is not necessary for EMT initiation.
In addition, we examined the phenotypes of embryos carrying epiblast-specific deletion of the small GTPase RhoA, the Guanine exchange factor Ect2, and the kinase Cdk1, which are involved in cytoskeleton regulation, cytokinesis, and cell cycle regulation, respectively. Δepiblast embryos were obtained from the combination of RhoA, Ect2 and Cdk1 conditional alleles39,40,41 and a Cre-recombinase under the dependency of the Sox2 promoter, which is turned on in the prospective epiblast at E3.542 (Fig. 5d, e, Supplementary Fig. 4).
Deletion of RhoA in the epiblast led to disruption of the pseudostratified arrangement, detectable from E6.5 onwards (Supplementary Fig. 4a–g). We found a slightly higher mitotic index at E6.5, possibly due to delays in cytokinesis (Supplementary Fig. 4c, d). Basement membrane degradation was detectable in the posterior region, and cells with a mesenchymal phenotype accumulated at the streak and into the lumen (Supplementary Fig. 4e). This is compatible with an adequate initiation of EMT but a defect in cell migration away from the PS, like what was observed in Rac1 epiblast-specific mutants22, or Rac1 and RhoA mesoderm-specific mutants43. By E7.5, RhoAΔepiblast mutants exhibited severe morphological defects. Lumens could be spotted within the accumulation of cells on the posterior side of the embryo, suggesting a disruption of polarity (Supplementary Fig. 4f). Similarly, epiblast-specific deletion of Cdk1 or the cytokinesis regulator Ect2 both resulted in a severe phenotype with developmental arrest (Fig. 6e, Supplementary Fig. 4h, i). In RhoAΔepiblast, as well as CDK1Δepiblast and ECT2Δepiblast mutants, mesoderm identity was specified, as shown by the expression of the PS and nascent mesoderm markers Brachyury (T), and Eomesodermin, (Fig. 5d), and the mesenchymal marker Vimentin (Fig. 5e).
a Scatterplot of the mean percentage of nuclei basal positioning in tested scenarios for epiblast and PS cells. Error bars: SD. b Legend for tested scenarios. Squares correspond to the region of the epiblast and circles to the PS. c Scatter plot of the percentage of non-apical mitoses and mean rate of nuclei basal positioning in tested scenarios for the epiblast and PS. d Scatterplot of the mean rate of basal extrusion and basal positioning in tested scenarios for the epiblast and the PS. e Scatter plot of the mean rate of basal extrusion and the percentage of non-apical mitosis in tested scenarios for the epiblast and PS. For each condition, 100 simulations with 19 PS cells surrounded by 40 control cells.
While these data underscore the critical role for RhoA, CDK1 and ECT2 in maintaining epiblast integrity and promoting cell migration, they suggest that changes in actomyosin contractility and cell cycle parameters are not sufficient to trigger or block EMT progression.
Non-apical mitosis frequency correlates with delamination
In view of the results from pharmacological screening and in silico modeling, we conclude that relaxation of cytoplasmic actomyosin, a faster cell cycle, and a shortening of the G2 phase collectively contribute to the increased incidence of non-apical mitoses at the PS during gastrulation. Given that in vivo time-lapse imaging showed that cells undergoing non-apical mitosis primarily gave rise to basally delaminating mesodermal cells13, we wondered if those mechanisms might also be upstream of basal extrusion.
Interestingly, computational modeling of EMT in a tall pseudostratified epithelium using the same model indicated that the position of nuclei correlates with the directionality of extrusion21. In particular, basal positioning (defined as more basal than the average position of nuclei) specifically favors basal extrusion. Therefore, some of the experimental and simulated experimental conditions we tested might have a broader impact on nuclei distribution in the tissue than the position of mitoses. However, in a comparatively flat epithelium like the gastrulating epiblast it is unclear whether basal positioning has any importance for delamination given that all cells have their nuclei relatively close to the basement membrane at any time point.
To assess this, we plotted the rate of basal positioning of nuclei in epiblast and PS for all simulated conditions (Fig. 6a, b). There was a high rate of basal positioning in control PS cells. Preventing detachment from the basal line or enforcing normal actomyosin contractility (Fig. 6a, PS experimental conditions 2 to 5) prevented basal positioning of nuclei in PS cells. By contrast none of the treatments affecting contractility or cell cycle parameters were able to promote basal positioning in epiblast cells, where only a forced detachment from the basal line was sufficient to generate a high rate of basal positioning (Fig. 6a, epiblast experimental condition 2).
Next, we wondered if overall, basal positioning of nuclei and the occurrence of non-apical mitoses were correlated (Fig. 6c). This revealed two different patterns: in epiblast cells, non-apical mitoses occurred in the absence of basal positioning unless loss of basal attachment was enforced (grey square with black outline). In PS cells both parameters were strongly correlated unless we imposed a relaxation of the springs throughout the tissue, which completely canceled basal positioning but only lowered the rate of non-apical mitoses (Fig. 6c, circles in hot colors). These data indicate that non-apical mitoses and overall basal positioning of nuclei can be uncoupled. This means that non-apical mitoses can occur even in absence of a global redistribution of nuclei in the tissue.
Next, we looked at a putative correlation between basal positioning and basal extrusion (Fig. 6d) and found a similar relationship: there was a strong positive correlation in PS cells unless contractility was affected (circles in hot colors), and no correlation in the epiblast unless epiblast cells were forced to detach from the basal line (grey square with a black outline). Finally, we looked at the relationship between non-apical mitoses and basal extrusion. Here, the data were not split in two groups, and we found an overall positive correlation between the two factors (Pearson r = 0.59, p = 0.0157).
These analyses revealed that non-apical mitoses can specifically occur in absence of overall tissue disorganization leading to massive basal positioning. In addition, non-apical mitoses are positively correlated with basal extrusion. However, it is important to note that extrusion itself only occurs significantly if cells detach from the basal line or if all springs or apical springs are relaxed. This means that actual delamination requires a condition in which epithelial integrity is impaired.
Proliferation favors delamination in epiblast spheroids
To further assess the putative relationship between the frequency/position of mitoses and exit from PS, we turned to a stem-cell-based epiblast model44. This allows for the study of specific events in a system where the chemical environment is controlled, and internal and external mechanical and biological constraints are reduced. Here, we used spheroids, three-dimensional cultures of self-renewing pluripotent epithelial cells (3D EpiSCs) that recapitulate the main transcriptional and architectural features of the gastrulating epiblast9. In this system, cells organize into a sphere lined by a short pseudostratified epithelium, from which cells spontaneously delaminate basally, then turn on the expression of Brachyury. As a large number of spheroids can easily be monitored overtime, they provide a unique opportunity to quantify delamination, which is very challenging in embryos.
First, we checked if non-apical mitoses were occurring in mature epiblast spheroids. The spheroids were stained for Podocalyxin-1 (lumen), Phospho-Histone H3 (mitosis), Brachyury (PS), F-actin (phalloidin), and DAPI (nuclei) (Supplementary Fig. 5a). Well-organized spheroids presenting a pseudo-stratified epithelium around a Podocalyxin-1 positive lumen were selected for quantification. Non-apical mitoses accounted for approximately 20% of all mitoses (Supplementary Fig. 5b). Interestingly, out of all Brachyury-positive mitoses, the percentage of non-apical mitoses (64%) was higher than that of apical mitoses (34%) (Supplementary Fig. 5c), indicating that non-apical mitoses are associated with PS-like features in this model as well.
3D EpiSCs were cultured with either control vehicle, 0.2 µM of Wee1 inhibitor or 30 µM of ROCK inhibitor, for 4 hours (Fig. 7a, a’). We analyzed the mitotic and non-apical mitotic indexes, as well as the proportion of epithelial and delaminated cells (Fig. 7b–e’). While Wee1 inhibition increased both the mitotic and non-apical mitotic indexes (Fig. 7b, c), ROCK inhibition did not affect the mitotic index, but raised the non-apical mitotic index (Fig. 7b’, c’). These data are consistent with the effects of the inhibitors on embryos. Furthermore, in 3D EpiSCs exposed to both drugs, the proportion of epithelial cells (Fig. 7d, d’) decreased to the benefit of cells that had exited the epithelium (“delaminated” cells, Fig. 7e, e’), particularly for spheroids treated with ROCK inhibitor (Fig. 7d’, e’). To explore this further, we performed live imaging of 3D EpiSCs spheroids expressing LifeAct-GFP, cultured in presence of a nuclear dye and either control vehicle or 0.2 µM of Wee1 inhibitor. This dynamic view illustrated the correlation between cell division and cell delamination (Supplementary Movie 4). In addition, while apical mitoses mostly gave rise to epithelial cells (Supplementary Fig. 5d; Supplementary Movie 5), non-apical mitosis could also lead to the direct exclusion of a daughter cell after cytokinesis (Supplementary Fig. 5e, f; Supplementary Movie 6). This is in line with what is observed in vivo at the PS13. These experiments show that increasing the mitotic and non-apical mitotic indexes favors the delamination of cells from the epithelium in 3D EpiSCs and suggests a similar functional role of proliferation in the mouse embryo at gastrulation.
3D EpiSCs cultured 4 hours with vehicle, 0.2 µM of Wee1 inhibitor (a) or 30 µM of ROCK inhibitor (a’), stained for F-actin (phalloidin, grey) and mitosis (Phh3, magenta). White arrowheads indicate non-apical mitosis and red arrowheads indicate delaminated cells. Scale bars: 10 µm. b—c’ Violin plots representing the mitotic (b, b’) and non-apical mitotic index (c, c’) (%) in epithelial cells of 3D EpiSCs cultured 4 hours with control vehicle, 0.2 µM of Wee1 inhibitor (b, c) or 30 µM of ROCK inhibitor (b’, c’). Inner doted lines indicate third quartile, median, and first quartile. d-e’) Boxplots representing the ratio (%) of epithelial (d, d’) and delaminated (e, e’) cells in 3D EpiSCs cultured 4 hours with control vehicle, 0.2 µM of Wee1 inhibitor (d, e) or 30 µM of ROCK inhibitor (d, e’). Boxplots span from the lower quartile to the upper quartile, with the inner line representing the median; whiskers extend to the minimum and maximum values. Normality was assessed using a Shapiro–Wilk test followed by a two-tailed Mann-Whitney or unpaired t-test. Wee 1 inhibitor incubations: control n = 29, Wee1 inhibitor 0.2 µM n = 37. ROCK inhibitor incubations: control n = 36, ROCK inhibitor 30 µM n = 46.
Discussion
At gastrulation, two novel germ layers are generated by delamination of cells from the epiblast. This process necessarily involves an increase in cell number at the site of cell exit that is superior to the one required for homogeneous embryo growth. In chick embryos, global movements in the epiblast precede gastrulation and position PS precursors along the antero-posterior axis45. The mouse PS, however, was shown to arise in situ, without large scale displacement of epiblast cells4. A higher rate of cell division at the streak therefore appears as a suitable mechanism to fuel gastrulation EMT12,13. In addition to its quantitative input, mitosis may also play a qualitative role, notably through the mechanical impact of mitotic rounding on epithelial architecture46. The apicobasal position of mitosis may be of importance, as one would instinctively postulate that cells dividing closer to the basal pole have an advantage for subsequent delamination, even if they only detach apically at the late steps of cytokinesis.
Neural crest delamination from the chick neuroepithelium is a well-studied example of EMT occurring in a middle size pseudostratified epithelium, where the impact of non-apical mitosis on cell exit was clearly demonstrated18. There, biological and computational data highlighted a role for the reduction of INM in basal positioning and subsequent basal delamination21. The correlation between alteration of INM and non-apical mitosis is however not straightforward: for instance, in chick embryo neuroepithelium, downregulation of vinculin leads to defects in apical ward nuclei movement, without causing the appearance of non-apical mitosis47. Loss of INM in EMT cells increases the probability of basal positioning, while maintenance of INM in surrounding non EMT cells results in apical crowding and could mechanically favor their delamination21. Interestingly, we find a similar pattern in the epiblast, a short pseudostratified epithelium where all cells have their nuclei relatively close to the basal pole. An additional point of similarity between those EMT events lies in the temporal regulation of INM. Indeed, non-apical mitoses were detected prior to the initiation of EMT in the pre-streak posterior epiblast13 and the prospective neural crest cells19,21. These conserved relationships between INM, mitosis position and delamination in two different epithelia presenting drastically different morphology and proliferation rates strongly suggest that it could be relevant in multiple configurations.
Apart from the acceleration of the cell cycle (particularly the G2/M phase), we found that a major factor promoting non-apical mitosis at the PS is relaxation of the actomyosin cytoskeleton. In silico simulations allowed to separately modify the tension in distinct cellular regions, which showed that apical actomyosin played little role in INM and was rather important for tissue stability. Indeed, the integrity and organization of F-actin is maintained in epiblast apical junctions throughout gastrulation, notably via ASPP248, which prevents apical delamination. In this context, it is important to note that apical mitoses fight against apical contractility20. Therefore, maintenance of normal INM at the PS would likely prevent apical constriction and the acquisition of the bottle shape morphology and subsequently lead to some apical delamination.
Epiblast cells that ingress at the PS to become mesoderm adopt a bottle shape with a constricted apical surface4,8. Delamination is a progressive process that only involves a proportion of PS cells at a given time. The asynchronous shrinkage of apical junctions is regulated by a Crumbs2-regulated anisotropy of myosin II localization6,8, and is observed in cells that express high levels of aPKC9. Similarly, cellular heterogeneity in the neural crest appears to boost epithelial destabilization and thereby favor delamination21.
In addition to the position of nuclei, polarized protrusive activity mediated by the interaction between α4 and 5 integrins in neural crest cells and the extracellular matrix promotes basal delamination21. Likewise, nascent mesoderm displays numerous projections mostly oriented towards the visceral endoderm in the PS, then towards the front in the mesodermal wings. Those protrusions are required for directional migration, as demonstrated by the mesoderm migration defects identified in mouse embryos deficient for PTK7, Rac1, Nap1, or β-Pix22,49,50,51.
To circumvent the technical challenge of quantifying delamination in the mouse PS, we leveraged the flexibility of computational modelling and simplified stem cell-based models. We used 3D EpiSCs (“spheroids”), an epiblast model recapitulating most PS cells characteristics9, while being subjected to less regulation since it lacks extraembryonic tissues. Spheroids allowed establishing a role for proliferation-mediated crowding in the basal delamination of atypical Protein Kinase C (aPKC) high cells 9. Here, through pharmacological treatment of 3D EpiSCs, we confirmed a role for proliferation in favoring delamination, and added mechanistic insights by demonstrating that acceleration of the cell cycle and relaxation of actomyosin favor the appearance of non-apical mitosis, and ultimately cell exit from the epithelium.
Through combining ex vivo and in silico experiments, we found that gastrulation EMT coincides with relaxation of the cytoskeleton and changes in cell cycle regulation at the PS. These local changes disrupt the synchronization between apical nuclei movements and mitosis, leading to an elevated occurrence of non-apical mitoses and basal positioning of nuclei. This orchestration may ensure the proper progression of gastrulation while maintaining the integrity of the epithelium (Fig. 8) and may be an evolutionarily conserved strategy common to other events involving EMT in pseudo-stratified epithelia21.
Diagram representing the state of the cell cycle, actomyosin contractility, and basal attachment in the regions of the epiblast and the PS during gastrulation EMT in the mouse embryo. At the PS, faster cell cycle and shortened G2 duration, combined with weak cytoplasmic actomyosin contractility and basal detachment of cells, leads to a high frequency of non-apical mitosis that contribute to basal delamination.
Methods
Mouse breeding and genotyping
All experiments involving mice performed in Belgium included mouse colonies maintained in a certified animal facility in accordance with European guidelines. Experiments were approved by the local ethics committee (“Commission d’éthique et du bien-être animal”) under protocols 576 N, 725 N, 874 N and 893 N. Mouse lines were CD1 (Janvier Labs), mTmG52, Sox2-Cre42, RhoA39, ECT240, bred on a CD1 background, CDK141 mice were bred on a C57BL/6 background. FLRT323 mice were bred on C57BL/6 background and housed following proper guidelines in accordance with the Spanish National guidance and care for laboratory animals system and the guidelines set by Universitat de Lleida.
Mouse genomic DNA was isolated from ear biopsies treated for 1 h at 95 °C in 0.05 M NaOH to simultaneously genotype and identify animals.
For embryo genotyping, embryos were digested in Direct PCR mix (Viagen #102-T) with 1.5% Proteinase K (Quiagen #19133) for 2 hours at 56 C°, followed by 30 minutes at 90 C°.
The following sequences of primers were used for genotyping:
Sox2-cre: (F) TGCTGTTTCACTGGTTATGCTG, (R) TTGCCCCTGTTTCACTATCCAG.
mTmG: (1) AAA GTC GCT CTG AGT TGT TAT, (2) GGA GCG GGA GAA ATG GAT ATG, (3) TCA ATG GGC GGG GGT CGT T.
RhoA fl: (1) AGC CAG CCT CTT GAC CGA TTT A, (2) TGT GGG ATA CCG TTT GAG CAT.
RHOA KO: (1) AGG CAT GGA CCA CCA TGT CA, (2) ATG TGC TTC CCG TGT CTA GT.
ECT FL: (1) GCA CTC CAA TTA TGA AGC (2) CAA TAT GTT GGG TAG AGA GAT GGC.
ECT KO: (1) CAA TAT GTT GGG TAG AGA GAT GGC (2) TCC TCC GGG TGG ACC AGA G.
CDK1 FL: (F) CCA GGG TGA CCT TGT CGT, (R) AGC CTG CCT CCA CTT CCA.
CDK1 KO: (1) TTCTCCACGCTTGTCTCCAA (2) CAGCTTTAGGAGTGCAGGC.
For embryos from the FLRT3 heterozygous cross, genotyping was performed by immunofluorescence with an anti-FLRT3 antibody.
Antibodies
Antibodies for mouse embryo staining were: Goat anti-collagen IV (MERK #AB769, 1:500), rabbit anti-Phh3 (Sigma #SAB4504429, 1:500), rabbit anti-E-cadherin (Cell signaling #3195S, 1:500), rabbit anti-laminB1 (abcam #ab229025, 1:250), sheep anti N-cadherin (R&D #AF64216, 1/100), rabbit anti-vimentin (abcam #ab92547,1:200), goat anti-Brachyury (R&D #AF2085, 1/100), rabbit anti-caspase3 (R&D # AF835, 1:250), goat anti-Snail (R&D #AF3639, 1:100), goat anti-FLRT3 (R&D #AF2795, 1:250). F-actin was visualized using rhodamine phalloidin (abcam #ab235138, 1:1000), and nuclei using DAPI (Sigma; 1:1000). Secondary antibodies were anti-goat Alexa Fluor 647 (Invitrogen #A21447, 1:500), anti-rabbit Alexa Fluor 488 (Invitrogen #A21206, 1:500), anti-sheep Alexa fluor 647 (Invitrogen #A11016, 1:500), anti-rabbit Alexa Fluor 647 (Jackson #711-605-152, 1:500) and anti-goat Alexa Fluor 488 (Invitrogen #A32814, 1:500).
Antibodies for 3D EpiSCs staining were rabbit anti-Phh3 (Sigma 06-570 1:1000), goat anti-Brachyury (R&D #AF2085, 1/250), rat anti-Podocalyxin (R&D system MAB1556 1:400).
Embryo recovery, staging, and pharmacological treatment
Embryos were recovered at the appropriate time point after observation of a vaginal plug at day 0. E6.5 and E7.5 embryos were dissected in dissection medium using #5 forceps and tungsten needles under a transmitted light stereomicroscope. Dissection medium was composed of Dulbecco’s modified Eagle medium (DMEM) F-12 supplemented with 10 mM HEPES and L-glutamin (Thermofisher, #11039047) with 10% Fetal Bovine Serum (Thermofisher, #10270106) and 1% Penicillin/Streptomycin (P/S). Bright-field pictures of the litter or single embryo were taken before any manipulation to ensure adequate staging. Deciduae from heterozygous FLRT3 +/- crosses were extracted at E6.5 and directly fixed in PBS containing 4% paraformaldehyde (PFA) overnight.
For pharmacological treatment, E6.5 embryos were allowed to recover in equilibrated culture medium (50% DMEM F12 (Thermofisher,# 21041025), 50% rat serum (Janvier), 1% P/S) in an incubator (37 °C, 5% CO2) for 1 h. Embryos were then transferred to 15-wells ibidi (#81507) with 30 µl of equilibrated culture medium containing inhibitors or control vehicle depending on what chemical the inhibitor was diluted in. Bright-field pictures were taken at t0 and after the incubation with inhibitors to assess embryo growth. For the post-treatment survival test, medium was replaced with fresh culture medium and embryos were cultured over night at 37 C°.
Inhibitors were: PD166285 (Wee1 inhibitor, MECK #PZ0116, diluted in dH20), R03366 (CDK1 inhibitor, Sigma #217699 in DMSO), Y27632 (Rock inhibitor, Abcam #ab120129 in dH20 or DMSO), Prinomastat hydrochloride (Pan-MMPs inhibitor, Sigma #PZ198 in dH20), NSC405020 (MMP14 inhibitor, Tocris #4902/10 in ethanol).
Immunofluorescence on mouse embryos
For immunofluorescence, embryos were fixed in 4% PFA for 2 h at 4 C°. Embryos were cryopreserved in 30% sucrose, embedded in OCT and sectioned at 7 µm. Staining was performed in PBS containing 0.5% Triton X-100, 0.1% BSA and 5% heat-inactivated horse serum. For whole mount, embryos were imaged in PBS 1X. Alternatively, to enhance optical clarity, E6.5 and E7.5 were treated in 35 µl of Refraction Index Matching (RIM), as described in ref. 53. For proximal-distal oriented whole-mount, embryos were embedded in warm 0.5% low melting agarose dissolved in PBS 1X, in a 15-well ibidi. Sections and whole-mount embryos were imaged on a Zeiss LSM 780 microscope equipped with Plan-Apochromat 25×/0.8, C Achroplan 32×/0.85 and LD C Apochromat 40×/1.1 objectives.
3D EpiSC generation
3D epiblast spheroids were generated following the protocol described in ref. 9. Briefly, 40,000 mouse ESC were resuspended in 50 µL of 100% ice-cold growth factor-reduced Matrigel (356231, Corning) and plated on a 24 non-adherent multi-well plate (662 102, CellStar). The drops were incubated for 10 minutes at 37 °C and 600 μL of 3D EpiSC medium was added per well. 3D EpiSC medium (FAXN) contained N2B27 supplemented with 12 ng/ml of bFGF2 (Marko Hyvonen lab, University of Cambridge), 50 ng/ml of Activin-A (Marko Hyvonen lab, University of Cambridge), 5 μM XAV939 (X3004, Sigma) and 150 ng/ml of mouse noggin (78061, STEMCELL Technologies). N2B27 contained a 1:1 mix of DMEM F12 (21331-020, Thermo Fisher Scientific) and Neurobasal A (10888-022, Thermo Fisher Scientific) supplemented with 1% v/v B27 (10889-038, Thermo Fisher Scientific), 0.5% v/v N2 (homemade), 100 μM β -mercaptoethanol (31350-010, Thermo Fisher Scientific), penicillin-streptomycin (15140122, Gibco) and GlutaMAX (35050061, Thermo Fisher Scientific).
3D EpiSCs were passaged at a splitting ratio of 1 to 4. Matrigel was depolymerized by incubating the plates with 1 ml of dispase (07923, STEMCELL Technologies) at 37 °C for 20 minutes. Spheroids were dissociated by incubation with 1 mL of TrypLE (12604-021, Gibco) at 37 °C for 3 minutes. The resulting cells were resuspended in cold Matrigel.
Inhibitor treatment of 3D EpiSCs
Spheroids were dissociated and plated at a ratio of 1:4 on µ-Slide 8-well (80827, Ibidi) plates pre-coated with 40 µl of Matrigel (growth factor reduced phenol red free, Corning CLS356231-1EA) with 250 µl of N2B27 medium. After 15 minutes incubation at 37 C°, N2B27 medium was removed and replaced with 3D EpiSC medium supplemented with 5% of Matrigel and placed at 37 C°. After 48 hours in culture, 3D EpiSCs were treated with 0.2 µM of PD16685, 30 µM of Y27632 or an equivalent dose of DMSO for 4 hours. After the incubation, cells were washed three times with PBS 1X and fixed in PFA 4%.
Immunofluorescence on 3D EpiSC
Cells were fixed using PFA 4% (15710, Electron Microscopy Sciences) diluted in PBS for 20 minutes at room temperature and then washed three times with PBS plus 0.1% Tween. The permeabilization step was performed in PBS containing 0.3% Triton X-100 and 0.1 M glycine for 30 minutes at room temperature. The samples were incubated with primary antibodies overnight at 4 °C. The day after, samples were washed with PBS plus 0.1% Tween three times, followed by incubation with secondary antibodies Alexa 555 (1:500), Alexa 488 (1:500) Alexa 647 (1:500), Phalloidin 594 (1:500) and DAPI (1:1000), diluted in blocking solution for 2 hours at room temperature. The blocking solution contained 3% BSA and 0.1% Tween diluted in PBS. Images were acquired on an inverted SP8 confocal microscope (Leica Microsystems) with a Leica 40x/1.1NA Water objective.
Live imaging on LifeAct-GFP 3DEpiSCs
For live imaging, LifeAct-GFP Brachyury IRES H2B-mCherry mouse ES cells were used. 3D EpiSCs were plated on a well of a μ-Slide 8-well glass bottom plate (80827, Ibidi) using the protocol described above for inhibitor treatment. After 48 h, 3D EpiSCs were incubated with a nuclear dye (Spy 650 DNA) diluted at 1:1000 for 1 hour. The medium was then changed to apply a lower concentration of the dye (1:10000) as well as the pharmacological treatment (0.2 µM of PD16685 or an equivalent dose of DMSO), and 3D EpiSCs were recorded for 4 hours using a Nikon W1 Spinning Disk microscope (CSU-W1, Nikon) equipped with a 25x/1.05NA Silicone Oil lens and a heated incubation chamber at 37 °C and 5% CO2. Stacks were acquired every 5 minutes with a 2 µm Z step and analyzed using Image J Fiji software.
Image analysis
Images were processed using Arivis Vision4D v2.12.3 (Arivis, Germany), Image j Fiji and Icy software v2.5.2.0 (http://icy.bioimageanalysis.org).
Mitotic index and non-apical mitotic index statistical analysis were performed as described in ref. 13. Mitosis was considered “non-apical” when happening at least 10 µm away from the apical pole and not in the first pseudo-layer of nuclei lining the apical pole. Only cells still attached to the apical pole, and that did not cross the limit of basement membrane discontinuity, were considered in the measurements.
For transverse sections, anterior-posterior boundary was placed at mid-distance between the anterior and posterior poles. The PS region was defined by the area where the basement membrane was discontinuous, and the posterior region quantification excluded counts from the PS region. A cell was counted as being part of the PS region if at least 50% of its cell body was within the area where the basement membrane was non-ambiguously degraded, and if the cell retained its attachment to the apical pole (cell contours were defined by F-actin detection using Phalloidin). Phospho-histone H3 labelling was used to count cell in mitosis, normalised by the number of nuclei (DAPI staining) in each region. Non-apical mitosis ratio was calculated by dividing the number of non-apical Phh3+ nuclei by the total number of Phh3+ nuclei in a subregion of the epiblast. The percentage of basement degradation was calculated using Collagen IV labelling, through dividing the length of the area where the membrane was degraded by the total length of the basement membrane around the epiblast and conversion to percentage. Nuclei aspect ratio was calculated using Lamin B1 and DAPI labelling, by diving nuclei short axis by their long axis.
For FLRT3KO analysis, since FLRT3 deficiency may trigger ectopic degradation of the basement membrane, we relied on the position of the AVE (labeled with Cerberus 1) rather than collagen IV pattern to orient sections in the anteroposterior axis. The epiblast was divided into two regions: the anterior 2/3rd, approximating the anterior and posterior epiblast, adjacent to the Cerberus 1 positive VE region; and the posterior 1/3rd, opposite to the AVE, approximating the position of the PS. We then quantified the number of mitoses and non-apical mitoses in each region, normalized by the area in µm2. In sections where ectopic delamination sites were observed, we quantified the number of apical, non-apical, and delaminated mitoses, normalized by the area in µm2 of the ectopic delamination site. Delaminated mitoses are defined as Phh3-positive cells that have lost attachment to both the apical and basal poles and are no longer part of the pseudo-stratified epithelium.
For 3D EpiSCs analysis, cells were classified as epithelial when they were part of the main structure and connected to the apical and basal surfaces. A cell was considered delaminated if no clear connection to the apical and basal surfaces was observed, and such cells were excluded from the epithelial architecture. The epithelial mitosis ratio was calculated by dividing either the number of apical or non-apical mitoses by the total number of mitoses, and the result was converted into a percentage. Epithelial mitotic and non-apical mitotic indexes were calculated using the same method as employed for embryo samples.
The epithelial cell ratio was determined by dividing the number of epithelial cells by the total number of cells, including delaminated cells, and expressing it as a percentage. The delaminated cells ratio was calculated similarly.
For each population, normality was assessed using a Shapiro–Wilk test. According to the results of the precedent test, samples were compared using a non-parametric Mann–Whitney test or an unpaired t-test. For cross statistics, either Anova or Kruskall-Wallis followed by Dunn post-tests were performed. Ns: non-significant, *P-value ≤ 0.05, **P-value ≤ 0.01 and ***P-value ≤ 0.001. Graphs and statistics were obtained using Spyder environment (3.9) python code and prism 5.0.4.
In-Silico modeling
The complete details of the model are described in ref. 21. Parameters used to adjust the in silico tissue to the epiblast configuration are mentioned throughout the main text. Variability in simulations comes from multiple inputs. Briefly, the initial state of all simulations is generated as follows. First, the basal and apical points are equidistantly distributed with distance 7.5 µm and apical-basal distance 25 µm. Then, the nuclei positions are randomly seeded in a central area of size 50 µm ⨯ 8.325 µm. The pseudostratified organization emerges after a few simulation steps. We assign to each cell a random lifespan (depending on the cell cycle parameters) at simulation start. We also assign a random age within that lifespan to ensure that cells are asynchronous. When the lifespan is reached, a cell divides. In 95% of the cases division leads to one offspring and in 5% of the cases to two offspring, due to dimensional correction of only considering a 2D slice of a 3D tissue. To account for random forces, we add in each time-step a small amount of additive noise to the nuclei positions (white noise with σ² = 5 µm²/h). For PS cells that lose their basal attachment (event B), the time of occurrence is selected randomly between 6 h and 24 h after simulation start. Each spring (aa, aN, bN) and the soft repulsion (NN) is assigned a strength value. Strengths of springs are expressed in arbitrary unit. As such, they do not correspond to actual values and what matters is their relative strength to one another.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The link to access the online simulator for the model is indicated in the code availability section. Source data are provided with this paper.
Code availability
In silico simulations used a custom code which is open source at https://github.com/SteffenPL/sEMTor.jl. In silico data were analyzed with Julia (v1.6), the code used for analysis is available open source at https://doi.org/10.24433/CO.7746744.v1. The online simulator for the model is available at https://semtor.github.io/gastrulation/.
References
Nieto, M., Huang, R. Y., Jackson, R. A. & Thiery, J. P. Review EMT: 2016. Cell 166, 21–45 (2016).
Yang, J. et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020).
Nahaboo, W. & Migeotte, I. Cleavage and Gastrulation in the Mouse Embryo. eLs. (John Wiley & Son, Ltd: Chichester, 2018). https://doi.org/10.1002/9780470015902.a0001068.pub3.
Williams, M., Burdsal, C., Periasamy, A., Lewandoski, M. & Sutherland, A. Mouse primitive streak forms in situ by initiation of epithelial to mesenchymal transition without migration of a cell population. Developmental Dyn. 241, 270–283 (2012).
Kyprianou, C. et al. Basement membrane remodelling regulates mouse embryogenesis. Nature 582, 253–258 (2020).
Francou, A., Anderson, K. V. & Hadjantonakis, A.-K. A ratchet-like apical constriction drives cell ingression during the mouse gastrulation EMT. Elife 12, e84019 (2023).
Carver, E. A., Jiang, R., Lan, Y., Oram, K. F. & Gridley, T. The Mouse Snail Gene Encodes a Key Regulator of the Epithelial-Mesenchymal Transition. Mol. Cell Biol. 21, 8184–8188 (2001).
Ramkumar, N. et al. Crumbs2 Promotes Cell Ingression During the Epithelial-to-Mesenchymal Transition at Gastrulation. Nat. Cell Biol. 18, 1281–1291 (2016).
Sato, N. et al. Basal delamination during mouse gastrulation primes pluripotent cells for differentiation. Dev. Cell 59, 1–17 (2024).
Snow, M. H. L. & Tam, P. P. L. Is compensatory growth a complicating factor in mouse teratology? Nature 279, 555–557 (1979).
Lewis, N. E. & Rossant, J. Mechanism of Size Regulation in Mouse Embryo Aggregates. J. Embryol Exp. Morph. 72, 169–181 (1982).
Snow, M. H. L. Gastrulation in the mouse: Growth and regionalization of the epiblast. Development 42, 293–303 (1977).
Mathiah, N. et al. Asymmetry in the frequency and position of mitosis in the mouse embryo epiblast at gastrulation. EMBO Rep. 21, e50944 (2020).
Mohammed, H. et al. Single-Cell Landscape of Transcriptional Heterogeneity and Cell Fate Decisions during Mouse Early Gastrulation. Cell Rep. 20, 1215–1228 (2017).
MacAuley, A., Werb, Z. & Mirkes, P. E. Characterization of the unusually rapid cell cycles during rat gastrulation. Development 117, 873–883 (1993).
Strzyz, P. J., Matejcic, M. & Norden, C. Heterogeneity, Cell Biology and Tissue Mechanics of Pseudostratified Epithelia: Coordination of Cell Divisions and Growth in Tightly Packed Tissues. Int. Rev. Cell. Mol. Biol.325, 89–118 (2016).
Ichikawa, T., Nakazato, K., Keller, P. J., Kajiura-Kobayashi, H. & Stelzer, E. H. K. Live Imaging of Whole Mouse Embryos during Gastrulation: Migration Analyses of Epiblast and Mesodermal Cells. PLoS One 8, 64506 (2013).
Andrieu, C. et al. MMP14 is required for delamination of chick neural crest cells independently of its catalytic activity. Development 147, dev183954 (2020).
Ahlstrom, J. & Erickson, C. The neural crest epithelial-mesenchymal transition in 4D: a ‘tail’ of multipke non-obligatory cellular mechanisms. Development 136, 1801–1812 (2009).
Ferreira, A. M., Despin-Guitard, E., Duarte, F., Degondid, P. & Theveneauid, E. Interkinetic nuclear movements promote apical expansion in pseudostratified epithelia at the expense of apicobasal elongation. PLoS Comput Biol. 15, e1007171 (2019).
Plunder, S. et al. Modelling variability and heterogeneity of EMT scenarios highlights nuclear positioning and protrusions as main drivers of extrusion. BioXiv https://doi.org/10.1101/2023.11.17.567510 (2023).
Migeotte, I., Grego-bessa, J. & Anderson, K. V. Rac1 mediates morphogenetic responses to intercellular signals in the gastrulating mouse embryo. Development 138, 3011–3020 (2011).
Egea, J. et al. Genetic ablation of FLRT3 reveals a novel morphogenetic function for the anterior visceral endoderm in suppressing mesoderm differentiation. Genes Dev. 22, 3349–3362 (2008).
Haase, K. et al. Extracellular Forces Cause the Nucleus to Deform in a Highly Controlled Anisotropic Manner. Sci Rep 6, 21300 (2016).
Brandt, A. et al. Developmental control of nuclear size and shape by kugelkern and kurzkern. Curr. Biol. 16, 543–552 (2006).
Lee, H. O. & Norden, C. Mechanisms controlling arrangements and movements of nuclei in pseudostratified epithelia. Trends Cell Biol. 23, 141–150 (2013).
Sauer, F. C. The interkinetic migration of embryonic epithelial nuclei. J. Morphol. 60, 1–11 (1936).
Vergnes, L., Terfy, P., Bergo, M. O., Young, S. G. & Reue, K. Lamin B1 Is Required for Mouse Development and Nuclear Integrity. PNAS 101, 10428–10433 (2004).
Thorpe, S. D. & Lee, D. A. Dynamic regulation of nuclear architecture and mechanics—a rheostatic role for the nucleus in tailoring cellular mechanosensitivity. Nucleus 8, 287–300 (2017).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014).
Amano, M. et al. Phosphorylation and Activation of Myosin by Rho-associated Kinase (Rho-kinase). J. Biol. Chem. 271, 20246–20249 (1996).
Amin, E. et al. Rho-kinase: regulation, (dys)function, and inhibition HHS Public Access. Biol. Chem. 394, 1399–1410 (2013).
Meyer, E. J., Ikmi, A. & Gibson, M. C. Interkinetic Nuclear Migration Is a Broadly Conserved Feature of Cell Division in Pseudostratified Epithelia. Curr. Biol. 21, 485–491 (2011).
Lukaszewicz, A. I. & Anderson, D. J. Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. PNAS 108, 11632–11637 (2011).
Strzyz, P. J., Lee, H. O., Leung, L. C. & Norden, C. Interkinetic Nuclear Migration Is Centrosome Independent and Ensures Apical Cell Division to Maintain Tissue Integrity. Dev. Cell 32, 203–219 (2015).
Hashimoto, O. et al. Cell cycle regulation by the Wee1 Inhibitor PD0166285, Pyrido [2,3-d] pyimidine, in the B16 mouse melanoma cell line. BMC Cancer 6, 292 (2006).
Rose, J. T. et al. Inhibition of the RUNX1-CBFβ transcription factor complex compromises mammary epithelial cell identity: a phenotype potentially stabilized by mitotic gene bookmarking. Oncotarget 11, 2512–2530 (2020).
Zaidi, S. K., Lian, J. B., Vanwijnen, A. J., Stein, J. L. & Stein, G. S. Mitotic Gene Bookmarking: An Epigenetic Mechanism for Coordination of Lineage Commitment, Cell Identity and Cell Growth. Adv. Exp. Med. Biol. 292, 95–102 (2017).
Jackson, B. et al. RhoA is dispensable for skin development, but crucial for contraction and directed migration of keratinocytes. MBoC 22, 593–605 (2011).
Cook, D. R. et al. The Ect2 Rho Guanine Nucleotide Exchange Factor Is Essential for Early Mouse Development and Normal Cell Cytokinesis and Migration. Genes Cancer 2, 932–942 (2011).
Chaffee, B. R. et al. Nuclear removal during terminal lens fiber cell differentiation requires CDK1 activity: appropriating mitosis-related nuclear disassembly. Development 141, 3388–3398 (2014).
Hayashi, S., Lewis, P., Pevny, L. & McMahon, A. P. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Gene Expr. Patterns 2, 93–97 (2002).
Saykali, B. et al. Distinct mesoderm migration phenotypes in extra-embryonic and embryonic regions of the early mouse embryo. Elife 8, e42434 (2019).
Shahbazi, M. N. & Zernicka-Goetz, M. Deconstructing and reconstructing the mouse and human early embryo. Nat. Cell Biol. 20, 878–887 (2018).
Voiculescu, O. Movements of chick gastrulation. Curr. Top. Dev. Biol. 136, 409–428 (2020).
Despin-Guitard, E. & Migeotte, I. Mitosis, a springboard for epithelial-mesenchymal transition? Cell Cycle 20, 2452–2464 (2021).
Ochoa, A. et al. Vinculin is required for interkinetic nuclear migration (INM) and cell cycle progression. J. Cell Biol. 223, e2021106169 (2024).
Royer, C et al. ASPP2 maintains the integrity of mechanically stressed pseudostratified epithelia during morphogenesis. Nat. Com. 13, 941 (2022).
Omelchenko, T. Cellular protrusions in 3D: Orchestrating early mouse embryogenesis. Semin Cell Dev. Biol. 129, 63–74 (2022).
Rakeman, A. S. & Anderson, K. V. Axis specification and morphogenesis in the mouse embryo require Nap1, a regulator of WAVE-mediated actin branching. Development 133, 3075–3083 (2006).
Yen, W. W. et al. PTK7 is essential for polarized cell motility and convergent extension during mouse gastrulation. Development 136, 2039–2048 (2009).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, N. & Luo, L. A global double-fluorescent Cre reporter mouse. genesis 45, 593–605 (2007).
Despin-Guitard, E. et al. Regionally specific levels and patterns of keratin 8 expression in the mouse embryo visceral endoderm emerge upon anterior-posterior axis determination. Front. Cell Dev. Biol. 10, https://doi.org/10.3389/fcell.2022.1037041 (2022).
Acknowledgements
We wish to thank the Université Libre de Bruxelles/Erasme animal facility. We gratefully acknowledge the Université Libre de Bruxelles light microscopy (LiMiF) core facility (M. Martens and Pr. J-M. Vanderwinden) for help with confocal imaging. E.D.G. received a FRIA fellowship of the Fonds de la Recherche Scientifique (FNRS) and a grant from the “Fondation Alice and David Van Buuren” and “Fondation Jaumotte-Demoulin”. S.P. was partially funded by the Vienna Science and Technology Fund (WWTF) [10.47379/VRG17014]. K.V.S. is supported by a PDR grant of the FNRS, and E.N. by a FRIA fellowship. I.M. is a FNRS senior research associate. E.T. is a research director at the French National Research Center (CNRS). V.S.R is supported by a Milstein fellowship. This work was supported by PDR T.0084.16 and CDR J.0154.20 F from the FNRS to I.M., PJA22020060002084 from Association pour la Recherche contre le Cancer (ARC) and ANR Single Crest to E.T. Work in the Shahbazi lab was supported by the Medical Research Council, as part of United Kingdom Research and Innovation (MC_UP_1201/24) and the Engineering and Physical Sciences Research Council (Horizon Europe guarantee funding, EP/X023044/1).
Author information
Authors and Affiliations
Contributions
E.D.G., I.M. and E.T. conceptualized the study. E.D.G. performed and analyzed experiments on mouse embryos. S.P., S.M.A. and E.T. developed the computational model. E.T. designed, performed and analyzed simulations. S.P. developed the stand-alone EMT simulator. Experiments on 3D EpiSCs were designed by V.S.R. and M.S., performed by V.S.R. and analyzed by E.D.G. N.M. performed experiments on ECT2 and CDK1 epiblast specific mutant embryos, and N.M. and E.D.G. performed and analyzed experiments on RhoA epiblast-specific mutants. J.E. generated and provided embryos from the FLRT3KO line. E.D.G., K.V.S., and E.N. performed immunostaining on FLRT3 deficient embryos, and E.D.G. analyzed and quantified the images. E.D.G., I.M. and E.T. wrote the original draft, which was reviewed and edited by S.P., V.S.R., N.M., J.E. and M.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Despin-Guitard, E., Rosa, V.S., Plunder, S. et al. Non-apical mitoses contribute to cell delamination during mouse gastrulation. Nat Commun 15, 7364 (2024). https://doi.org/10.1038/s41467-024-51638-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51638-6