Abstract
Intermediate-length repeat expansions in ATAXIN-2 (ATXN2) are the strongest genetic risk factor for amyotrophic lateral sclerosis (ALS). At the molecular level, ATXN2 intermediate expansions enhance TDP-43 toxicity and pathology. However, whether this triggers ALS pathogenesis at the cellular and functional level remains unknown. Here, we combine patient-derived and mouse models to dissect the effects of ATXN2 intermediate expansions in an ALS background. iPSC-derived motor neurons from ATXN2-ALS patients show altered stress granules, neurite damage and abnormal electrophysiological properties compared to healthy control and other familial ALS mutations. In TDP-43Tg-ALS mice, ATXN2-Q33 causes reduced motor function, NMJ alterations, neuron degeneration and altered in vitro stress granule dynamics. Furthermore, gene expression changes related to mitochondrial function and inflammatory response are detected and confirmed at the cellular level in mice and human neuron and organoid models. Together, these results define pathogenic defects underlying ATXN2-ALS and provide a framework for future research into ATXN2-dependent pathogenesis and therapy.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disorder that is characterized by degeneration of lower and upper motor neurons (MNs), leading to progressive loss of motor function and ultimately death1,2,3,4. ALS is a heterogeneous disease, and as a result, the development of effective treatments has been challenging. However, the discovery of various pathogenic gene mutations has linked ALS to defects in different biological processes and has served as a starting point for drug development5,6.
About 5–10% of ALS patients can be classified as having familial (f)ALS because of the demonstration of direct inheritance. The remaining patients are classified as having sporadic (s)ALS7. Currently, mutations in >40 genes have been reported to explain a large proportion of fALS cases, and genetic defects are also found in 5–17% of sALS patients8,9,10,11. Pathogenic mutations include disease-causing genetic changes, but also those that confer increased risk or that have disease-modifying effects. Further, they range from single-point mutations to expansions of repeat sequences. Repeat expansions in different genes have been associated with ALS12,13,14,15,16, but, with the exception of hexanucleotide repeat expansions in C9ORF72, their contribution to ALS pathogenesis is poorly understood.
Intermediate-length repeat expansions in ATAXIN-2 (ATXN2) are one of the strongest genetic risk factors for ALS, reported to have a frequency of 4,7% in ALS patients12. ATXN2 repeat expansions were initially linked to spinocerebellar ataxia type 2 (SCA2)17,18 and were subsequently associated with other neurodegenerative diseases (including ALS19) depending on repeat length and codon usage20. ATXN2 is a ubiquitously expressed cytoplasmic protein that regulates cellular metabolism, growth, and stress21. It has a polyglutamine (polyQ) repeat sequence in its N-terminal region, which is 22–23 repeats long in healthy individuals. Expansion of this region to 27–33 repeats significantly and consistently enhances the risk of developing ALS, with risk increasing exponentially with allele repeat size12,19,22,23,24,25,26,27,28. While these diseases share a genetic defect, differences exist in how ATXN2 is affected, and how the repeat expansions affect downstream pathways29,30. Interestingly, experimental reduction of ATXN2 in transgenic TDP-43 mice extends lifespan, reduces ALS pathology, and improves motor function, and ATXN2-directed antisense oligonucleotides (ASOs) have strong therapeutic effects in both ALS and SCA2 mouse models31,32. These studies identify ATXN2 as a therapeutic target in ALS. However, ATXN2 is required for several physiological processes, and ATXN2 loss-of-function (LOF) may contribute to the disease process in ALS and FTLD-TDP31,33. Therefore, further insight into how ATXN2 and ATXN2 repeat expansions contribute to ALS pathogenesis is needed to refine therapeutic strategies.
ATXN2 regulates various molecular processes including RNA processing, stress granule (SG) dynamics, and metabolic homeostasis. SCA2-associated ATXN2 repeat expansions (>34 repeats) induce several LOF and toxic gain-of-function (GOF) phenotypes linked to these physiological functions34,35,36,37,38,39,40. Interestingly, ATXN2 intermediate repeats found in ALS genetically and biochemically interact with and influence several other ALS-associated proteins, including TDP-43, a protein central to ALS pathology. ATXN2 intermediate expansions enhance TDP-43 pathological modification and toxicity in vitro, and affect TDP-43-positive inclusions in ALS patients in vivo12,29,41,42,43. However, whether or how this interaction triggers ALS pathogenesis at the cellular and functional level is unknown.
Here, we combined iPSC-derived MNs and brain organoids derived from ALS patients and newly generated mouse models carrying ATXN2 intermediate expansions (ATXN2-ALS) to define the pathogenic mechanisms downstream of ATXN2 intermediate expansions. This work unveiled several molecular, cellular, and motor function defects, including changes in mitochondrial function and microglia biology. Together, our data show that ATXN2 intermediate expansions can uncover and exacerbate several ALS-relevant phenotypes providing a future framework for better understanding and targeting (mutant) ATXN2 in ALS.
Results
ATXN2-ALS hMNs display early signs of MN disease in vitro
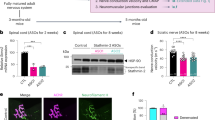
To define the pathogenic mechanisms underlying ATXN2 intermediate expansions in ALS, iPSCs were generated from different healthy control individuals (CTL) and ATXN2-ALS patients (Table S1; Fig. S1) and differentiated into hMN cultures (Figs. 1A–D, S2A–F). Previous studies using iPSC-derived MNs harboring different ALS mutations have described multiple ALS-specific phenotypes (for review see refs. 44,45). Therefore, cultures were analyzed using these observations as a starting point. No differences in soma size were observed between DIV12 CTL and ATXN2-ALS hMNs (Fig. S2G). However, ultrastructural analysis did reveal a significant increase in neurite damage in ATXN2-ALS cultures (Fig. 1E, F). ATXN2 is an RNA-binding protein that regulates SG assembly and recruitment of TDP-43 into SGs21. Therefore, SGs were assessed upon treatment with sodium arsenite (ARS) at DIV9. Cultures were treated with ARS for 1 hour, and allowed to recover for 2 hours. The number of hMNs with PABP+ SGs was similar between CTL and ATXN2-ALS cultures (Fig. S2H). However, 1 hour after ARS treatment ATXN2-ALS hMNs contained significantly fewer SGs (Fig. 1G, H). This difference was no longer observed after 2 hours of recovery (Fig. 1H). The total SG area was unchanged between CTL and ATXN2-ALS hMNs at 60 minutes (min) following ARS (Fig. 1I, Supplementary Data 1). However, whereas the total SG area in CTL neurons decreased in the recovery phase, total SG area in ATXN2-ALS neurons remained unchanged (Fig. 1I, Supplementary Data 1). Further analysis revealed that individual SGs were reduced in size in control neurons during recovery, while this effect was less pronounced in ATXN2-ALS neurons (Fig. 1I, J). As we, for technical reasons, were unable to generate isogenic control lines from the ATXN2-ALS lines (to generate patient lines carrying 22 instead of 33 repeats), we assessed whether hMNs carrying another ALS-associated genetic variant showed SG defects similar to those found in ATXN2-ALS hMNs. We selected hMNs carrying FUS mutations as a large number of studies have reported ALS-related phenotypes in FUS-ALS hMNs, including changes in SG dynamics46. Analysis of SG (dis)assembly in hMNs carrying ALS-associated mutations in FUS and CTL hMNs (Table S1) did not show changes in PABP+ SG number during treatment (assembly) or recovery (disassembly) (Fig. S2I, J). This lack of SG phenotypes, as compared to previous work46, may be explained by experimental differences, including patient mutations studied, species used, or the number of days in culture. Regardless, our data show that under similar experimental circumstances, ATXN2-ALS but not FUS-ALS hMNs show SG defects at DIV9. Given the SG defects detected in ATXN2-ALS hMNs, we also examined the subcellular distribution of TDP-43 and ATXN2. In line with the fact that protein mislocalization is generally not a prominent feature of hMN cultures, TDP-43 and ATXN2 localization were intact (Figs. 1K and S2K, L). Finally, to determine whether the different phenotypes detected in ATXN2-ALS hMNs have functional consequences, whole-cell patch clamping was performed. Interestingly, at DIV12 ATXN2-ALS hMNs were hypoexcitable, but at DIV24 hyperexcitable (Figs. 1L, M and S2M–P).
A, B Immunocytochemistry for TUJ1, CHAT, and ISL1 on human iPSC-derived hMN cultures. Scale bar is 200 µm. C Current-clamp recording of CTL hMNs following current injections at DIV12. D t-SNE map of scRNAseq experiments on hMN cultures showing individual cells and cluster numbers assigned by RaceID2, where 60% of the cells correspond to motor neurons. n = 384 cells divided in 4 libraries from 1 control (CTL_2) and 1 ATXN2-ALS (ALS_G) line (n = 1 differentiation). E, F Representative scanning electron microscopy images of DIV12 control (CTL) and ATXN2-ALS hMN cultures. CTL MNs contain smooth neurite thickenings (white arrowhead) whereas ATXN-ALS cultures show ruffled and damaged structures (black arrowhead). Scale bar is 2 µm. F Quantification of damaged neurites as in E. n = 149 neurites from 3 CTL lines and 308 neurites from 4 ATXN2-ALS lines (two-tailed upaired t test; *P = 0.02). Data show individual lines and mean ± SD. G Immunocytochemistry for PABP in DIV9 hMNs after 60 min of sodium arsenite (ARS) treatment. Scale bar is 5 µm. H Quantification of number of stress granules (SGs) per neuron. n = 100 cells per condition from 3 CTL and 4 ATXN2-ALS lines, 3 experimental replicates/line (Kruskal–Wallis test, CTL 60’ vs ATXN2-ALS 60’ *P = 0.0169; ALS-ATXN2 60’ vs ALS-ATXN2 180’ *P = 0.0135). Violin plot of individual cells. I Quantification of SG area. 3 CTL and 4 ATXN2-ALS lines, 3 experimental replicates/line (2-way ANOVA and Tukey’s multiple comparison, CTL 60’ vs CTL180’ *** P < 0.0001; CTL 180’ vs ATXN2-ALS 180’ **P = 0.0062). Bar plots show mean ± SEM (See Supplementary Data 1 for N, SEM, and all P values). J Distribution of SG area in DIV9 hMNs at 60 minutes after ARS and 120 min of recovery (180’). K Immunocytochemistry for ATXN2 in DIV12 hMN cultures. Phalloidin stains F-actin. Scale bar is 10 µm. L, M Current-clamp recording of hMN cultures from 2 CTL and 2 ATXN2-ALS lines. L At DIV12, ATXN2-ALS hMNs are hypoexcitable (n = 30 neurons; P = 0.049, P = 0.005, and P = 0.0001 for 30, 40, and 50 nA, respectively; 2-way ANOVA and Sidak’s multiple comparisons test) and M at DIV24 hyperexcitable as compared to CTL (n = 46 neurons; *P = 0.01 at 30 nA, ***P < 0.0001 at 40 and ***P = 0.0006 at 50 nA; 2-way ANOVA and Sidak’s multiple comparisons test). Data are mean ± SD. Source data are provided as a Source Data file for F, H, I, J, L, M.
To examine which molecular pathways are affected in ATXN2-ALS hMNs and may underlie the observed cellular phenotypes, RNA-seq was performed on hMN cultures at DIV12 to identify factors that may drive MN dysfunction rather than being a consequence of it. This revealed 25 DEGs between ATXN2-ALS and CTL hMNs (Fig. 2A; Table S2), some of which have reported functions in lipid metabolism (PLP1) and mitochondrial function (e.g., NNMT1, GPD1) (Fig. 2B). To test whether these gene expression changes may be specific to ATXN2-ALS or whether they reflect the general ALS background in the ATXN2-ALS hMNs, the expression of a selection of DEGs was examined in hMNs carrying ALS-associated variants in FUS (Table S1). With the exception of CBLN2 (which was increased in FUS-ALS hMNs), the expression of the selected genes was unchanged in FUS-ALS hMNs as compared to CTL hMNs (Fig. S3A). As the transcriptomic changes hinted at an effect of ATXN2 intermediate expansions on cellular metabolism and mitochondrial function, we assessed the bioenergetic state of hMN cultures using Seahorse assays. ATXN2-ALS hMN cultures displayed significantly reduced basal respiration, maximal respiration capacity, and ATP production (Fig. 2C–F). No changes in basal respiration, maximal respiration, and ATP production were found in FUS-ALS hMNs, as reported previously47 (Fig. S3B–D). In addition, glycolysis and glycolytic capacity were reduced in ATXN2-ALS hMN cultures (Fig. 2G–I).
A Hierarchical clustering of DEGs in DIV12 control (CTL) and ATXN2-ALS iPSC-derived motor neuron (hMN) cultures (P < 0.05, FDR < 0.05). Color legend indicates logFC values. B Volcano plot of the quantified transcripts as in A. Colored dots, statistically significant hits (P values adjusted for multiple testing were calculated using Benjamini Hochberg false discovery rate (FDR), significantly differentially expressed genes at FDR < 0.05). A few selected hits are shown. C–I Representative kinetics graphs of C mitochondrial oxygen consumption and G extracellular acidification rates in DIV12 hMN cultures. Quantification of D basal respiration, **P = 0.0014, E maximal respiration, ***P < 0.0001, F ATP production, **P = 0.0022, H glycolysis, **P = 0.0002 and I glycolytic capacity, **P = 0.0027. 3 CTL and 2 ATXN2-ALS lines were used per experiment. D–F n = 2 independent experiments for CTL_2 and CTL_4, n = 1 for CTL_1, n = 4 for ALS_P and n = 5 for ALS_N. H, I n = 3 independent experiments for all lines. Two-tailed unpaired t test. Data are mean ± SD. Individual datapoints correspond to the experimental replicate of one iPSC line. Source data are provided as a Source Data file for D–F, H, I.
Together, these experiments show that hMNs from ATXN2-ALS patients exhibit specific gene expression changes and defects in neuronal integrity, SG dynamics, excitability, cellular metabolism, and mitochondrial function.
Generation of ATXN2 BAC transgenic mice
To next study the effect of ATXN2 intermediate expansions in ALS in vivo, BAC transgenic mice were generated carrying the full human ATXN2 gene with either the Q22 repeat sequence (most common variant in healthy individuals) or the Q33 repeat expansion (associated with ALS) (Fig. 3A). Two founder lines were obtained per genotype and assessed (Fig. S4A–E). Two lines were selected for subsequent experiments (ATXN2Q22(2) and ATXN2Q33(2)) based on total ATXN2 expression and distribution. The selected lines (referred to as ATXN2Q22/+ and ATXN2Q33/+) displayed similar levels of total ATXN2 expression (Fig. 3B, C) and comparable ATXN2 distribution, in line with endogenous mouse ATXN2 expression (non-transgenic, NTg) (Fig. 3D–F, S4F). Next, repeat size and integrity were confirmed by PCR and sequencing (Fig. 3G, Table S17), and transgene copy number was estimated with quantitative (q)RT-PCR (Fig. 3H). The integrity and integration site of the BAC transgenes was assessed by targeted locus amplification (TLA) (Fig. 3I). The ATXN2Q22 transgene was integrated into an intronic region between exons 2 and 3 of Ddhd1 and the ATXN2Q33 transgene was integrated in the third exon of 4930578E11Rik (Fig. 3J, K). We were unable to detect 4930578E11Rik in adult brain and spinal cord tissue and did not observe altered Ddhd1 expression in ATXN2Q22/+ mice (Fig. S4G, H). Interestingly, ATXN2Q33/+, but not ATXN2Q22/+, mice displayed altered germ line segregation as reported for ataxin2−/− mice34,35 (Table S3). Together, these results show that we have generated mouse models that display near-endogenous expression and distribution (compared to Atxn2) of human ATXN2Q22 and ATXN2Q33 in vivo.
A BAC clone containing the human ATXN2 locus with flanking genomic sequence. B, C Western blot for total ATXN2 (mouse and human) in cortical lysates of 6-months-old mice. Black arrowhead, human transgene. White arrowhead, mouse ATXN2. C Quantification of western blots as in B. Each datapoint corresponds to one cortical lysate obtained from one animal (n = 8 for NTg, n = 7 for Q22/+ and n = 6 for Q33/+). D–F Immunohistochemistry for ATXN2 (mouse and human) in 6-months-old mice. G PCR on mouse genomic DNA to confirm ATXN2 CAG repeat integrity. H qRT-PCR to determine ATXN2 transgene copy number. Q22/+ and Q33/+ mice contain six and four copies, respectively. Each datapoint corresponds to one animal (n = 3 for NTg, n = 3 for Q22/+ and n = 4 for Q33/+). I–K Targeted Locus Amplification: high throughput sequencing reads covering the complete transgene sequence (primer set 2). J, K Identification of transgene integration sites. Bar graphs show individual mice and mean ± SD, n ≥ 3 animals per genotype. Scale bar is 40 D, 30 E, and 100 µm F. Source data are provided as a Source Data file for C, H.
ATXN2-Q33 causes motor deficits in a mutant TDP-43M337V background
ATXN2 intermediate expansions confer increased risk for ALS and act as modifiers of TDP-43 toxicity12. Therefore, to study these repeat expansions in an ALS-relevant background, ATXN2 mice were crossed with transgenic mice harboring a human TDP-43M337V transgene (TDP-43Tg). This model was chosen as a mild, sensitized genetic ALS background48. The longer disease process in these mice provides a unique opportunity for studying the deleterious effects of risk factors, such as ATXN2, on ALS-related phenotypes.
To assess the in vivo effect of ATXN2 intermediate expansions, survival, motor coordination and muscle strength were tested. No significant changes in survival were observed in this study in contrast to what was reported previously in TDP-43Tg/+ mice48 (Fig. S5A). Similarly, no overt motor coordination (accelerating rotarod) and muscle strength (grip strength) deficits were found in TDP-43Tg/+ mice. However, analysis of ATXN2Q33/+;TDP-43Tg/+ mice showed motor phenotypes (Fig. 4A, B). First, reduced motor coordination was found in ATXN2Q33/+;TDP-43Tg/+ as compared to ATXN2Q22/+;TDP-43Tg/+ mice up to 60 weeks-of-age (Fig. 4A, Supplementary Data 2). Failure to detect this difference at later stages likely results from loss of statistical power due to experimental dropout. Motor coordination was unchanged in other group comparisons (Fig. 4A). Second, a significant repeat length-dependent decrease in grip strength was observed in ATXN2Q33/+;TDP-43Tg/+ mice. ATXN2Q22/+;TDP-43Tg/+ mice also showed a reduction in grip strength, but only in the first 35 weeks-of-age (Fig. 4B, Supplementary Data 3). Together, these results show that ATXN2-Q33 influences in vivo motor phenotypes through interactions with mutant TDP-43.
A, B Behavioral data presented using a generalized (non-)linear mixed model, corrected for sex, weight and longitudinal acquisition. A Accelerating rotarod analysis reveals reduced latency to fall in Q33/+;TDP-43Tg/+ mice as compared to Q22/+;TDP-43Tg/+ mice. B Q33/+;TDP-43Tg/+ mice show grip strength deficits as compared to NTg control and Q22/+;TDP-43Tg/+ mice. Curves reflect the mean and confidence interval bands correspond to 2x SD. Statistical significance is indicated by the gray areas. n ≥ 8 mice for each genotype and sex. See Supplementary Data 2 and 3 for details on N, SD and P value. C–G Quantification of lumbrical NMJs in 24-months-old Q33/+;TDP-43Tg/+and Q22/+;TDP-43Tg/+male mice. Individual panels show C NMJ number, D representative immunohistochemistry (IHC) images of NMJs with α-bungarotoxin (BTX), E NMJ area, F NMJ perimeter, and G overlap between pre- and post-synaptic parts of the synapse. All quantifications were performed with BTX (C, E, F) or BTX combined with SV2/2H3 IHC G. n = 3 mice for each genotype. Two-tailed unpaired t test (**P = 0.0044 (E, F)). Data are mean ± SD. D Scale bar is 10 µm. H Calbindin (CALB1) immunohistochemistry in the cerebellum. Inserts show Purkinje cells (PC) at higher magnification. Scale bar is 250/50 µm. I Quantification of PC density (midsagittal cerebellum sections, lobules VI–VIII) shows reduced PC density in Q33/+;TDP-43Tg/+ mice as compared to NTg controls. n = 7 mice for NTg, n = 8 for TDP-43Tg/+, n = 4 for Q22/+;TDP-43Tg/+ and n = 9 for Q33/+;TDP-43Tg/+. NTg vs Q33/+;TDP-43Tg/+ *P = 0.019 (one-way ANOVA with Dunnett’s multiple comparison test). Data are mean ± SD. Source data are provided as a Source Data file for C, E–G, I.
ATXN2-Q33 causes mutant TDP-43-dependent NMJ changes and PC loss
ALS is characterized by MN loss and SCA2 by degeneration of cerebellar Purkinje cells (PCs), both leading to motor disability. Cerebellar involvement has been reported for ALS cases carrying ATXN2 variants49,50,51. Therefore, both spinal MNs and PCs were examined. No evidence for loss of ChAT+ spinal MNs was found in TDP-43Tg/+ mice up to 24 months, as reported previously48, nor for the other genotypes (Fig. S5B–D). No differences in neuromuscular junction (NMJ) number were observed in the lumbrical muscle at 24 months, but NMJ area and perimeter were significantly reduced in male ATXN2Q33/+;TDP-43Tg/+ as compared to ATXN2Q22/+;TDP-43Tg/+ mice (Fig. 4C–G). This phenotype had previously been reported for TDP-43Tg/Tg but not TDP-43Tg/+ mice48. Innervation of lumbrical NMJs, quantified as overlap of presynaptic SV2/2H3 over postsynaptic BTX, was unchanged in ATXN2Q33/+;TDP-43Tg/+ mice and no defects were observed in NMJs of the gastrocnemic muscle, as reported previously48 (Fig. S5E–G).
Immunohistochemistry for calbindin revealed a loss of PCs in 24-months-old ATXN2Q33/+;TDP-43Tg/+ mice (Figs. 4H, I and S5H). Next, the distribution of TDP-43 and ATXN2 were assessed. Nuclear depletion and cytoplasmic aggregation of TDP-43 is a pathological hallmark of ALS, while ATXN2 can also localize to cytoplasmic inclusions. However, no mislocalization of TDP-43 or ATXN2, nor changes in detergent insolubility and cleavage of human TDP-43 were found in MNs of ATXN2Q33/+;TDP-43Tg/+ mice in vitro or in vivo (Fig. S5I–O).
Thus, ATXN2-Q33 and TDP-43M337V interactions induce changes at the NMJ and degeneration of PCs in vivo.
ATXN2-Q33-TDP-43M337V interactions trigger robust transcriptomic changes in vivo
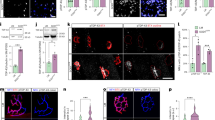
To explain the motor and neuronal phenotypes found in ATXN2Q33/+;TDP-43Tg/+ mice at the molecular level, we performed RNA-seq on 10-months-old spinal cords. Spinal cord is a primary site of ALS pathogenesis and at 10 months early molecular pre-symptomatic changes may be detected in the absence of robust cellular phenotypes. Unsupervised hierarchical clustering revealed that ATXN2Q33/+;TDP-43Tg/+ samples clustered separately, in contrast to other samples (Figs. 5A and S6A). Indeed, many differentially expressed genes (DEGs) were detected between ATXN2Q33/+;TDP-43Tg/+and NTg control or ATXN2Q22/+;TDP-43Tg/+ mice, in contrast to other comparisons (Fig. 5B–D and S6B–E,Table S4–7, Supplementary Data 4). 3395 DEGs were detected in ATXN2Q33/+;TDP-43Tg/+ as compared to ATXN2Q22/+;TDP-43Tg/+ littermates (1517 downregulated and 1878 upregulated) (Fig. 5D, Tables S6, 7, Supplementary Data 4). In line with the role of TDP-43 and ATXN2 in RNA processing, relative exon usage was also affected in ATXN2Q33/+ and ATXN2Q33/+;TDP-43Tg/+ mice (Fig. S6E, F). We validated the RNAseq data by qRT-PCR and confirmed the downregulation of Camk2a and C4b (Fig. S6G–J). Further analysis showed that several ALS-associated genes displayed mild but significant changes, similar to observations in ATXN2Q72 SCA2 mice52 (e.g., Tia1, Sod1, C9orf72) (Supplementary Data 4). Gene Set enrichment analysis (GSEA) revealed enrichment of DEGs in several different molecular pathways and cellular processes with strong links to mitochondrial function and inflammatory response (Fig. 5E, F; Tables S8–11). Genes associated with mitochondrial function and cellular metabolism, such as oxidative phosphorylation or fatty acid metabolism, were generally upregulated, while genes implicated in the inflammatory response were most frequently downregulated (Fig. 5E, F). Interestingly, deregulation of genes linked to cellular metabolism and mitochondrial function was also observed in ATXN2-ALS hMNs (Fig. 2). To study whether ATXN2Q33/+;TDP-43Tg/+ MNs, similar to ATXN2-ALS hMNs, display ALS-specific phenotypes and mitochondrial defects, spinal MN cultures were prepared from E13 mice. These cultures did not reveal obvious differences in neuronal morphology or number of MNs with SGs (Fig. S7A–C). However, ATXN2Q33/+;TDP-43Tg/+ MNs contained fewer SGs at 60 min after ARS treatment as compared to NTg, but not ATXN2Q22/+;TDP-43Tg/+, cultures (Fig. S7D, E). These data suggest that ATXN2-Q33 affects SG dynamics in both mouse and human ALS MNs. To examine whether mitochondrial function was perturbed in ATXN2Q33/+;TDP-43Tg/+ mice, respiratory capacity was evaluated. Similar to ATXN2-ALS hMNs, significantly reduced basal and maximal respiratory capacity and ATP production were detected in ATXN2Q33/+;TDP-43Tg/+, but not ATXN2Q22/+;TDP-43Tg/+, mice as compared to NTg control (Fig. S7F–H).
A Unsupervised hierarchical cluster analysis on DESeq2 log transformed raw counts of 10-months-old spinal cord samples based on all genes after removal of common genes (FDR < 0.05, sum of raw read counts > 0) between samples. n = 3 mice per genotype. B–D Volcano plots of the quantified transcripts in B Q22/+;TDP-43Tg/+ versus NTg control littermates; C Q33/+;TDP-43Tg/+ versus NTg control littermates, and D Q33/+;TDP-43Tg/+ versus Q22/+;TDP-43Tg/+ littermates. Colored dots, statistically significant hits (Padj < 0.05 derived from differential expression analysis using DESeq2, adjusted for multiple testing with Benjamini and Hochberg method). Dashed lines, ± 1 log2FC. A few selected top hits are shown. See Supplementary Data 4 for a complete DEG list of each pair-wise comparison. E, F Gene Set Enrichment Analysis (GSEA) in E Q33/+;TDP-43Tg/+ versus NTg control spinal cords and F Q33/+;TDP-43Tg/+ versus Q22/+;TDP-43Tg/+ spinal cords. Upregulated pathways (NES > 0) are shown in green; downregulated pathways (NES < 0) are shown in red.
Together, these data show synergistic effects of ATXN2 intermediate expansions and mutant TDP-43 on the expression of genes related to mitochondrial function in mouse and human spinal MNs. This combined with altered SG dynamics in mouse ATXN2Q33/+;TDP-43Tg/+ MNs, motor function defects in vivo and strong spinal cord transcriptome changes indicates that ATXN2-Q33 compromises MN function in an (mutant TDP-43) ALS background.
ATXN2-Q33 changes microglia gene expression and morphology
In addition to changes in cellular metabolism and mitochondrial function, RNA-seq also indicated an altered inflammatory response in ATXN2Q33/+;TDP-43Tg/+ mouse spinal cord (Fig. 5E, F). Two main immune-related processes were overrepresented: chronic inflammation (e.g., Aplnr53, Sema4D54) and immune cell activation (e.g., Rgs1655, c-Met56, Slc7a2, Kcna3) (Supplementary Data 4). Further, deregulation of several pro-inflammatory cytokine receptors was detected (e.g., Il6st, Il20ra, Il17rb, Il1rap, Il6ra) (Fig. 5, Supplementary Data 4). Microglia are the resident immune cells of the CNS and are linked to various neurodegenerative disorders, including ALS57,58. In this study, we sought to identify whether the basal state of spinal cord microglia (as found in NTg control tissue, hereby referred to as “homeostatic microglia”) is altered in ATXN2Q33/+;TDP-43Tg/+ mice. Therefore, we assessed expression of genes related to microglial homeostasis and (neurodegenerative) disease-associated states (as previously defined in other studies59). We found several deregulated homeostatic genes, including Csf1r and Cx3cr1, in ATXN2Q33/+;TDP-43Tg/+ spinal cords as compared to Ntg and/or ATXN2Q22/+;TDP-43Tg/+ (Supplementary Data 4). Although no clear signature of disease-associated microglia (DAM) genes60 was detected, several genes described to be upregulated in neurodegeneration-related DAM were deregulated in ATXN2Q33/+;TDP-43Tg/+ mouse spinal cord as compared to NTg and/or ATXN2Q22/+;TDP-43Tg/+ (Clec7a and Spp1) (Supplementary Data 4). In order to assess whether ATXN2Q33/+;TDP-43Tg/+ microglia exhibit specific phenotypic changes as compared to their basal state, microglia morphology and number were assessed in spinal cord tissue sections. This revealed a significant reduction in the area covered by the microglia marker IBA1 in ATXN2Q33/+;TDP-43Tg/+ mice as compared to NTg control (Fig. 6A, B and S8A, B). Interestingly, this reduction was already detectable at 6 months and IBA1+ area was also reduced in ATXN2Q33/+ mice at 24 months (Fig. S8A). No significant changes in GFAP+ area, marking astrocytes, was detected (Fig. S8C, D). The decrease in IBA1+ area was caused by the more ameboid morphology of microglia in ATXN2Q33/+;TDP-43 Tg/+ spinal cord in the absence of changes in microglia number (Fig. 6C–E). Next, we sought to determine if morphological changes also occur in an ATXN2 repeat length-dependent manner. Morphology was assessed in 6 months-old microglia in ATXN2Q22/+;TDP-43Tg/+ and ATXN2Q33/+;TDP-43Tg/+ spinal cords, which revealed that ATXN2Q33/+;TDP-43Tg/+ microglia acquire a more round cell shape (Fig. S8E). To examine whether ATXN2 is expressed in microglia, we performed immunohistochemistry for ATXN2 and IBA1. ATXN2 was not only expressed in neurons but also in microglial cells in the adult mouse spinal cord (Fig. 6F), suggesting that ATXN2-Q33 effects can be exerted in a cell-autonomous and non-cell autonomous manner. The ameboid morphology of microglia can be an indication of an activated state61 and to assess this at the molecular level, spinal cord microglia were isolated from 7-months-old NTg and ATXN2Q33/+;TDP-43Tg/+ mice (Fig. S8F) and subjected to RNA-seq. Only 7 DEGs were found in ATXN2Q33/+;TDP-43 Tg/+ microglia as compared to NTg (Fig. 6G, H and Table S12). All DEGs were (in)directly related to inflammation and toxicity, and differentially expressed in microglia in various neurodegenerative disease contexts62. Pathway analysis revealed that 5 DEGs were part of the phagosome pathway (Table S13), while 3 DEGs were involved in chemotaxis (Fig. 6H). To examine whether gene expression changes were more pronounced at later stages in ATXN2Q33/+;TDP-43 Tg/+ mice, microglial cells were isolated from 16-months-old NTg and ATXN2Q33/+;TDP-43Tg/+ spinal cords. Gene expression changes of several homeostatic and DAM genes59 were assessed by qRT-PCR, revealing an absence of an altered homeostatic gene signature in ATXN2Q33/+;TDP-43 Tg/+ microglia as compared to NTg (Fig. 6I, J). Interestingly, one deregulated DAM gene in 7-months-old ATXN2Q33/+;TDP-43 Tg/+ microglia was also deregulated at 16 months (Atp6v1e1) (Fig. 6K), whereas other DAM genes (Spp1, Apoe) were only upregulated at later stages (Fig. 6L, M).
A Immunohistochemistry for IBA1 (red) on ventral spinal cord of 24-months-old NTg and Q33/+;TDP-43Tg/+ mice. DAPI stains nuclei. Line demarcates ventral gray matter. Scale bar is 400 µm. B Quantification of IBA1+ area in Q33/+;TDP-43Tg/+ ventral spinal cord normalized to NTg littermates (*P = 0.0275 at 24 months; *P = 0.032 at 10 months and *P = 0.0464 at 6 months, Brown-Forsythe and Welch ANOVA with Dunnett’s multiple comparison test). n ≥ 7 ventral horns per mouse; n = 4 mice per genotype and timepoint (except n = 3 per genotype at 24 months). Mean ± SD. C Representative images of microglia in ventral spinal cord. Scale bar is 10 µm. D Quantification of microglia circularity in the ventral horn spinal cord of 6-months-old NTg and Q33/+;TDP-43Tg/+ mice. (P < 0.0001, two-tailed unpaired t test). n ≥ 80 cells per mouse; n ≥ 3 mice per genotype. Violin plot of individual cells. E Quantification of microglia cell density in the ventral spinal cord of 6-months-old NTg and Q33/+;TDP-43Tg/+ mice. (ns; two-tailed unpaired t test). n ≥ 8 ventral horns per mouse; n = 3 mice for NTg and n = 4 for Q33/+;TDP-43Tg/+. Data are mean ± SD. F Immunohistochemistry for ATXN2 and IBA1 of ventral horn spinal cord microglia of 6-months-old NTg and Q33/+;TDP-43Tg/+ mice. Scale bar is 50 µm. G Volcano plot of quantified transcripts in microglia isolated from 7-months-old Q33/+;TDP-43Tg/+ spinal cord compared to NTg littermates. Colored dots, statistically significant hits (Padj < 0.05 derived from differential expression analysis using DESeq2, adjusted for multiple testing with Benjamini and Hochberg method). Dashed lines indicate ± 1 log2FC. A few selected hits are shown. H GO analysis of non-adjusted P < 0.05 DEGs in Q33/+;TDP-43Tg/+ spinal cord microglia using the metascape platform126. P < 0.05 derived from differential expression analysis using DESeq2, not adjusted for multiple testing. I–M RT-qPCR analysis of selected homeostatic (P2Y12, TMEM119) and disease-associated microglia (ATP6V1E1, SPP1, APOE) genes of spinal cord microglia from 16-months-old NTg and Q33/+;TDP-43Tg/+ mice. (*P = 0.0129 K; *P = 0.0339 L; *P = 0.0102 M, two-tailed unpaired t test). n = 4 mice per genotype; each sample consists of 3 pooled spinal cords. Data are mean ± SD. Source data are provided as a Source Data file for B, D, E, I–M.
In certain disease contexts such as neurodegeneration, microglia have been found to respond differentially to pro-inflammatory stimuli63. Therefore, the effect of in vivo LPS injection on microglia-secreted cytokine gene expression was studied. Five hours after intraperitonial LPS injection, spinal cord microglia were collected and analyzed by qRT-PCR. Basal expression of IL1b, IL6, TNFa and TGFb mRNA was similar in NTg and ATXN2Q33/+;TDP-43Tg/+ mice. Upon LPS exposure, cytokine expression levels increased in both samples as compared to basal levels. However, no significant differences were detected in the LPS response of NTg and ATXN2Q33/+;TDP-43Tg/+ mice (Fig. S8G).
Several studies indicate that mouse and human microglia can differ significantly64. Therefore, to examine the molecular changes observed in mouse microglia in human ATXN2-ALS microglia, iPSCs were differentiated into cerebral organoids that innately develop microglia65 (Figs. 7A, B and S9A, B). At DIV60, organoids were collected for IBA1 immunostaining or dissociated for microglia isolation by MACS followed by RNA extraction. IBA1+ microglia were detected in all organoids and ATXN2 expression was observed in microglia as well as in surrounding neurons, similar to our observation in mouse spinal cord (Figs. 6G, 7C). No obvious differences in microglia morphology were detected between control and ATXN2-ALS organoids (Fig. 7D, E). Next, we isolated microglia and performed RT-qPCR to examine the expression of a few homeostatic and DAM genes, some of which were differentially expressed in mouse microglia (ATP6V1E1, SPP1, APOE). Expression of the selected homeostatic genes59 was not different between control and ATXN2-ALS organoid-derived microglia (oMG) (Fig. 7F, S9C). In contrast, the expression of APOE, a key protein in activation of the DAM program66, was upregulated in ATXN2-ALS oMGs. Similarly, both SPP1 and ATPV1E1 were upregulated in ATXN2-ALS oMGs. Expression of TREM2 was also enhanced but this effect was not statistically significant (Fig. 7F). To test whether these gene expression changes may be specific to ATXN2-ALS or whether they reflect a more general ALS background in the ATXN2-ALS microglia, the expression of a selection of genes was examined in DIV60 oMGs carrying C9ORF72-HREs (Table S1). C9ORF72-HREs are the most common genetic cause of ALS and have been extensively linked to microglial phenotypes57. In contrast to ATXN2-ALS oMGs, microglia derived from C9-ALS lines did not show significant differences in the expression of the selected DAM genes, with the exception of TREM2 (Fig. 7G). Finally, organoids were treated with LPS followed by microglia isolation and qRT-PCR to assess the expression of pro-inflammatory cytokines65. This showed that although oMGs from control and ATXN2-ALS organoids showed enhanced expression of pro-inflammatory cytokines in response to LPS, no difference in response between the two groups (CTL and ATXN2-ALS) was present (Fig. S9D).
A Summary of the protocol used to generate microglia-containing cerebral organoids65. B Immunohistochemistry for MAP2 and CTIP2 in cryosections of DIV60 cerebral organoids from control (CTL) and ATXN2-ALS iPSC lines. Scale bar is 100 µm. C Immunohistochemistry for ATXN2, IBA1 and MAP2 in cryosections of DIV60 cerebral organoids from control (CTL) and ATXN2-ALS iPSC lines. Arrowheads indicate ATXN2+IBA1+ cells. Scale bar is 25 µm. D Immunohistochemistry for IBA1 in DIV60 cerebral organoids showing 3 distinct microglial morphologies used for quantification in E. Scale bar is 10 µm. E Semi-quantitative analysis of organoid microglia morphology using 3 pre-defined categories (ramified, rod-like and ameboid) using 2 CTL (CTL_1, CTL_7) and 2 ATXN2-ALS (ALS_A, ALS_B) iPSC lines. n = 100 cells per line from 1 experimental replicate, n ≥ 3 organoids. Data are mean ± SD. F qRT-PCR for homeostatic (P2Y12) and disease-associated genes (APOE, SPP1, ATPV1E1, TREM2) in microglia isolated from DIV60 cerebral organoids from 3 CTL (CTL_1, CTL_7, CTL_4) and 2 ATXN-ALS (ALS_A, ALS_B) iPSC lines (n = 3 independent experimental replicates per line, except n = 2 for ALS_A in ATPV1E1 plot). Two-tailed upaired t test (*P = 0.0102 for APOE, *P = 0.0148 for SPP1, **P = 0.0094 for ATPV1E1). Data are mean ± SD. G qRT-PCR for homeostatic (P2Y12) and disease-associated genes (APOE, SPP1, ATPV1E1, TREM2) in microglia isolated from DIV60 cerebral organoids from 4 CTL (CTL_1, CTL_7, NL1, NL2.1) and 4 C9-ALS (ND06769, ND10689, CS29, ND12099) iPSC lines (P2Y12/TREM2 – n = 5 for CTL_1, n = 7 for CTL_3, n = 2 for NL1, n = 3 for NL2, n = 6 for ND06769, n = 5 for ND12099, n = 4 for ND10689, n = 2 for CS29; APOE/SPP1/ATPV1E1 – n = 4 for CTL_1, n = 4 for CTL_3, n = 2 for NL1, n = 3 for NL2, n = 4 for ND06769 (except n = 3 in SPP1 plot), n = 4 for ND10689, n = 3 for ND12099, n = 2 for CS29). *P = 0.0046 for TREM2 (two-tailed upaired t test). Data are mean ± SD. F, G Individual datapoints correspond to an experimental replicate of one iPSC line. Source data are provided as a Source Data file for E–G. A was created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Thus, ATXN2-Q33 is linked to molecular and morphological microglial changes associated with microglial activation and altered phagocytosis.
Discussion
In this study, patient-derived and mouse models were generated to examine the pathogenic effects of ATXN2 intermediate expansions. hMN from ATXN2-ALS patients showed SG defects in addition to neurite damage and electrophysiological changes. Similarly, expression of ATXN2-Q33 in TDP-43Tg-ALS mice caused reduced motor function accompanied by NMJ changes, PC degeneration, and altered SG dynamics. Finally, effects on gene expression were detected that highlighted altered mitochondrial function and inflammatory response, including microglia-related responses, both of which were confirmed at the cellular level in our mouse and human model systems. These data provide insight into the pathogenic effects of ATXN2 intermediate expansions and a framework and tools for further studies into their mechanism-of-action and therapeutic potential.
The best characterized and reported pathogenic effect of ATXN2 intermediate expansions is their ability to enhance TDP-43 toxicity in vitro12. To establish the consequences of these interactions in vivo, ATXN2 transgenic mice were crossed with TDP-43Tg/+ BAC transgenic mice48. In contrast to previous results, no changes in survival or motor function were observed in TDP-43Tg/+ mice in our study48. Nevertheless, usage of the TDP-43Tg/+ model revealed that ATXN2-Q33 causes various pathogenic phenotypes in a mutant TDP-43 background in vivo (e.g., motor dysfunction, PC loss or muscle-specific NMJ changes). No changes in MN number or TDP-43 distribution were reported in TDP-43Tg/+ mice, and ATXN2-Q33 did not unveil these phenotypes. However, TDP-43Tg/Tg, but not TDP-43Tg/+, mouse MNs and spinal cord showed an increase in insoluble TDP-43 in vitro and in vivo, respectively. It is therefore plausible that in a more severe TDP-43-ALS model (e.g., TDP-43Tg/Tg) ATXN2-Q33 may affect survival, TDP-43 pathology and/or cause MN loss.
Impaired motor function in ALS is generally attributed to visible changes in MN number or NMJs. Despite motor deficits, e.g. reduced grip strength, no MN number or gastrocnemius NMJ changes were detected in ATXN2Q33/+;TDP-43Tg/+ mice, nor in TDP-43Tg/+ mice48. However, lumbrical NMJ shrinkage and PC degeneration were observed in ATXN2Q33/+;TDP-43Tg/+ mice, both occurring in a repeat length-dependent manner. A decrease in hindlimb lumbrical NMJ area was previously reported in TDP-43Tg/+ mice48, a phenotype known to contribute to motor impairment, including reduced grip strength. Emerging evidence indicates that NMJ disruption at the postsynaptic level can precede denervation and MN loss, where NMJ changes represent a primary pathogenic event that progresses in a “dying back fashion” (see review67). Further, primary muscle pathology may contribute to abnormal NMJ68,69. In addition, PC degeneration may contribute to the motor phenotypes detected in this study. Although whole-brain imaging of ATXN2-ALS patients did not show significant cerebellar degeneration, histopathological analysis revealed PC loss49,50,51. The cerebellum affects motor functions such as precision grip force70 and PC loss could, therefore, contribute to the reduced grip strength found in ATXN2Q33/+;TDP-43Tg/+ mice. Finally, altered MN function rather than loss could cause or contribute to changes in motor behavior. Robust transcriptomic changes were found in the spinal cord in addition to altered SG dynamics, mitochondrial function, neuronal excitability and neurite damage in MNs. Together, our results suggest that in a mild TDP-43-M337V background ATXN2-Q33 causes motor deficits by eliciting subtle molecular and physiological, rather than overt degenerative, changes. These changes may represent early stages of the disease.
ATXN2-ALS hMNs contained larger SGs during disassembly, which may indicate a higher propensity of ATXN2-Q33 to aggregate. Nevertheless, ATXN2 and TDP-43 expression and localization appeared intact in mouse and human models in this study. It is possible that small changes in the distribution or post-translational modification of these proteins were below the detection threshold of the methods used. However, disease phenotypes can occur in the absence of clear mislocalization or altered expression of these proteins, especially at early disease stages. For example, SCA2 pathogenesis has been shown in the absence of ATXN2 aggregation71 and some ALS-linked phenotypes occur without TDP-43 aggregation or nuclear depletion72,73. Nevertheless, the robust effects of ATXN2-Q33 in the presence of TDP-43-M337V confirms a crucial role for TDP-43 pathogenesis in the effects of ATXN2 intermediate expansions. A possible site of convergence of ATXN2-Q33 and (mutant) TDP-43 are SGs. Both TDP-43 mutations and ATXN2 depletion have been linked to disturbed SG formation21,31,74,75,76. In both ATXN2-ALS and ATXN2Q33/+;TDP-43Tg/+ MNs, fewer SGs assembled upon oxidative stress. Our data show that ATXN2-Q33 and TDP-43 cooperate to elicit SG-associated phenotypes, which could, for example, depend on altered protein expression or changes in the (shared) interactome of both proteins, which contain several SG-associated proteins30.
How the expansion of the polyQ region of ATXN2 causes phenotypes such as in SG dynamics is unknown. As reported here, and in other studies focusing on SCA2, phenotypes can be repeat length-specific29,71. For example, the severity of some of the changes observed in ATXN2Q22/+;TDP-43Tg/+ mice and MN cultures lies in between those found in non-transgenic and ATXN2Q33/+;TDP-43Tg/+ mice (e.g., reduced grip strength or mitochondrial dysfunction). It is, therefore, tempting to speculate that the ATXN2-Q33 protein is more stable or has an altered interactome leading to sustained downstream effects on TDP-43 and the subsequent phenotypes discovered in this study29. Although the transgenic ATXN2 mice in this study only expressed moderate levels of human ATXN2, in addition to endogenous mouse ATXN2 (lacking polyQ repeats), differences in germ line segregation were detected in ATXN2Q33/+ mice. Even though polyQ expansions are generally associated with toxic GOF phenotypes77, germ-line segregation differences were previously reported in ataxin-2−/− mice34,35. Therefore, LOF and GOF mechanisms might co-exist in ATXN2Q33/+;TDP-43Tg/+ mice.
The most robust change observed in ATXN2Q33/+;TDP-43Tg/+ mice was at the transcriptomic level. GO analysis revealed deregulation of genes associated with inflammatory response, PI3K/AKT/mTOR signaling, oxidative phosphorylation and mitochondrial function, in part overlapping with previous observations in other ATXN2-polyQ models38,52,78,79. Expression changes in ATXN2Q33/+;TDP-43Tg/+ mice did not always show the same direction as in other ATXN2 mouse models, which may be attributed to the interaction of ATXN2-Q33 and TDP-43-M337V. Such processes included upregulation of adipogenesis and fatty acid metabolism, which are downregulated in ATXN2 LOF models35, and downregulation of the immune response, found to be upregulated in SCA2 spinal cord52,80. Interestingly, DEGs and deregulated pathways were not only shared with previously published ATXN2Q72 mouse spinal cord but also cerebellum datasets38, which is in line with the PC degeneration observed in ATXN2Q33/+;TDP-43Tg/+ mice. Finally, the expression of several ALS-associated genes was altered, as observed in ATXN2Q100 mice52, which may contribute to the observed phenotypes and relates to reported genetic and functional interactions between ATXN2 intermediate expansions and other ALS-associated mutations81,82.
Similar to ATXN2Q33/+;TDP-43Tg/+ spinal cord samples, RNA-seq of ATXN2-ALS hMNs revealed DEGs related to lipid metabolism and mitochondrial function. Mitochondrial dysfunction is a common phenotype in ALS and contributes to the selective vulnerability of MNs83. Our results show that several bioenergetic parameters are reduced in mouse and hMNs carrying ATXN2-Q33 expansions, including basal and maximal mitochondrial respiration. This is in line with reported effects of TDP-43 mutations on mitochondrial function84, with the previously described role of ATXN2 in the regulation of bioenergetic pathways in mitochondria40,85, and the effects of SCA2-related polyQ expansions86. Future work will undoubtedly focus on exploiting the transcriptomic data to further link ATXN2-Q33 and mitochondrial dysfunction.
Analysis of ATXN2Q33/+;TDP-43Tg/+ spinal cord also revealed a change in the inflammatory response, with an overrepresentation of genes related to chronic inflammation and immune cell activation. Neuroinflammation is increasingly associated with ALS and contributes to disease severity and progression87. While the spinal cord contains different types of immune cells, several DEGs related to microglia homeostasis and neurodegenerative disease states59 were found. In ALS and other neurodegenerative diseases, microglia shift from a homeostatic to a disease state which may have beneficial or detrimental effects58. In ATXN2Q33/+;TDP-43Tg/+ spinal cord, microglia acquired a more ameboid morphology, indicative of an activated state, as reported in other ALS mouse models88,89. This morphological change was also observed in ATXN2Q33/+ mice, and ATXN2CAG100 mice80, although at later stages. This suggests that ATXN2 intermediate expansions might be sufficient to alter microglia, but that in the presence of TDP-43-M337V the microglial phenotype is exacerbated. This is in line with previously described effects of mutant TDP-43 on microglia90. Neuron-specific TDP-43 pathology is sufficient to induce microglia activation91,92, which suggests that the microglial phenotypes in ATXN2Q33/+;TDP-43Tg/+ spinal cord may originate from cell-autonomous and/or non-cell-autonomous effects. To explore whether early microglia-specific transcriptomic changes drive immune system dysfunction in ATXN2Q33/+;TDP-43Tg/+ spinal cord, RNA-seq was performed on isolated microglia. This only revealed a small number of DEGs but showed deregulation of the phagosome pathway. Different ALS-associated mutations are linked to altered microglial phagocytosis93, indicating that disturbed microglial phagocytosis is generally altered in ALS. Additionally, gene expression analysis at a later disease stage revealed that ATXN2Q33/+;TDP-43Tg/+ microglia acquire a transcriptomic disease state encompassing upregulation of core DAM genes59 (APOE, SPP1). The presence of morphological changes starting at an early disease stage coupled with DAM gene regulation appearing at later stages might suggest that non-cell-autonomous mechanisms alter ATXN2Q33/+;TDP-43Tg/+ microglia basal states. Likewise, numerous studies have reported time-dependent changes in microglia state as (ALS) disease pathogenesis progresses94. While gene expression changes hinted at loss of microglial homeostasis, LPS treatment showed that microglia were not in a primed state in our models. However, it is possible that the transcriptomic and morphological changes observed represent a microglial state that precedes primed states occurring at later stages of the pathogenic process.
Mouse and human microglia can differ significantly, even within the same disease context95,96,97,98. The use of a human brain organoid model that contains microglia65 showed upregulation of several DAM genes in microglia, including APOE and SPP1, in ATXN2-ALS organoids in the absence of obvious morphological differences. Interestingly, these core DAM genes were also found to be upregulated in ATXN2Q33/+;TDP-43Tg/+ microglia and were not changed in organoid microglia derived from ALS lines harboring other disease mutations. The upregulation of DAM genes is necessary to shift microglia from homeostatic to distinct disease states99. Together with our mouse data, these observations suggest that ATXN2-Q33 alters the microglia basal state at early disease stages, which become more pronounced as pathogenesis progresses. Further research is needed to dissect the underlying mechanisms and pathogenic consequences.
In this study, we newly generated patient-derived and mouse models to identify pathways and cellular processes that underly the pathogenic effects of ATXN2 intermediate expansions in ALS. Our work shows that ATXN2 intermediate expansions can uncover and exacerbate several ALS-relevant molecular, cellular and behavioral phenotypes, in part through synergistic interactions with mutant TDP-43. ATXN2-Q33 has pathogenic effects in the absence of clear TDP-43 pathology, perhaps reflecting early disease phenotypes. Overall, our data provide insights into the pathogenic consequences of ATXN2 intermediate expansions and present a framework for future research into the mechanism-of-action and therapeutic potential of ATXN2 and associated repeat expansions.
In contrast to previous studies (e.g. ref. 48), we did not detect changes in survival or motor function in the TDP-43Tg/+ mouse model48 used in our study. This apparent discrepancy may have different causes, e.g. both genetic background and environmental factors have been shown to impact the variability and expressivity of phenotypes in mice100,101. Further, considerable variation in the onset of phenotypes or pathology across mouse models with similar transgenes or even for the same transgene, e.g. TDP-43Q331K, in different labs is consistently reported72,102,103,104,105.Therefore, future studies should determine whether in a more severe TDP-43-ALS model (e.g., TDP-43Tg/Tg) ATXN2-Q33 also affects survival, induces TDP-43 pathology and/or causes MN loss. In this study, we have used a random integration strategy to generate ATXN2 BAC transgenic mice that show comparable transgene expression in ALS relevant brain and spinal cord regions. However, we cannot rule out that because of this approach differential transgene expression exists in other regions or tissues. In future studies, we therefore hope to employ a ‘genomic safe harbor’ approach to generate next generation ATXN2 transgenic mice with guaranteed stable transgene expression.
Another limitation is that we were unable to generate isogenic controls for the ATXN2-ALS iPSC lines. This leaves open the possibility that a few of the observed phenotypes in hMNs represent general, ALS-associated changes rather those specific to ATXN2-Q33. Most of the phenotypes reported in this study are observed both in human and mouse models, or only mouse models, for which ATXN2-Q22 control data are presented (e.g., SG dynamics, mitochondria, and immune cells). Further, the absence of several of the molecular and cellular changes, and presence of different changes, in hMN or microglia derived from familial ALS patients carrying gene mutations in genes other than ATXN2 (i.e. in FUS or C9ORF72) suggest that, rather than representing general ALS-associated changes, these molecular and cellular defects are likely linked to ATXN2-Q33. Nevertheless, it will be important to generate isogenic control lines in future studies.
Lastly, we acknowledge that the number of DEGs that were detected in ATXN2-ALS hMNs by RNA-seq is rather modest, especially in comparison to the mouse data. Stringent analysis and limited sequencing depth may have contributed to this. In further analyses, the rigor of this experiment could be increased by including more experimental replicates or iPSC lines and by employing a higher sequencing depth.
Methods
Mouse lines
All animal experiments in this study were approved by the (CCD) Centrale Commissie Dierproeven of Utrecht University (CCD license: AVD 1150020171565 and AVD 11500202216085) and were in accordance with Dutch law (Wet op de Dierproeven 2014) and European regulations (guideline 2010/63/EU). Mice (Charles Rivers) were housed at 22 ± 1 °C on a wood-chip bedding supplemented with tissue on a 12 hour/12 hour day/night cycle. Pregnant mothers were housed individually from the moment of observation of the plug (E0.5). Animals were fed ad libitum and mice from both sexes were used for all experiments unless stated differently. C57BL/6 J mice were obtained from Charles River Laboratories (Wilmington, Massachusetts, USA) and TDP-43M337V BAC transgenic mice were as reported previously48 (The Jackson Laboratory; JAX#029266).
ATXN2 BAC-transgenic mice were generated at InnoSer (Leiden, The Netherlands) by microinjection of purified intact BAC DNAs. The BAC clone used (RP11-798L15) contained the 147 kb human ATXN2 locus (NG_011572.2) in addition to 25 kb of 5’ and 4 kb of the 3’ flanking sequence. The BAC constructs were created using a two-step BAC modification with PLD53.SC-AB vector, as previously described106. Two different ATXN2 lines were engineered (1) carrying the most common polyQ repeat in the healthy human population (ATXN2Q22; (CAG)13-CAA-(CAG)8), or (2) carrying an ALS-associated expanded polyQ repeat (ATXN2Q33; (CAG)24-CAA-(CAG)8). Following microinjection at Polygene (Switzerland), founder mice (n = 3 per line) were backcrossed to C57BL/6 J mice for a minimum of three generations.
Genotyping of ATXN2 BAC-transgenic mice was performed using primers to detect the human ATXN2 transgene and flanking regions of the transgenes (Table S14). DNA fragments were amplified using Firepol mix (Solis BioDyne). Detection of the human TDP-43 transgene was performed as described previously48. To confirm repeat length and sequence by Sanger sequencing (BaseClear, The Netherlands), ATXN2 repeat sequences were amplified using Long PCR Enzyme Mix (ThermoFisher) and primers flanking the repeat (Table S14).
For experiments using embryos and animals younger than P10, the sex of the animals was not considered. For phenotypic comparisons, same sex littermates and/or mice of similar ages (in case of females only) were randomly assigned to experimental groups. Control animals were age-matched littermates
Generation, characterization, and maintenance of iPSCs
All subjects have provided written informed consent and generation of iPSC lines was approved by the Ethical Medical Committee of the University Medical Center Utrecht. All experiments were performed under license METC 12-267/O and Deelbiobank 16/436. Patients were diagnosed according to the diagnostic criteria for ALS (revised El Escorial). Controls were donors without a psychiatric or neurologic diagnosis (Table S1). iPSC lines CTL_665; NL1 and NL2107; ND12099, ND06769 and ND10689108; CS29 (Cedars-Sinai); FUS2109 and FUS2/244 were reported previously. iPSC lines CTL_1, CTL_2, CTL_3, CTL_4, CTL_7, ALS_A, ALS_B, ALS_G, ALS_N and ALS_P were generated in this study as described previously110. Briefly, low passage (P < 5) human skin fibroblasts were plated at a density of 10,000 cells per well in a 6-well plate in DMEM GlutaMAX (Thermo-Fisher, 31966-021), 10% fetal bovine serum (FBS; Sigma, F7524), 1% Penicillin/Streptomycin (P/S; Life Technologies, 15140122) and cultured at 37 °C with 5% CO2. After 24 hours, cells were incubated with a lentivirus expressing OCT4/SOX2/c-MYC/KLF4. After 5 days, human fibroblasts were dissociated and seeded on top of gamma-irradiated mouse embryonic fibroblasts and cultured in human embryonic stem cell (huES) medium (DMEM-F12, 20% Knockout serum replacement (Life Technologies, 10828028), 0.5% P/S, 1% non-essential amino acids (NEAA; Life Technologies, 11140035), 1% L-Glutamine (Life Technologies, 25030024), 496 mM β-mercaptoethanol (Life Technologies, 21985-023), 20 ng/mL human basic FGF (Peprotech, 100-188). After 3–6 weeks, iPSC colonies were manually picked for expansion and characterization. Line CTL_1.2 was generated in this study by reprogramming dermal fibroblasts with the CytoTune®-iPS 2.0 Sendai Reprogramming Kit (Invitrogen, Waltham, MA, USA, Cat. No. A16517), using Sendai Virus Vectors encoding OCT3/4, SOX2, KLF4, and c-MYC, according to the manufacturer’s protocol. For every line generated in this study, stemness of the clones was analyzed by immunostaining using the StemLight kit (Cell Signaling, 9656S) and quantitative reverse transcription PCR (qRT-PCR) relative to hUES6 line (Harvard University, RRID: CVCL_B194). Cells were karyotyped by G band staining and pluripotency was evaluated by embryoid body formation followed by 5 weeks of spontaneous differentiation in minimal medium (DMEM-GlutaMax; 10% FBS). Successfully reprogrammed clones were frozen and stored in liquid nitrogen. iPSCs were maintained in feeder-free conditions at 37 °C with 5% CO2 and passaged once a week. For passaging, cells were washed with PBS and incubated with 0.5 mM EDTA (Thermo Fisher Scientific, 15-575-020) for 2 minutes at 37 °C. EDTA was removed and cells were sprayed with 1 mL of StemFlex medium supplemented with 10 mM ROCK inhibitor (Y27632, Axon Medchem, AXON 1683) to generate small iPSC aggregates. Aggregates were transferred to Geltrex-coated dishes in StemFlex medium (Thermo Fisher Scientific, A3349401) with 10 µM Y27632. The following day medium was changed to StemFlex and subsequently medium was changed every other day. Cells were tested monthly for potential mycoplasma infection (Lonza Bioscience, LT07-318).
Differentiation of iPSC-derived human spinal MNs
Two different protocols were used to generate iPSC-derived spinal motor neurons (hMN). The first protocol was performed as described previously with minor modifications111. In brief, iPSCs were dissociated into a single cell suspension in mTeSR1 medium (StemCell Technologies, 85857) supplemented with 50 mM Y27632 and 300-400 cells were seeded in microwells to form embryoid bodies (EB) overnight (ON). The next day, medium was replaced with mTeSR1 containing 50 mM Y27632, 10 mM SB431542 (Axon Biochemicals, AXON1661) and 0.2 mM LDN (Miltenyi Biotec, 130103925). At day 3, medium was changed to neural induction medium (NIM; DMEM-F12, 1% NEAA, 1% L-Glutamine, 0.5% P/S, 1% N2 supplement, 0.16% D-glucose) containing 10 mM SB431542 and 0.2 mM LDN. Between day 5 and 7 (depending on EB size), EBs were gently flushed and plated in non-coated petri dishes. Between days 5 and 7, EBs were cultured in NIM containing 10 mM SB431542, 0.2 mM LDN, 1 mM retinoic acid (RA, Sigma-Aldrich, R2625)) and 10 ng/mL of brain-derived neurotrophic factor (BDNF, STEMCELL Technologies, 78005). From days 7 to 16, EBs were kept in NIM supplemented with 1 mM RA, 10 ng/mL BDNF and 1 mM smoothened agonist (SAG, Merck Chemicals, 566660). At day 17, medium was switched to neural differentiation medium (Neurobasal, 1% NEAA, 1% L-Glutamine, 0.5% P/S, 1% N2 supplement, 2% B27 without vitamin A supplement, 0.16% D-glucose) containing 1 mM RA, 1 mM SAG, 10 ng/mL BDNF, 10 ng/mL glial-derived neurotrophic factor (GDNF), 10 ng/mL ciliary neurotrophic factor (CNTF) (STEMCELL technologies, 78058/78010) and 10 ng/mL IGF-1 (STEMCELL technologies, 78022). On day 21, EBs were dissociated with papain and DNAse (Antonides, LK003172 and LK003178) and plated in PDL-laminin (Sigma-Aldrich, P0899/ L2020) coated coverslips in hMN medium (Neurobasal, 1% NEAA, 1% L-Glutamine, 0.5% P/S, 1% N2 supplement, 2% B27 without vitamin A supplement) supplemented with 10 ng/ml of BDNF, GDNF and CNTF. The following day coverslips were transferred to 12-well plates containing mouse primary glia and were kept in sandwich co-culture for 12 (DIV12) or 24 days (DIV24). Half medium change was performed every 3 days. At the start of hMN differentiation, mouse pups (P2-P3) were sacrificed by decapitation, and whole brains were isolated in cold glia medium (Alpha-MEM, 1% P/S, 10% FBS, 0.6% D-Glucose), mechanically dissociated using a 20 G needle and plated in 15 cm dishes in glia medium. Mouse glial cultures were passaged once a week for 3 weeks using Trypsin-EDTA (Thermo Fisher Scientific, 25300062). To create height for the sandwich co-culture paraffin bumps were made in wells of 12-well plates. After 3 weeks, 36,000 cells were seeded per well in 12-well plates. At the time of hMN dissociation, the medium was switched to hMN medium supplemented with 1 mM SAG for 24 hrs. Before transferring the coverslips with hMN, the medium was switched to hMN supplemented with 10 ng/ml of BDNF, GDNF, and CNTF. hMN purity was above 60% as estimated by scRNA-seq based on multiple markers (ISL1, ISL2, LMO4, FOXP1, and ALADH1A2) (see Figs. 1 and Figs. S2). For treatment with sodium arsenite, a separate protocol previously published elsewhere was followed, reported to yield an 80% hMN purity112. Human iPSCs were plated in 6-well plates and kept in StemFlex medium for 48 hours. Medium was switched to neural differentiation medium (NDM, DMEM-F12: Neurobasal (1:1) with 1% P/S, 1% NEAA, 1% L-Glutamine, 1% Sodium pyruvate, 1% N2 supplement, 2% B27 supplement) supplemented with ascorbic acid (AA), SB431542, LDN, CHIR99021 (Sigma-Aldrich, SML1046) (NDM1). From day 7 to 12, NDM2 (NDM supplemented with AA, cAMP (Sigma-Aldrich, A6885), SB431542, LDN, CHIR, RA, Puromorphine) was added. At day 13, cells were washed once with PBS and incubated with Dispase (Bio-Connect, 07923) for 3–4 minutes at 37 °C. Dispase was removed and cells were sprayed with 1 mL of NDM3 (NDM supplemented with AA, RA, Puromorphine, and cAMP) in order to obtain small cell clumps. Clumps were then transferred to non-coated 10 cm Petri dishes (2–3 wells of a six-well plate per 10 cm plate) in NDM3 medium. At day 17, aggregates in suspension were dissociated with Accutase (Innovative Cell Technologies, AT104) and 120,000 neurons were plated per well in a 12-well plate in NDM4 (NDM supplemented with AA, BDNF, CNTF, GDNF, BrainXcell supplement). hMN were kept in culture up to DIV9. For every experiment, 3 independent differentiations were generated per line. hMN from other ALS lines (ALS-FUS and respective controls) were obtained following the latter differentiation protocol112 and kept in culture up to DIV7.
Mouse spinal MN cultures
Spinal cord MN cultures were prepared using a modified version of a published protocol113. In short, E13 embryos were collected and spinal cords were isolated and transferred to cold DPBS (Thermo Fisher Scientific, 14190144). Next, spinal cords were trypsinized in 0.1% trypsin solution (Applichem, A3964) followed by DNAse treatment in 50 μg/mL DNase solution (Applichem, A3778). Cells were seeded at a density of 60,000 cells/13 mm coverslip coated with 20 μg/mL PDL (Sigma-Aldrich, P0899) and 10 μg/mL laminin (Sigma-Aldrich, L2020) in plating medium (High glucose DMEM 1×, 5% FBS, 33 mM glucose, 1% P/S/A) supplemented with 0.1 ng/mL BDNF, CTNF and GDNF (STEMCELL Technologies, 78005/78010/78058). After 24 hours, coverslips were transferred to a maintenance medium (Neurobasal, 4% B27, 20 mM glucose, 2 mM L-glutamine, 2% P/S/A) supplemented with 0.1 ng/mL BDNF, CTNF, and GDNF. Three days after plating, 5 μM AraC was added to the medium to inhibit glia proliferation. Half of the medium was refreshed every other day.
Cerebral organoids
Cerebral organoids were generated using a modified version of a previously published protocol65,114. In short, iPSCs were dissociated into a single cell suspension with Accutase (Innovative Cell Technologies, AT104) and 3.5 × 106 cells were seeded per well in an Aggrewell800 microwell plate (StemCell Technologies, 27865) in 2 mL of stem cell medium (20% KOSR, 3% FBS, 2 mM L-glutamine, 1% MEM-NEAA and 7 μl/L 55 mM β-mercaptoethanol in DMEM-F12) supplemented with 4 ng/mL bFGF (Peprotech, 100-18B) and 48.2 μM ROCK-inhibitor Y-27632 (Axon Medchem, AXON 1683). After 48 h, EBs were transferred to ultra-low attachment 96-well plates (Corning, 3474). Medium was replaced at day 4 by stem cell media without factors and at day 6 by neural induction medium (1% N2, 2mM L-glutamine, 1% MEM-NEAA and 0.5 μg/ml heparin in DMEM-F12). At day 13, organoids were embedded in matrigel (Corning, 356234) and kept in organoid differentiation medium (0.5% N2, 1%B27 without RA, 2mM L-glutamine, 1% P/S, 0.5% MEM-NEAA, 2.5 μg/mL insulin, β-mercaptoethanol, in 1:1 DMEMF12:Neurobasal). Four days later, organoids were transferred to an orbital shaker (3.5 speed; Sigma Aldrich, Z768545) and organoid differentiation medium containing B27 with RA. Medium was changed every 2–3 days. Three batches per line were generated. Each batch had a minimum of 16 organoids. All experiments were performed with 3 control lines (CTL_1, CTL_4, CTL_7) and 2 ATXN2-ALS lines (ALS_A, ALS_B) unless stated otherwise.
Protein extraction and western blotting
Mice were sacrificed by cervical dislocation and dissected tissue was snap frozen on dry ice and stored at -80 °C. Samples were collected in ice-cold lysis buffer (20 mM Tris, pH 8, 150 mM NaCl, 10% glycerol, 1% Triton X-100 and complete protease inhibitor cocktail (Roche)). Mouse tissue was homogenized using a micropestle and a syringe. Cells were harvested and lysed directly in the well using a cell scraper. Lysates were incubated on ice for 10 min and in a rotor at 4 °C for 15 minutes followed by centrifugation at 16,000 rcf for 20 min at 4 °C. Supernatant was collected and protein concentration determined by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 10741395). Western blotting was performed using 50 µg of total protein per lane and NuPAGE LDS sample buffer (Invitrogen, NP0007) containing 10% β-mercaptoethanol. Samples were boiled for 5 min at 90 °C, separated in a NuPAGE 3–8% Tris-Acetate gradient gel (Invitrogen) and transferred onto a 0.45 µm nitrocellulose membrane (Amersham Hybond-C Extra, GE Healthcare). After blocking with 5% milk powder, 0.01% Tween 20 in Tris-buffered saline, membranes were incubated ON at 4 °C. Blots were probed with peroxidase-conjugated secondary antibodies for 1 hour at room temperature (RT) and developed with SuperSignal West Dura Extended Duration Substrate (Pierce, Thermo Fisher Scientific). Images were acquired using a FluorChem M imaging system (Protein Simple). Signal quantification was performed using Image Studio Lite (LI-COR) software. Individual band intensities were measured and normalized to ACTINB or TUBB levels.
Soluble and insoluble TDP-43 protein quantification was performed as previously reported48. In short, spinal cord tissue was homogenized in RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP40, 1% Sodium deoxycholate, 0.1% SDS, complete protease inhibitor cocktail and Phosphatase Inhibitor Cocktail 2 (Sigma-Aldrich)) using a motorized pestle and a syringe. Lysates were centrifuged at 16,000 rcf for 20 min at 4 °C. After supernatant collection (soluble fraction), pellets were washed with RIPA buffer and homogenized in UREA buffer (30 mM Tris-HCl pH 8.5, 7 M Urea, 2 M Thiourea, 4% CHAPS) using a motorized pestle. Lysates were centrifuged at 16,000 rcf for 30 minutes at RT and supernatants were collected (insoluble fraction). Protein concentration quantification and blotting were performed as described above using 30 µg of total protein per lane (soluble fraction) and 10% polyacrylamide gels. Membranes were blocked with supermix solution (50 mM Tris-HCl pH 7.4, 0.25% gelatin, 0.9% NaCl, 0.5% TX100) and incubated ON at 4 °C. Blots were probed with fluorescent-dye conjugated secondary antibodies (AB_621843, LI-COR Biosciences) for 1 hour at RT. Images were acquired using an Odyssey DLx imaging system (LI-COR Biosciences). Signal quantification was performed using Image Studio Lite (LI-COR Biosciences) software. Individual band intensities were measured and normalized to total protein levels using Revert™ 700 Total Protein Stain Kit (LI-COR Biosciences, 926-11010). Primary MN protein samples were obtained using the same protocol, followed by vacuum centrifugation until dry and resuspended in MiliQ to increase protein concentration. 8 µg of total protein were loaded (soluble fraction). Individual band intensities were measured and normalized to ACTINB levels. Uncropped and unprocessed scans of the most important blots are provided in the Source Data file.
RNA extraction, cDNA synthesis and quantitative RT-PCR
Fresh frozen mice tissue was collected in RNA lysis buffer and total RNA was isolated using the miRNeasy Mini kit (QIAGEN), according to the manufacturer’s instructions. To assess transgene levels in mice, cDNA was synthesized using MultiScribe Reverse Transcriptase (ThermoFisher). For the other qRT-PCR analyses, cDNA synthesis was performed using Superscript IV kit according to manufacturer’s instructions. iPSC and hMN samples were collected in Trizol and briefly vortexed before storing at −80 °C. For RNA extraction, samples were thawed on ice and kept at RT for 5 minutes. Chloroform was added followed by vigorous shaking for 15 seconds and high-speed centrifugation (12,000 × g) for 15 minutes at 4 °C. The aqueous phase was transferred to a new tube and mixed with 1.5 volumes of absolute ethanol. Samples were loaded onto mRNAeasy mini kit columns and extraction was performed according to manufacturer’s instructions. RNA quality and purity were analyzed in a NanoDrop. cDNA synthesis was conducted using the Superscript IV kit according to manufacturer’s instructions. Samples were spiked with 30 ng of C. elegans RNA to correct for cDNA synthesis efficiency. Quantitative real-time (q)PCR was performed with FastStart Universal SYBR Green Master (Rox) in a QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems). Expression levels were determined using the ddCT method. Expression levels of target genes were normalized to Gapdh, Hmbs, Hk2 and/or Tbp (mouse tissue), ACTINB and GAPDH (iPSC), TBP and GAPDH (iPSC-MN), Tbp and Rpl13 (spinal cord microglia) and TBP and RPII (organoid microglia) (see Table S15). Microglia homeostatic and disease-associated (DAM) genes were selected according to a source study59.
Tissue fixation and immunostaining
Mice were deeply anesthetized with Euthanimal (Alfasan) and intracardially perfused with ice-cold phosphate buffered saline (PBS, pH 7.4) followed by 4% Paraformaldehyde (PFA) in PBS. Brain, spinal cord and gastrocnemius muscle were dissected and fixed ON in 4% PFA at 4 °C. Fixed tissue was cryoprotected in 10% sucrose for 24 hours and then in 30% sucrose at 4 °C until saturation. The tissue was snap-frozen in isopentane and stored at −80 °C. Brain and spinal cord tissue (20 μm sections) and gastrocnemius muscle (30 μm sections) were obtained using a Leica CM1950 cryostat and mounted on SuperFrost Plus slides (Thermo Fisher Scientific). For some antibodies (see resources table), antigen retrieval was performed by boiling the sections in 10 mM citrate buffer, pH 6.0, in a microwave. For diaminobenzidine (DAB) stainings, endogenous peroxidases were quenched with 0.3% H2O2 in PBS-0.5% Triton-X-100 for 30 minutes followed by blocking in 1% BSA, 0.5% Triton-X-100 and 5% normal goat serum (NGS) in PBS for 30 minutes. Primary antibodies were incubated ON at 4 °C in blocking solution. Sections were incubated with secondary biotinylated antibodies for 1 hour at RT followed by 1 hour incubation in ABC reagent (Vector Laboratories). Slides were washed in Tris 50 mM, pH7.6, and developed with DAB reagent (Sigma). Sections were dehydrated in an Ethanol-Xylene series and mounted in Entallan® New (EDM Millipore). For immunofluorescent stainings, brain and spinal cord tissue sections were blocked in 1% BSA, 0.5% Triton-X-100 and 5% NGS in PBS for 1-2 hours at RT. For gastrocnemius muscle, tissue was first permeabilized for 30 minutes in 2% Triton-X-100 in PBS and then blocked in 3% BSA, 10% NGS and 1% Triton X-100 in PBS for 1 hour at RT. For TDP-43 immunostaining, blocking was performed with 5% Triton-X-100 to increase permeability. Primary antibodies were incubated ON at 4 °C in blocking buffer. Alexa Fluor-conjugated secondary antibodies (1:400/1:250 for muscle) were incubated in blocking buffer for 1-2 hours at RT. Bugarotoxin-488 (1:250) and Hoechst (1:1000, ThermoScientific, 33342) were incubated in blocking buffer for 2 hours at RT. Slides with muscle tissue were washed six times for 20 minutes in 0.5% Triton-X-100 in PBS after each antibody incubation. For lumbrical muscle (left hindlimb), tissue was dissected in ice- cold PBS and fixed in 4% PFA for 10 minutes. Whole muscle was permeabilized for 30 minutes with 2% Triton X-100 in PBS followed by blocking with PBSGT (0.2% gelatin + 0.5% Triton X-100 in PBS) and ON primary antibody incubation at 4 °C in PBSGT. Muscles were washed 3 × 30 min with 0.1% Triton X-100 in PBS (PBS-T) at RT prior to ON incubation at 4 °C with secondary antibodies and Bugarotoxin-488 in PBS. Muscles were washed 3 × 30 min in PBS-T at RT and mounted on microscope slides in FluorSave Reagent (Sigma-Aldrich).
Coverslips with human iPSC, hMN or mouse primary MN were fixed in 4% PFA at 4 °C for 15 minutes and then transferred to PBS and stored at 4 °C. Samples were blocked using a 10% Normal Donkey Serum, 3% BSA in 1% Triton-X-100 PBS solution for 30 minutes followed by incubation with primary antibodies ON at 4 °C. The following day, coverslips were washed and incubated with the secondary antibodies for 1 hour at RT.
Organoids were fixed in 4% PFA for 1 hour, washed in PBS and transferred to 30% sucrose solution at 4 °C for at least 24 hours. Next, organoids were transferred to base molds (M475-1, Simport Scientific), embedded in O.C.T (23-730-625, Fisher Scientific), snap frozen in isopentane and stored at –80 °C. 20 μm thick sections were obtained as specified above for mouse tissue. For immunostaining, organoid sections were blocked in 3% BSA, 1% Triton-X-100 and 10% Normal Donkey Serum in PBS for 45 minutes at RT. Primary antibodies were incubated ON at 4 °C in blocking buffer. Alexa Fluor-conjugated secondary antibodies (1:1000) were incubated in blocking buffer for 1 hour at RT.
4,6-diamidino-2-phenylindole (DAPI) or Hoechst (1:1000) were used to stain nuclei. All slides and coverslips were mounted with FluorSave reagent (345789-20,VWR), left to dry at RT and stored at 4 °C.
In situ hybridization
Fresh frozen spinal cord and brain tissue from adult mice were cut in 20 μm sections as described above and stored at -80 °C. Non-radioactive in situ hybridization was performed using an alkali-hydrolyzed digoxigenin-labeled cRNA probe transcribed from human ATXN2 (355 bp fragment corresponding to nucleotides 277–737). The probe was labeled using a DIG RNA labeling SP6/T7 kit (Roche, Basel, Switzerland). Sections were fixed in 4% PFA for 10 minutes at RT and then acetylated for 10 minutes at RT. Prehybridization was done for 2 hours at RT incubating the slides in hybridization mix (50% deionized formamide, 5× SSC, 5× Denhardt’s solution, 250 g/ml tRNA from bakers’ yeast (Millipore Sigma) and 500 mg/ml sonicated salmon sperm DNA). Probes (400 ng/ml) were incubated at 85 °C for 10 minutes in hybridization mix. Sections were hybridized ON at 68 °C followed by a quick wash in 2× SSC (68 °C) and a 2 hour wash in 0.2× SSC (68 °C). Then, sections were blocked with 10% FCS in B1 buffer (0.1 M Tris, pH 7.4, 0.15 M NaCl) for 1 hour at RT and incubated with anti-digoxigenin-AP Fab fragments (1:5000, Roche Diagnostics) in 1% FCS in B1 buffer ON at 4 °C. Slides were treated with BCIP and NBT substrates (NBT/BCIP stock solution, Roche Diagnostics) in 1× Levamisole (Sigma), 0.1 M Tris, pH 9.5, 0.1 M NaCl, 50 mM MgCl2 at RT. Color reaction was stopped with T10E5 (Tris 10 mM and EDTA 5 mM, pH 8.0) and sections were mounted using Vectashield (Vector Labs). Sense probes and NTg tissue were used to confirm specificity. Images were taken on an Axio Imager M2 microscope (Zeiss) and processed with ImageJ.
Estimation of transgene copy number
Transgene copy number was established by qRT-PCR on genomic DNA isolated from mouse ear punches using primers amplifying both human ATXN2 and mouse Ataxin-2. TATA box binding protein (tbp) and hexokinase 2 (hk2) were used as reference genes (see Table S15). DNA samples from 3 mice per genotype were amplified in duplicate in a QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems) using FastStart Universal SYBR Green Master (Rox) reaction mix. Quantification of Ataxin-2 (human and mouse) was calculated using the ΔΔCT method and the comparison between the transgenic lines and the non-transgenic (NTg) animals was used to estimate copy number.
Targeted locus amplification
Bone marrow samples were isolated from adult mouse hind limb bones. Dissected bones were crushed in a small mortar and the dissociated tissue was collected in PBS and centrifuged at 250 rcf for 5 min to collect bone marrow cells. The pellet was resuspended in cryopreserving freezing medium (10% DMSO and 10% FCS in PBS), frozen at 1 °C/min in a cryo-freezing container and stored at −80 °C. TLA was used to selectively amplify and identify the insertion sites (Cergentis B.V; The Netherlands)115. Three primer sets on the transgene were designed and used in individual TLA amplifications (see Table S16). PCR products were purified, library prepped using the Illumina Nextera flex protocol and sequenced in an Illumina sequencer. Reads were mapped using BWA-SW2 (version 0.7.15-r1140, settings bwasw −b 7). Next-generation sequencing reads were aligned to the transgene sequence and the host reference mouse mm10 genome. Integration sites were detected based on coverage peaks in the genome and the identification of fusion-reads between the transgene sequence and host genome.
Behavioral testing
All behavioral tests were performed on ≥8 mice per sex and genotype for each time point. Testing cohorts were age-matched, sex-matched and composed of mice of different genotypes. The observer was blinded for the genotype. Mice were weighed weekly. Grip strength was assessed every 4 weeks using a grip strength meter (Bioseb, Vitrolles, France). The maximum grip force G was measured 5 times for each mouse for both front and hind paws. Motor function was tested every 4 weeks on an accelerating rotarod apparatus (47600 model, Ugo Basile, Varese, Italy) at four rounds per minute (rpm) to 40 rpm. The latency to fall (s) was recorded 5 consecutive times in 180 seconds trials. One day prior to the test, mice were trained 2 times at a low acceleration mode (4–20 rpm in 120 s) and then three times at high acceleration mode (4–40 rpm in 180 s), with at least a 15 minutes break between the two settings. Mice that rotated passively were scored as fallen.
Spinal cord microglia isolation by magnetic automated cell sorting (MACS)
Mice were deeply anesthetized with Euthanimal (Alfasan) and intracardially perfused with ice-cold HBSS (14175095, ThermoFisher Scientific) followed by spinal cord collection on ice-cold HBSS. Tissue was sliced in small pieces using surgical blades and transferred into ice-cold HBSS. Next, samples were centrifuged for 1 minute, pelleted tissue was dissociated by gentle pipetting and centrifuged again. Pellet was resuspended in 3 mL of Papain (LK003178, Worthington) and DNAse (LK003172, Worthington) solution (1 vial Papain, 1 vial DNAse in 6 mL DMEMF12) and incubated for 45 minutes at 37 °C on a shaking platform (additional DNAse solution was added after 30 minutes). Next, samples were centrifuged for 10 minutes and pellet was resuspended in cold GNK/BSA solution (0.44% glucose, 0.66% BSA in PBS) followed by transfer to a 100 mm strainer (921008-950, VWR). Filtered cells were centrifuged for 10 minutes and resuspended in cold GNK/BSA solution. Next, Percoll gradient was performed by adding 1/3 of the final volume of Percoll (17-0891, VWR) followed by centrifugation for 30 minutes at RT. The resulting bottom layer was collected, cold GNK/BSA solution was added and samples were centrifuged for 10 minutes. All centrifugation steps were performed at 300 rcf at 4 °C unless specified.
CD11b+ cells were isolated by MACS according to the manufacturer’s protocol (130049601, Miltenyi). In short, the pellet was resuspended in cold MACS (5 mM EDTA, 10% FBS in PBS) buffer and centrifuged for 5 minutes. Single cell suspension was incubated with CD11b-coated beads (561015, BD Biosciences) for 20 minutes at 4 °C and magnetic separation was performed by passing cells through a 70um strainer (734-0003, VWR) onto LS columns (130-042-401, Miltenyi Biotec). CD11b+ fraction was collected and centrifuged at 300 rcf for 10 minutes. Pellets were collected for RNA isolation in lysis buffer.
Cerebral organoid microglia isolation by MACS
Organoids were dissociated into a single cell suspension as previously described with minor modifications65. In short, 10 to 12 organoids were washed in DPBS (ThermoFisher, 14190094) and dissociated in 6 mL of papain/DNAse solution (1 vial Papain, 1 vial DNAse in 6 mL DMEMF12). Cell suspension was passed through a 100 μm strainer before magnetic isolation of microglial cells. CD11b+ fraction was isolated by MACS, as described above, and according to the manufacturer’s protocol (130049601, Miltenyi). Pellets were collected for RNA isolation in lysis buffer.
Lipopolysaccharide (LPS) treatment of cerebral organoids
Organoids were treated with 100 ng/mL LPS from Escherichia coli (L4391, Sigma-Aldrich) for 24 hours. Next, microglial cells were isolated by MACS and collected in RNA lysis buffer for qRT-PCR analysis as described above.
LPS injections
7-months-old mice were intraperitoneally injected with 1 mg/kg LPS from Escherichia coli (0111:B4, InvivoGen) or PBS. 4.5 hours after injection, mice were deeply anesthetized with Euthanimal (Alfasan) and intracardially perfused with ice-cold HBSS followed by spinal cord collection on ice-cold HBSS. Subsequently, microglial cells were sorted using MACS and collected in RNA lysis buffer for qRT-PCR analysis as described above. LPS concentration and incubation time were chosen according to a published study116.
RNA isolation, library preparation, sequencing, and analysis
Total RNA was isolated from 10-months-old spinal cord samples (n = 3 mice per genotype) or 7-months-old spinal cord microglia samples (n = 3 samples per genotype, 3 spinal cords per sample pooled) using QIAzol Lysis reagent (Qiagen) and miRNeasy mini kit (Qiagen) with DNA digestion on-column, according to the manufacturer’s instructions. RNA concentration and integrity was validated using the RNA 6000 Nano Kit on a 2100 Bioanalyser (Agilent), with RNA integrity (RIN) values for all extractions >7.5. Samples were rRNA depleted and prepared for sequencing using SMARTer Stranded Total RNA Sample Prep Kit - HI Mammalian (Takara), using 350 ng per sample. Library size and purity control was performed on a 2100 Bioanalyzer using a High-Sensitivity DNA-Chip (Agilent). Library concentration was measured using a KAPA Library Quantification Kit (Roche). Prepared libraries were sequenced on three lanes PE100 with PhiX in lane control on the HiSeq4000 Illumina platform. Data was filtered and trimmed using trim_galore (v 0.4.1). Adaptor sequences were removed and added as part of the strand switching library preparation method used. The number of filtered reads per sample was between 67-113 M (average of 87 M reads/sample). For spinal cord microglia samples, a similar strategy was followed except for the kit used to prepare samples for sequencing (SMARTer Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian (Takara)). Input RNA was 10 ng and the number of filtered reads per sample was between 63-96 M (average of 72 M reads/sample).
Total RNA was isolated from DIV12 human iPSC-derived MN samples from 5 controls (CTL_1, CTL_2, CTL_4, CTL_5, CTL_6) and 5 ATXN2-ALS (ALS_A, ALS_B, ALS_G, ALS_N, ALS_P) lines. RNA (1μg in 5 μl) was isolated and extracted using the RNeasy Micro Kit (Qiagen) with DNA digestion on-column, according to the manufacturer’s instructions. RIN was verified in an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, Ca, USA), and only samples with RIN ≥ 8.0 were used for RNA sequencing. rRNA was depleted using the Ribo-Zero Magnetic Kit (human/mouse/rat; Epicenter) before library preparation. Stranded, paired-end sequencing libraries were prepared (Genome Analysis Facility, Groningen, The Netherlands) and sequenced in the Illumina HiSeq2500 platform (Illumina, San Diego, California). On average, sequencing depth was 10 M reads/sample.
Electrophysiology
Coverslips with human iPSC-derived MN cultures were placed in a recording chamber and continuously perfused at RT with artificial cerebrospinal fluid containing 120 mM NaCl, 3.5 mM KCl, 1.3 mM MgSO4, 1.25 mM NaH2PO4, 2.5 mM CaCl2, 10 mM D-glucose, and 25 mM NaHCO3, gassed with 95% O2 and 5% CO2, pH 7.4. Individual hMNs were selected for whole cell current clamp recordings using an upright microscope (Axoiskop, Zeiss). Patch pipettes for recording were produced from borosilicate glass (1.5 mm outer diameter, 0.86 mm inner diameter; Harvard Apparatus Limited; pipette resistance ~4–5 MΩ) on a P-97 Flaming/Brown micropipette puller (Sutter Instruments) and filled with pipette solution containing: 140 mM K-methanesulfonate, 10 mM HEPES, 0.1 mM EGTA, 4 mM MgATP, 0.3 mM NaGTP, pH 7.4, adjusted with KOH. Whole cell current clamp recordings were performed using an Axopatch 200B (Molecular Devices) amplifier. The responses were filtered at 5 kHz and digitized at 10 kHz using Digidata 1322 A (Axon Instruments, USA). Data was analyzed using pClamp 9.0 and Clampfit 9.2 (Axon Instruments). Recordings with a series resistance of less than 2.5 times the pipette resistance were accepted for analysis. If necessary, with a small holding current, hMNs were kept at −65 mV before the start of the current protocol. Cells were depolarized to induce spike trains in 10 steps of 10 nA with an interval of 30 seconds and duration of 500 ms. Hyperpolarizing current steps of −10 nA were included to study hyperpolarizing membrane properties. Two control (CTL_1, CTL_6) and 2 ATXN2-ALS (ALS_A, ALS_G) lines were used for electrophysiology experiments.
Single cell RNA sequencing
DIV12 human iPSC-derived MN cultures from 1 control (CTL_2) and 1 ATXN2-ALS (ALS_G) line were dissociated with trypsin (Sigma) and sorted into single wells, where fluorescent-activated cell sorting (FACS) into 384-well plates was combined with automated single cell RNA sequencing (SORT-seq pipeline). RNA was extracted by 2× heat lysis (65 °C) in 384 well and immediately used for downstream single-cell RNA sequencing reactions (Celseq2) as previously described117,118. Material was purified using Ampure beads (Beckman Coulter, #A63881) and amplified by in vitro transcription ON using the MEGAscript T7 transcription kit (ThermoFisher Scientific, # AMB1334). Amplified RNA was measured using the Agilent RNA 6000 pico chips (Agilent # 5067-1513), reverse-transcribed using random hexamer primers that introduce Truseq Small RNA kit RP1 primer binding sites (Illumina) and finally converted into DNA libraries using custom rpi primers (RNA PCR Primer Index) adapted from the Truseq Small RNA kit (Illumina). 96 cells were pooled to generate a single library. Following two rounds of Ampure bead clean up and quality control using the Agilent High Sensitivity DNA Kit (5067–4626), paired-end sequencing of libraries was performed by Nextseq500 (USEQ, Life Sciences faculty, Utrecht University).
Scanning electron microscopy
DIV12 human iPSC-derived MN cultures were fixed using 4% PFA followed by serial dehydration by incubation steps of 15 minutes in 12.5% EtOH/PBS, 25% EtOH/PBS, 50% EtOH/PBS, 75% EtOH/H2O, 90% EtOH/H2O, 100% EtOH, followed by incubation in 50% EtOH/50% hexamethyldisilizane (HDMS) and finally 100% HDMS. Samples were mounted onto aluminum specimen mounts (Agar Scientific), followed by coating with 4 nm gold using a Quorum Q150R S Rotary-Pumped Sputter Coater. Samples were examined with a FEI Nova NanoSEM 200 scanning electron microscope operated with an accelerated voltage of 10 kV at a magnification of 7500× using a Phenom Pro desktop scanning electron microscope and Pro Suite software (PhenomWorld). 3 control (CTL_1, CTL_3 and CTL_6) and 4 ATXN2-ALS (ALS_A, ALS_G, ALS_N, ALS_P) lines were used.
SGs analysis
DIV9 human iPSC-derived MNs plated on coverslips were transferred into new plates containing fresh NDM medium with 0.5 mM sodium arsenite for 60 minutes. After 60 minutes, coverlips were either fixed or transferred back into the initial wells containing NDM4 for another 120 minutes. Experiments with ATXN2-ALS lines were performed in 3 control (CTL_1, CTL_3, CTL_4) and 4 ATXN2-ALS lines (ALS_A, ALS_B, ALS_G, ALS_P), and 3 independent differentiations were performed per line. Experiments with FUS-ALS lines were performed in 2 control (CTL_1.2 and CTL_4) and 2 FUS-ALS lines (FUS2 and FUS2/2) in DIV7 cultures.
At day 7 after plating (DIV7), primary MN cultures were treated with 0.05 mM sodium arsenite for 30 and 60 minutes in a maintenance medium supplemented with 0.1 ng/mL BDNF, CTNF, and GDNF. After 30 or 60 minutes, coverslips were either fixed or transferred to fresh medium for another 120 minutes. 3 independent experiments per line were performed. Each culture contained 3–5 pooled E13.5 embryo spinal cords.
Seahorse
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were analyzed in a XF24-3 Extracellular Flux Analyzer (Seahorse Biosciences). Human iPSC-derived MNs from ATXN2-ALS lines (and respective controls) were plated in 24-well V7-PS plates (Seahorse Biosciences) at a density of 150,000 cells per well and used at DIV12. Human iPSC-derived MNs from FUS-ALS lines (and respective controls) were plated at a density of 350,000 cells per well and used at DIV7. Primary MNs were plated at a density of 70,000 cells per well and cultured for 7 days. Two wells from each plate were left blank as background controls. For measurements, cells were gently rinsed with 0.5 ml/well assay medium (20 mM glucose, 2 mM glutamine, 0.5 mM NaOH), put into fresh assay medium for at least 30 minutes, and assayed. For OCR analysis, three baseline recordings were made, followed by sequential injection of the ATP synthase inhibitor oligomycin, the mitochondrial uncoupler p-triflouromethoxyphenylhydrazone, and the mitochondrial complex 1 inhibitor rotenone and complex 3 inhibitor antimycin. For ECAR analysis, three baseline recordings were made, followed by sequential injection of glucose, oligomycin, and 2-Deoxy-D-glucose (2-DG). After each assay, individual wells were examined to ensure major cell death did not occur during the assay and cells remained evenly distributed. Wells that did not fulfill these criteria were excluded for the analysis. Raw oxygen consumption rate and extracellular acidification rate values were used to calculate different bioenergetic parameters as described previously119,120 and according to the manufacturer’s guidelines. For ATXN2-ALS experiments, 3 control (CTL_1, CTL_2, CTL_4) and 2 ATXN2-ALS (ALS_P and ALS_N) lines were used. For FUS-ALS experiments, 2 control (CTL_1.2 and CTL_4) and 2 FUS-ALS lines (FUS2 and FUS2/2) were used.
Quantification and statistical analysis
General statistical analysis
Statistical analysis was performed in GraphPad Prism version 9.1.2. (except for the scRNA-seq, RNA-seq and behavioral analysis). Datasets were tested for normality and differences in variance between groups to determine the appropriate statistical test. Tests applied for each experiment are stated in the figure legends as well as N. A test was considered significant when P < 0.05 unless specified otherwise. All statistical significance was ranked as the following: *P < 0.05; ** P < 0.01; and *** P < 0.001.
Behavioral analysis
Statistical analysis for animal behavioral assays was carried out using R software (http://www.r-project.org). Generalized (non-)linear mixed models were used to account for repeated measurements and other sources of correlation within the longitudinal data and to provide a correct model in case of missing data. For the behavioral analysis, body weight and sex were used as covariates. Survival and offspring genotype ratios were analyzed with GraphPad Prism 9.0 (GraphPad Software Inc). Kaplan–Meier curves were compared with the Log-rank (Mantel–Cox) test for the survival analysis. Chi-squared test was used to compare the observed offspring genotype ratios with the expected ratios.
Image analysis
All image analysis was performed using ImageJ/FIJI and blinded for the genotype or experimental condition. For the mouse histological analysis, n ≥ 3 mice per genotype per time point were included and both male and female mice were used, unless stated differently. For MN counting, n ≥ 10 spinal cord sections (L1–L6) per animal were imaged with an Axio Imager M2 microscope (Zeiss). CHAT+ neurons with a clear nucleus were manually counted. Gastrocnemius muscle NMJ were imaged with a FV1000 Olympus confocal microscope. Maximum intensity projections of z-stack images (1.00 μm interval) of at least n ≥ 10 NMJ post synapses were analyzed as previously described121. In brief, maximum intensity projections of AChR clusters stained by BTX were thresholded and outlined with the “create selection” tool and the area was measured. The endplate area was measured consecutively by subtracting the background (rolling ball radius at 50.0 pixels) and using the “create selection” tool to delimit the endplate. Lumbrical muscle NMJ were imaged using Axio Imager M2 (Zeiss) microscope with ×10 objective and were counted manually (BTX stained). For measurements of post-synaptic AChR (BTX stained) area/perimeter, all postsynaptic endplates were imaged using Olympos FV1000 confocal microscope with ×40 objective through the thickness of the muscle containing NMJs and maximum intensity projections of the z-stacks of images were used for measurements. NMJs with entire structure visible (‘en-face’; as described elsewhere122) were selected for measurement using at least 60 NMJs per mouse. The images were then processed with FIJI software as follows, Images were made 8-bit (Image-Type-8-bit), tresholded (Image-Adjust-Treshold-Default), NMJs selected and measured (area/perimeter). For measurement of presynaptic-postsynaptic (SV2/2H3-BTX) ‘overlap’, NMJs with clearly visible pre-synaptic axons and ‘en-face’ NMJ structure were imaged with Zeiss LSM880 confocal microscope using ×63 objective using following settings–8 bit depth, 512 × 512 frame size and 2 zoom, with sequential image acquisition to minimize bleed through. All image analysis was performed on maximum intensity projections of the z-stacks, using FIJI software following workflow of NMJ-morph platform described previously121 and ‘overlap’ of SV2/2H3 staining and AChR (BTX) staining was quantified on at least 15 NMJs per mouse. Lumbrical muscle quantifications were performed in male mice.
For PC density quantification, midsagittal cerebellum sections were imaged using an Axio Imager M2 microscope (Zeiss). 5 × 16-bit images were sharpened and background was subtracted. Next, images were thresholded using Yen AutoThreshold and converted into a mask. For cell counting, images were randomized and blinded and CALB+ cells were counted using the cell counter plugin in Fiji. Lobule perimeter was quantified by manually drawing a line on the PC layer.
To analyze TDP-43 mislocalization in the spinal cord and hMNs (same control and ATXN2-ALS lines as for SGs analysis were used, as specified before), z-stack images (1.0 μm interval for hMN; 1.5 μm interval for spinal cord sections) were acquired on a FV1000 Olympus confocal microscope. Relative TDP-43 expression in nucleus and cytoplasm was quantified in maximum intensity projections images by measuring integrated intensity (IntDen) of the separate compartments after manually delineating their respective area. The same images were used to manually determine the soma size by manually delineating the area in a blinded fashion.
SGs were visualized by immunocytochemistry for PABP. To determine the percentage of neurons containing SGs, coverslips were imaged using a Zeiss AxioScopeA1 fluorescence microscope and manually quantified in a blinded fashion. For analysis of the area and number of granules per neuron, cells were imaged in a FV1000 Olympus confocal microscope. Z-stacks were obtained and images were quantified using the particle analyzer function on FIJI. Briefly, z-stacks were transformed into maximum intensity projection, and turned into 8-bit images. Then background subtraction was performed followed by sharpening. Lastly, pictures were tresholded using the AutoThreshold function (MaxEntropy for ATXN2/CTL hMN; Intermodes for primary MN; Shanbhag for FUS/CTL hMN), converted into a mask and analyzed using particle analyzer (0.1–4 μm). For quantification of the number of SGs per MN, cells were selected manually, and the steps described above were performed for each individual neuron.
Neurite damage was assessed based on the degree of structural disruption observed in SEM images as previously described110,123. Quantitative analysis of neurite damage was performed using ImageJ software. For each condition (control vs ALS), at least 15 fields of view were randomly selected and neurons were analyzed (n = 15 for control, n = 42 for ALS). The extent of neurite damage was measured and quantified as a percentage of total damaged neurites per viewfield.
To assess inflammation, images of n ≥ 15 ventral horns (gray area) of the spinal cord were taken with a Zeiss AxioScopeA1 fluorescence microscope. Ventral horns were manually delimited using the selection tool. GFAP mean gray value was measured after background subtraction and IBA1+ area was measured after image thresholding. To calculate microglial cell density (cells/mm2), IBA1+ cells and nuclei were manually counted and the area of the ventral horn was measured. For the spinal cord microglia morphological analysis, n ≥ 25 IBA1+ cells were measured. Images were thresholded and circularity of the cells was measured after the cell perimeter was delimited with the “create selection” tool. Microglia roundness was measured by first applying a mask to the images (unsharp, radius 5 and mask 0.7) followed by a despeckle filter. Next, threshold was manually set to cover all IBA1+ cells and the “Analyze particles” function was used. Cells bigger than 10 μm and with a visible nucleus (excluding cells on the edge of the image) were selected from the ROI Manager (fragmented ROIs were combined with the OR command) and used for analysis using the “Measure” function with shape descriptors as parameters. n > 60 IBA1+ cells per animal were analyzed. For the organoid microglia morphology analysis, n ≥ 100 IBA1+ cells from n ≥ 3 organoids per line were assessed. Morphology categories were defined prior to the analysis and cells were blindly assigned to each category.
Spinal cord (microglia) RNA-seq
Filtered reads were mapped to the mouse genome (mm10) using Tophat2 and quantified with featureCounts software using gene annotation from Gencode release M19. Counts were normalized and differential expression analysis was performed using DESeq2 in R with an FDR < 0.05. All plotting was done in R. For the spinal cord dataset, GSEA was performed following author’s guidelines124. Default parameters from the website tool were used and gene lists were mapped against MSigDB hallmark mouse collection (2022.1 Mm version). Differential exon usage between groups was tested using DEXSeq as previously described125 with an FDR < 0.1. For the spinal cord microglia dataset, GO analysis was performed using metascape platform126.
Human iPSC-derived MN RNA-seq
Reads (2×100 bp) were demultiplexed and converted to FASTQ format using CASAVA software from Illumina. FASTQ files were mapped to the hg19/GCh37 reference human genome (iGenomes) with TopHat2 (version 2.0.13)50, using the ‘fr-firststrand’ option for strand orientation to generate BAM-formatted genomic coordinates. Count data for genes were analyzed in R using the Bioconductor package edgeR version 3.12.152 with the trimmed mean of M323 values (TMM) normalization method 53.
A generalized linear model was used to test the null hypothesis of absence of differential expression between the two groups. Gene expression levels were corrected for sex effect by including sex as a covariate in the model. P values adjusted for multiple testing were calculated using Benjamini Hochberg false discovery rate (FDR) and only genes at FDR < 0.05 were considered significantly differentially expressed. Volcano plots and expression MA scatter plots were generated in R using the ggplot2 library. DEGs at FDR < 0.1 were entered into the GO-term R package goseq54 to correct for bias due to transcript length. Gene set tests were conducted using the fry function55 that runs an infinite number of rotations to test whether a set of genes is differentially expressed by assessing the entire set of genes as a whole.
Human iPSC-derived MN sc RNA-seq
Quantification of paired-end reads was performed as previously described127 with the following exceptions. Reads that did not align, or aligned to multiple locations were discarded (MapAndGo). RaceID2 was used to analyze the single cell transcriptome data127. Unique molecular identifier (UMI) corrected reads were used to exclude PCR artifacts during library preparation117. Cells with less than 10,000 unique reads were filtered out; 306 (out of 384 cells) passed our filtering criteria, yielding a median of 20,434 unique reads and ~6000 genes per cell. Overall our database encompassed 16,846 genes. For cluster analysis, lowly expressed genes were excluded; only genes expressed in multiple cells and with at least five molecules detected in one cell were used for cluster analyses (5478 genes in total). Gap statistics were used to define an optimal number of six clusters, which were revealed using K-means clustering of the Pearson correlation of the whole transcriptome of cells. MN, interneuron, and progenitor clusters were assigned based on marker gene expression.
Resources table
REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
Antibodies | ||
Mouse anti-Ataxin-2 | BD Biosciences | Cat#611378; RRID: AB_398900 |
Mouse anti-β-actin | Sigma-Aldrich | Cat#A5316; RRID:AB_476743 |
Goat anti-ChAT | RnD systems | Cat#AF3447; RRID:AB_2079603 |
Rabbit anti-GFAP | Abcam | Cat#ab7260; RRID:AB_305808 |
Rabbit anti-IBA1 | Wako | Cat#019-19741; RRID:AB_839504 |
Rabbit anti-TDP-43 | ProteinTech | Cat#10782-2-AP; AB_615042 |
Rabbit anti-FUS | Sigma-Aldrich | Cat#HPA008784;RRID:AB_1849181 |
Mouse anti-ISLET1 | Hybridoma Bank | Cat#40.2D6; RRID:AB_528315 |
Mouse anti-TUJ1 | Covance | Cat#MMS-435P; RRID:AB_2313773 |
Rabbit anti-TUJ1 | Sigma-Aldrich | Cat#T2200; RRID:AB_262133 |
Mouse anti-TUJ1 | Biolegend | Cat#801201; AB_2313773 |
Mouse anti-TUJ1 | Sigma-Aldrich | Cat# T8328, RRID:AB_1844090 |
Mouse anti-β-Tubulin | Sigma-Aldrich | Cat#T5168; RRID:AB_477579 |
Rabbit anti-PABP | Abcam | Cat#ab21060; RRID:AB_777008 |
Mouse anti-G3BP | Abcam | Cat#ab56574; AB_941699 |
Mouse anti-SMA | Sigma-Aldrich | Cat#A2547; AB_476701 |
Rabbit anti-AFP | ProteinTech | Cat#14550-1-AP;RRID:AB_2223933 |
Mouse anti-Calbindin-D-28K | Sigma-Aldrich | Cat# C9848; RRID:AB_476894 |
Rat anti-Ctip2 | Abcam | Cat# ab18465, RRID:AB_2064130 |
Chicken anti-MAP2 | Abcam | Cat# ab92434, RRID:AB_2138147 |
Chicken-anti-Vimentin | Sigma-Aldrich | Cat# ab5733, RRID:AB_11212377 |
Rabbit-anti-PAX6 | ThermoFisher | Cat#42-6600, RRID:AB_2533534 |
Mouse-anti-NF-H (SH3) | Dshb | Cat# ab531793, RRID N/A |
Mouse-anti-SV2 | Dshb | Cat# SV2, RRID:AB_2315387 |
StemLightTM Pluripotency Antibody kit | Cell Signaling | Cat#9656S, RRID N/A |
anti-α-Bungarotoxin, Alexa FluorTM, 488 | ThermoFisher | Cat#B13422, RRID N/A |
Donkey anti-mouse Alexa FluorTM, 568 | ThermoFisher | Cat#A10037, RRID:AB_2534013 |
Donkey anti-mouse Alexa FluorTM, 555 | ThermoFisher | Cat#A31570, RRID:AB_2536180 |
Donkey anti-mouse Alexa FluorTM, 488 | ThermoFisher | Cat#A21202, RRID:AB_141607 |
Donkey anti-rabbit Alexa FluorTM, 568 | ThermoFisher | Cat#A10042, RRID:AB_2534017 |
Donkey anti-rabbit Alexa FluorTM, 488 | ThermoFisher | Cat#A21206, RRID:AB_2535792 |
Donkey anti-goat Alexa FluorTM, 488 | ThermoFisher | Cat#A11055, RRID:AB_2534102 |
Goat anti-rabbit Alexa FluorTM, 488 | ThermoFisher | Cat#A11034, RRID:AB_2576217 |
Goat anti-mouse Alexa FluorTM, 594 | ThermoFisher | Cat#A11032, RRID:AB_2534091 |
IRDye 800CW Goat anti-Rabbit | LI-COR Biosciences | Cat# 926-3221;RRID;AB_621843 |
Secondary biotinylated antibodies | ||
Horse anti-goat, Biotinylated | Vector Laboratories | Cat# BA-9500, RRID:AB_2336123 |
Goat anti-mouse, Biotinylated | Vector Laboratories | Cat# BA-9200, RRID:AB_2336171 |
Anti-digoxigenin-AP Fab fragments | Roche Diagnostics | Cat#11093274910, RRID:AB_2313640 |
Biological samples | ||
RNA from C. elegans | Mike Boxem | N/A |
RNA from hUES6 cell line (RRID: CVCL_B194) | Niels Gijsen | N/A |
Chemicals, peptides, and recombinant proteins | ||
Sodium Arsenite | Fisher Scientific | Cat# 11963203 |
CD11b microbeads, human and mouse | Miltenyi Biotec | Cat#130-049-601 |
Lipopolysaccharides (LPS) from Escherichia coli 0111:B4 strain | Sigma-Aldrich | Cat#L4391 |
Ultrapure LPS from Escherichia coli 0111:B4 strain | InvivoGen | Cat#tlrl-3pelps |
Critical commercial assays | ||
Seahorse XF Cell Mito Stress Test Kit | Agilent | Cat#103015-100 |
Pierce™ BCA Protein Assay Kit | Thermo-Fisher | Cat#23225 |
MycoAlert Mycoplasma detection kit | Lonza Bioscience | Cat#LT07-318 |
Revert 700 Total Protein Stain Kit | LI-COR Biosciences | Cat#926-11010 |
DIG RNA labeling kit (SP6/T7) | Roche | Cat#1175025910 |
SMARTer Stranded Total RNA Sample Prep kit-HI Mammalian | Takara Bio | Cat#634873 |
miRNeasy mini kit | Qiagen | Cat#217004 |
miRNeasy micro kit | Qiagen | Cat#217084 |
Experimental models: Cell lines | ||
iPSC cell line: CTL_1 | This paper | N/A |
iPSC cell line: CTL_1.2 | This paper | N/A |
iPSC cell line: CTL_2 | This paper | N/A |
iPSC cell line: CTL_3 | This paper | N/A |
iPSC cell line: CTL_4 | This paper | N/A |
iPSC cell line: CTL_6 | Ormel et al. 65 | N/A |
iPSC cell line: CTL_7 | This paper | N/A |
iPSC cell line: CTL_13 | This paper | N/A |
iPSC cell line: NL1 | Meyer et al. 107 | N/A |
iPSC cell line: NL2 | Meyer et al. 107 | N/A |
iPSC cell line: ATXN2-ALS-A | This paper | N/A |
iPSC cell line: ATXN2-ALS-B | This paper | N/A |
iPSC cell line: ATXN2-ALS-G | This paper | N/A |
iPSC cell line: ATXN2-ALS-N | This paper | N/A |
iPSC cell line: ATXN2-ALS-P | This paper | N/A |
iPSC cell line: ND12099 | Shi et al. 108 | N/A |
iPSC cell line: ND06769 | Shi et al. 108 | N/A |
iPSC cell line: ND10689 | Shi et al. 108 | N/A |
iPSC cell line: CS29 | Cedars-Sinai | CS29iALS-C9nxx |
iPSC cell line: FUS2 | Naujock et al. 109 | N/A |
iPSC cell line: FUS2/2 | Guo et al. 44 | N/A |
Experimental models: Organisms/strains | ||
Mouse: C57BL/6 J (JAX® Mice Strain) | Charles River Laboratories | Cat#632 |
Mouse: TDP-43M337V BAC knock-in | Jackson Laboratory | Cat#JAX029266 |
Mouse: ATXN2Q22/+ | This paper | N/A |
Mouse: ATXN2Q33/+ | This paper | N/A |
Mouse: ATXN2Q22/+; TDP-43M337V/+ | This paper | N/A |
Mouse: ATXN2Q33/+; TDP-43M337V/+ | This paper | N/A |
Oligonucleotides | ||
Genotyping and ATXN2 polyQ length primers (Table S14) | This paper | N/A |
TLA primers (Table S15) | This paper | N/A |
RT-qPCR primers (Table S16) | This paper | N/A |
Software and algorithms | ||
Fiji version 2.0.0-rc-69/1.52o & version 1.53f51 | Fiji | RRID:SCR_002285. https://imagej.net/Fiji |
Graphpad Prism version 9.1.1 | GraphPad Software | RRID:SCR_002798. https://www.graphpad.com/ |
trim_galore version 0.4.1 | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
Tophat2 version 2.0.13 | Johns Hopkins University CCB | |
R-Studio | R project for statistical computing | |
MapAndGo | github | https://github.com/anna-alemany/transcriptomics/tree/master/mapandgo |
DESeq2 (versionXXX) | ||
GO-term R package (goseq54) | ||
RaceID2 | ||
BWA-SW2 version 0.7.15-r1140, settings bwasw -b 7 | ||
Bioconductor package edgeR version 3.12.152 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
CASAVA software | zhanglabtools | |
Gene Set Enrichment Analysis GSEA 4.3.2 | Broad Institute | |
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA-seq and single cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are as follows: GSE224578 (iPSC hMN RNAseq), GSE224580 (Mouse spinal cord RNAseq), GSE224581 (Mouse spinal cord microglia RNAseq), GSE224582 (iPSC hMN single cell RNAseq).Given their complexity and diversity microscopy data are available on request. Source data are provided with this paper.
References
Brown, R. H. & Al-Chalabi, A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172 (2017).
van Es, M. A. et al. Amyotrophic lateral sclerosis. Lancet 390, 2084–2098 (2017).
Masrori, P. & Van Damme, P. Amyotrophic lateral sclerosis: a clinical review. Eur. J. Neurol. 27, 1918–1929 (2020).
Taylor, J. P., Brown, R. H. & Cleveland, D. W. Decoding ALS: from genes to mechanism. Nature 539, 197–206 (2016).
Van Damme, P., Robberecht, W. & Van Den Bosch, L. Modelling amyotrophic lateral sclerosis: progress and possibilities. Dis. Model. Mech. 10, 537–549 (2017).
Kim, G., Gautier, O., Tassoni-Tsuchida, E., Ma, X. R. & Gitler, A. D. ALS genetics: gains, losses, and implications for future therapies. Neuron 108, 822–842 (2020).
Byrne, S. et al. Rate of familial amyotrophic lateral sclerosis: a systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 82, 623–627 (2011).
Kenna, K. P. et al. Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. J. Med. Genet. 50, 776–783 (2013).
Gibson, S. B. et al. The evolving genetic risk for sporadic ALS. Neurology 89, 226–233 (2017).
Zou, Z.-Y. et al. Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 88, 540–549 (2017).
Nguyen, H. P., Van Broeckhoven, C. & van der Zee, J. ALS genes in the genomic era and their implications for FTD. Trends Genet. 34, 404–423 (2018).
Elden, A. C. et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-Linked ALS-FTD. Neuron 72, 257–268 (2011).
Blauw, H. M. et al. NIPA1 polyalanine repeat expansions are associated with amyotrophic lateral sclerosis. Hum. Mol. Genet. 21, 2497–2502 (2012).
Tazelaar, G. H. P. et al. ATXN1 repeat expansions confer risk for amyotrophic lateral sclerosis and contribute to TDP-43 mislocalization. Brain Commun. 2, fcaa064–fcaa064 (2020).
Lorenzetti, D., Bohlega, S. & Zoghbi, H. Y. The expansion of the CAG repeat in ataxin-2 is a frequent cause of autosomal dominant spinocerebellar ataxia. Neurology 49, 1009–1013 (1997).
Scoles, D. R. & Pulst, S. M. Spinocerebellar ataxia type 2 BT - polyglutamine disorders. In: C. Nóbrega and L. Pereira de Almeida, eds. (Springer International Publishing), pp. 175–195 (2018).
Sproviero, W. et al. ATXN2 trinucleotide repeat length correlates with risk of ALS. Neurobiol. Aging 51, 178.e1–178.e9 (2017).
Laffita-Mesa, J. M., Paucar, M. & Svenningsson, P. Ataxin-2 gene: a powerful modulator of neurological disorders. Curr. Opin. Neurol. 34, 578–588 (2021).
Ostrowski, L. A., Hall, A. C. & Mekhail, K. Ataxin-2: from RNA control to human health and disease. Genes (Basel) 8, 2–21 (2017).
Ross, O. A. et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum. Mol. Genet. 20, 3207–3212 (2011).
Borghero, G. et al. ATXN2 is a modifier of phenotype in ALS patients of Sardinian ancestry. Neurobiol. Aging 36, 2906.e1–2906.e29065 (2015).
Chiò, A. et al. ATXN2 polyQ intermediate repeats are a modifier of ALS survival. Neurology 84, 251–258 (2015).
Van Damme, P. et al. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology 76, 2066–2072 (2011).
Wang, M.-D., Gomes, J., Cashman, N. R., Little, J. & Krewski, D. Intermediate CAG repeat expansion in the ATXN2 gene is a unique genetic risk factor for ALS-a systematic review and meta-analysis of observational studies. PLoS One 9, e105534–e105534 (2014).
Neuenschwander, A. G., Thai, K. K., Figueroa, K. P. & Pulst, S. M. Amyotrophic lateral sclerosis risk for spinocerebellar ataxia type 2 ATXN2 CAG repeat alleles: a meta-analysis. JAMA Neurol. 71, 1529–1534 (2014).
Glass et al. ATXN2 intermediate expansions in amyotrophic lateral sclerosis. Brain 145, 2671–2676 (2022).
Hart, M. P. & Gitler, A. D. ALS-associated ataxin 2 PolyQ expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications. J. Neurosci. 32, 9133–9142 (2012).
Blokhuis, A. M. et al. Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol. 132, 175–196 (2016).
Becker, L. A. et al. Therapeutic reduction of ataxin 2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367 (2017).
Scoles, D. R. et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 544, 362–366 (2017).
Watanabe, R. et al. Intracellular dynamics of Ataxin-2 in the human brains with normal and frontotemporal lobar degeneration with TDP-43 inclusions. Acta Neuropathol. Commun. 8, 176 (2020).
Kiehl, T.-R. et al. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem. Biophys. Res. Commun. 339, 17–24 (2006).
Lastres-Becker, I. et al. Insulin receptor and lipid metabolism pathology in ataxin-2 knock-out mice. Hum. Mol. Genet. 17, 1465–1481 (2008).
Damrath, E. et al. ATXN2-CAG42 sequesters PABPC1 into insolubility and induces FBXW8 in cerebellum of old ataxic knock-in mice. PLoS Genet. 8, e1002920 (2012).
Jiménez-lópez, D. & Guzmán, P. Insights into the evolution and domain structure of ataxin-2 proteins across eukaryotes. BMC Res. Notes 7, 453 (2014).
Dansithong, W. et al. Ataxin-2 regulates RGS8 translation in a new BAC-SCA2 transgenic mouse model. PLoS Genet. 11, e1005182 (2015).
Fittschen, M. et al. Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics 16, 181–192 (2015).
Meierhofer, D. et al. Ataxin-2 (Atxn2) -knock-out mice show branched chain amino acids and fatty acids pathway alterations*. Mol. Cell Proteomics 15, 1728–1739 (2016).
Hart, M. P., Brettschneider, J., Lee, V. M. Y., Trojanowski, J. Q. & Gitler, A. D. Distinct TDP-43 pathology in ALS patients with ataxin 2 intermediate-length polyQ expansions. Acta Neuropathol. 124, 221–230 (2012).
van den Heuvel, D. M. A., Harschnitz, O., van den Berg, L. H. & Pasterkamp, R. J. Taking a risk: a therapeutic focus on ataxin-2 in amyotrophic lateral sclerosis? Trends Mol. Med. 20, 25–35 (2014).
Auburger, G., Sen, N.-E., Meierhofer, D., Başak, A.-N. & Gitler, A. D. Efficient prevention of neurodegenerative diseases by depletion of starvation response factor ataxin-2. Trends Neurosci. 40, 507–516 (2017).
Guo, W. et al. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat. Commun. 8, 861 (2017).
Pasteuning-Vuhman, S., de Jongh, R., Timmers, A. & Pasterkamp, R. J. Towards advanced iPSC-based drug development for neurodegenerative disease. Trends Mol. Med. 27, 263–279 (2021).
Cui, Q., Liu, Z. & Bai, G. Friend or foe: the role of stress granule in neurodegenerative disease. Neuron 112, 2464–2485 (2024).
Vandoorne, T. et al. Differentiation but not ALS mutations in FUS rewires motor neuron metabolism. Nat. Commun. 10, 4147 (2019).
Gordon, D. et al. Single-copy expression of an amyotrophic lateral sclerosis-linked TDP-43 mutation (M337V) in BAC transgenic mice leads to altered stress granule dynamics and progressive motor dysfunction. Neurobiol. Dis. 121, 148–162 (2019).
Tan, R. H. et al. Cerebellar neuronal loss in amyotrophic lateral sclerosis cases with ATXN2 intermediate repeat expansions. Ann. Neurol. 79, 295–305 (2016).
Bede, P. et al. Genotype-associated cerebellar profiles in ALS: focal cerebellar pathology and cerebro-cerebellar connectivity alterations. J. Neurol. Neurosurg. Psychiatry 92, 1197–1205 (2021).
Kabiljo, R., Iacoangeli, A., Al-Chalabi, A. & Rosenzweig, I. Amyotrophic lateral sclerosis and cerebellum. Sci. Rep. 12, 12586 (2022).
Scoles, D. R. et al. ALS-associated genes in SCA2 mouse spinal cord transcriptomes. Hum. Mol. Genet. 29, 1658–1672 (2020).
Wang, X. et al. Apelin/APJ system in inflammation. Int. Immunopharmacol. 109, 108822 (2022).
Maleki, K. T., Cornillet, M. & Björkström, N. K. Soluble SEMA4D/CD100: a novel immunoregulator in infectious and inflammatory diseases. Clin. Immunol. 163, 52–59 (2016).
Tian, M. et al. Functions of regulators of G protein signaling 16 in immunity, inflammation, and other diseases. Front. Mol. Biosci. 9, 962321 (2022).
Benkhoucha, M. et al. c-Met enforces proinflammatory and migratory features of human activated CD4+ T cells. Cell. Mol. Immunol. 18, 2051–2053 (2021).
Lall, D. & Baloh, R. H. Microglia and C9orf72 in neuroinflammation and ALS and frontotemporal dementia. J. Clin. Invest. 127, 3250–3258 (2017).
Clarke, B. E. & Patani, R. The microglial component of amyotrophic lateral sclerosis. Brain 143, 3526–3539 (2020).
Paolicelli, R. C. et al. Microglia states and nomenclature: a field at its crossroads. Neuron 110, 3458–3483 (2022).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017).
Colonna, M. & Butovsky, O. Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 35, 441–468 (2017).
Vincenti, J. E. et al. Defining the microglia response during the time course of chronic neurodegeneration. J. Virol. 90, 3003–3017 (2015).
Lull, M. E. & Block, M. L. Microglial activation and chronic neurodegeneration. Neurotherapeutics 7, 354–365 (2010).
Lloyd, A. F. et al. Deep proteomic analysis of human microglia and model systems reveal fundamental biological differences of in vitro and ex vivo cells. bioRxiv https://www.biorxiv.org/content/10.1101/2022.07.07.498804v1 (2022).
Ormel, P. R. et al. Microglia innately develop within cerebral organoids. Nat. Commun. 9, 4167 (2018).
Krasemann, S. et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9 (2017).
Verma, S. et al. Neuromuscular junction dysfunction in amyotrophic lateral sclerosis. Mol. Neurobiol. 59, 1502–1527 (2022).
Wong, M. & Martin, L. J. Skeletal muscle-restricted expression of human SOD1 causes motor neuron degeneration in transgenic mice. Hum. Mol. Genet. 19, 2284–2302 (2010).
Loeffler, J.-P., Picchiarelli, G., Dupuis, L. & Gonzalez De Aguilar, J.-L. The role of skeletal muscle in amyotrophic lateral sclerosis. Brain Pathol. 26, 227–236 (2016).
Prodoehl, J., Corcos, D. M. & Vaillancourt, D. E. Basal ganglia mechanisms underlying precision grip force control. Neurosci. Biobehav. Rev. 33, 900 (2009).
Huynh, D. P., Figueroa, K., Hoang, N. & Pulst, S.-M. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 26, 44–50 (2000).
Arnold, E. S. et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl Acad. Sci. 110, E736–E745 (2013).
Austin, J. A. et al. Disease causing mutants of TDP-43 nucleic acid binding domains are resistant to aggregation and have increased stability and half-life. Proc. Natl Acad. Sci. 111, 4309–4314 (2014).
Li, Y. R., King, O. D., Shorter, J. & Gitler, A. D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013).
Ling, S.-C., Polymenidou, M. & Cleveland, D. W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013).
Nonhoff, U. et al. Ataxin-2 interacts with the DEAD/H-Box RNA helicase DDX6 and interferes with p-bodies and stress granules. Mol. Biol. Cell 18, 1385–1396 (2007).
Michalik, A. & Van Broeckhoven, C. Pathogenesis of polyglutamine disorders: aggregation revisited. Hum. Mol. Genet. 12, R173–R186 (2003).
Liu, J. et al. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 2. J. Neurosci. 29, 9148–9162 (2009).
Arsović, A. et al. Mouse ataxin-2 expansion downregulates camkii and other calcium signaling factors, impairing granule—purkinje neuron synaptic strength. Int. J. Mol. Sci. 21, 6673 (2020).
Canet-Pons, J. et al. Atxn2-CAG100-KnockIn mouse spinal cord shows progressive TDP43 pathology associated with cholesterol biosynthesis suppression. Neurobiol. Dis. 152, 105289 (2021).
Farg, M. A. et al. Ataxin-2 interacts with FUS and intermediate-length polyglutamine expansions enhance FUS-related pathology in amyotrophic lateral sclerosis. Hum. Mol. Genet. 22, 717–728 (2013).
Sellier, C. et al. Loss of C9 ORF 72 impairs autophagy and synergizes with polyQ Ataxin‐2 to induce motor neuron dysfunction and cell death. EMBO J. 35, 1276–1297 (2016).
Nijssen, J., Comley, L. H. & Hedlund, E. Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathol. 133, 863–885 (2017).
Wang, W. et al. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nat. Med. 22, 869–878 (2016).
Seidel, G. et al. Quantitative global proteomics of yeast PBP1 deletion mutants and their stress responses identifies glucose metabolism, mitochondrial, and stress granule changes. J. Proteome Res. 16, 504–515 (2017).
Cornelius, N. et al. Evidence of oxidative stress and mitochondrial dysfunction in spinocerebellar ataxia type 2 (SCA2) patient fibroblasts: Effect of coenzyme Q10 supplementation on these parameters. Mitochondrion 34, 103–114 (2017).
Mccauley, M. E. & Baloh, R. H. Inflammation in ALS/FTD pathogenesis. Acta Neuropathol. 137, 715–730 (2019).
Zhao, W. et al. Extracellular mutant SOD1 induces microglial-mediated motoneuron injury. Glia 58, 231–243 (2010).
Zhou, Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020).
Bright, F., Chan, G., van Hummel, A., Ittner, L. M. & Ke, Y. D. TDP-43 and inflammation: implications for amyotrophic lateral sclerosis and frontotemporal dementia. Int. J. Mol. Sci. 22, 7781 (2022).
Spiller, K. J. et al. Microglia-mediated recovery from ALS-relevant motor neuron degeneration in a mouse model of TDP-43 proteinopathy. Nat. Neurosci. 21, 329–340 (2018).
Hunter, M. et al. Microglial transcriptome analysis in the rNLS8 mouse model of TDP-43 proteinopathy reveals discrete expression profiles associated with neurodegenerative progression and recovery. Acta Neuropathol. Commun. 9, 140 (2021).
Hickman, S., Izzy, S., Sen, P., Morsett, L. & El Khoury, J. Microglia in neurodegeneration. Nat. Neurosci. 21, 1359–1369 (2018).
Trias, E. et al. Emergence of microglia bearing senescence markers during paralysis progression in a rat model of inherited ALS. Front. Aging Neurosci. 11, 42 (2019).
Gosselin, D. et al. An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222 (2017).
Geirsdottir, L. et al. Cross-species single-cell analysis reveals divergence of the primate microglia program. Cell 179, 1609–1622.e16 (2019).
Masuda et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019).
Fattorelli, N. et al. Stem-cell-derived human microglia transplanted into mouse brain to study human disease. Nat. Protoc. 16, 1013–1033 (2021).
Deczkowska, A. et al. Disease-associated microglia: a universal immune sensor of neurodegeneration. Cell 173, 1073–1081 (2018).
Kafkafi, N. et al. Reproducibility and replicability of rodent phenotyping in preclinical studies. Neurosci. Biobehav. Rev. 87, 218–232 (2018).
Doetschman, T. Influence of genetic background on genetically engineered mouse phenotypes BT - gene knockout protocols: 2nd Edn. In: W. Wurst and R. Kühn, eds. (Humana Press), pp. 423–433 (2009).
Shan, X., Chiang, P.-M., Price, D. L. & Wong, P. C. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc. Natl Acad. Sci. 107, 16325–16330 (2010).
Mitchell, J. C. et al. Wild type human TDP-43 potentiates ALS-linked mutant TDP-43 driven progressive motor and cortical neuron degeneration with pathological features of ALS. Acta Neuropathol. Commun. 3, 36 (2015).
White, M. A. et al. TDP-43 gains function due to perturbed autoregulation in a Tardbp knock-in mouse model of ALS-FTD. Nat. Neurosci. 21, 552–563 (2018).
Alhindi, A., Boehm, I. & Chaytow, H. Small junction, big problems: neuromuscular junction pathology in mouse models of amyotrophic lateral sclerosis (ALS). J. Anat. 241, 1089–1107 (2022).
Gong, S., Yang, X. W., Li, C. & Heintz, N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 12, 1992–1998 (2002).
Meyer, K. et al. REST and neural gene network dysregulation in iPSC models of Alzheimer’s disease. Cell Rep. 26, 1112–1127.e9 (2019).
Shi, Y. et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat. Med. 24, 313–325 (2018).
Naujock, M. et al. 4-Aminopyridine induced activity rescues hypoexcitable motor neurons from amyotrophic lateral sclerosis patient-derived induced pluripotent stem cells. Stem Cells 34, 1563–1575 (2016).
Harschnitz et al. Autoantibody pathogenicity in a multifocal motor neuropathy induced pluripotent stem cell–derived model. Ann. Neurol. 80, 71–88 (2016).
Amoroso, M. W. et al. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 33, 574–586 (2013).
Du, Z. W. et al. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 6, 1–9 (2015).
Gonçalves, IdoC. G. et al. Neuronal activity regulates DROSHA via autophagy in spinal muscular atrophy. Sci. Rep. 8, 7907 (2018).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
de Vree, P. J. P. et al. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat. Biotechnol. 32, 1019–1025 (2014).
Raj, D. D. A. et al. Priming of microglia in a DNA-repair deficient model of accelerated aging. Neurobiol. Aging 35, 2147–2160 (2014).
Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 1–7 (2016).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e3 (2016).
Divakaruni, A. S., Rogers, G. W. & Murphy, A. N. Measuring mitochondrial function in permeabilized cells using the seahorse XF analyzer or a clark-type oxygen electrode. Curr. Protoc. Toxicol. 60, 25.2.1–25.2.16 (2014).
Zhang, L. & Trushina, E. Respirometry in neurons BT - techniques to investigate mitochondrial function in neurons. In: S. Strack and Y. M. Usachev, eds. (Springer New York), pp. 95–113 (2017).
Jones, R. A. et al. NMJ-morph reveals principal components of synaptic morphology influencing structure-function relationships at the neuromuscular junction. Open Biol. 6, 160240 (2016).
Mejia Maza, A. et al. NMJ-analyser identifies subtle early changes in mouse models of neuromuscular disease. Sci. Rep. 11, 12251 (2021).
Brüll, M., Geese, N., Celardo, I., Laumann, M. & Leist, M. Preparation of viable human neurites for neurobiological and neurodegeneration studies. Cells 13, 24 (2024).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102, 15545–15550 (2005).
Anders, S., Reyes, A. & Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 (2012).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Grün, D. et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015).
Acknowledgements
We thank Aaron Gitler, Stefan Pulst, and Renzo Mancuso for reading the manuscript. Marina de Wit, Liset Rietman, and Christiaan van der Meer for technical support. Niels Geijsen and Mike Boxem for providing reagents. This project was financially supported by The Netherlands Organization for Scientific Research (NWO-VICI), Stichting ALS Nederland (ProjectMINE, TOTALS and ALS-on-a-Chip), the MAXOMOD and INTEGRALS consortia (E-Rare-3, the ERANet for Research on Rare Diseases), the EU joint Program Neurodegenerative Diseases (JPND; TRIAGE) (to R.J.P.), the Adessium Foundation (2014-16; to L.v.d.B.), Horizon 2020 Marie Skłodowska-Curie grant agreement no. 721890 (circRTrain) (to J.K., R.J.P.), European Research Council (ERC) (grant agreement no. 772376 (EScORIAL) (to J.H.V.).
Author information
Authors and Affiliations
Contributions
Conceptualization: R.V.d.S., E.S.L., M.C.L., O.H., R.J.P.; methodology: R.V.d.S., E.S.L., M.C.L., D.v.d.H., O.H., H.-J.W., E.M., O.B., F.G., M.T.V., D.G., K.T., J.K.; experimental studies: R.V.d.S., E.S.L., M.C.L., D.v.d.H., O.H., S.K., D.V., H.K., L.B., S.V.-M., D.K.S., M.M., M.H.B., N.K., Y.A., V.R.V., R.d.J., T.L.-M., L.P., S.S., T.V., G.K., T.C.P., E.M., O.B., M.T.V., P.Z.; clinical data & patient material: O.H., M.A.v.E., J.H.V., L.H.v.d.B.; writing–original draft: R.V.d.S., E.S.L., M.C.L., R.J.P.; writing–review, all; resources: D.G., K.T.; supervision: J.K., R.J.P.; funding acquisition, R.J.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vieira de Sá, R., Sudria-Lopez, E., Cañizares Luna, M. et al. ATAXIN-2 intermediate-length polyglutamine expansions elicit ALS-associated metabolic and immune phenotypes. Nat Commun 15, 7484 (2024). https://doi.org/10.1038/s41467-024-51676-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51676-0
This article is cited by
-
Targeting biomolecular condensates: beyond dissolution
BMC Biology (2026)
-
Hydrogen bond breakage in DNA molecules induced by nonuniform energy distribution
Russian Physics Journal (2025)
-
Spinocerebellar ataxia type 2 followed by amyotrophic lateral sclerosis due to a pure CAG repeat expansion in ATXN2: a case report and literature review
Neurological Sciences (2025)
-
Distinct patterns of cerebral and spinal pathology along the spectrum of ATXN2-related disorders
Journal of Neurology (2025)
-
An observational study of pleiotropy and penetrance of amyotrophic lateral sclerosis associated with CAG-repeat expansion of ATXN2
European Journal of Human Genetics (2025)