Abstract
Near-infrared (NIR) probes are highly sought after as fluorescent tags for multicolor cellular and in vivo imaging. Here we develop small NIR fluorescent nanobodies, termed NIR-FbLAG16 and NIR-FbLAG30, enabling background-free visualization of various GFP-derived probes and biosensors. We also design a red-shifted variant, NIR-Fb(718), to simultaneously target several antigens within the NIR spectral range. Leveraging the antigen-stabilizing property of the developed NIR-Fbs, we then create two modular systems for precise control of gene expression in GFP-labeled cells. Applying the NIR-Fbs in vivo, we target cells expressing GFP and the calcium biosensor GCaMP6 in the somatosensory cortex of transgenic mice. Simultaneously tracking calcium activity and the reference signal from NIR-FbLAGs bound to GCaMP6 enables ratiometric deep-brain in vivo imaging. Altogether, NIR-FbLAGs present a promising approach for imaging and manipulating various processes in live cells and behaving animals expressing GFP-based probes.
Similar content being viewed by others
Introduction
Near-infrared (NIR) fluorescent proteins (FPs) are useful for deep-tissue imaging. Compared with a visible spectral range (~450–650 nm), a NIR optical window (~650–900 nm) has the advantages of less phototoxicity, less absorption by tissue pigments, and less light-scattering1. NIR FPs can also be combined with FPs, biosensors, and optogenetic tools operating in visible spectral range without spectral crosstalk in multiplexing experiments2. Moreover, NIR FPs and their fusions can be used as optical guide stars for light focusing and correcting aberrations of visible-light FPs and biosensors in fluorescence2,3 and photoacoustic deep tissue imaging4,5.
Recently, a series of small (17 kDa) NIR FPs, termed miRFPnanos6,7,8,9, have been designed. Because of their small size and closely located N- and C-termini, miRFPnanos are ideal probes for insertion into proteins as internal tags. An insertion of miRFP670nano3 into GFP-binding nanobody (Nb; 15 kDa) resulted in a fluorescent Nb, termed NIR-FbGFP7. Importantly, while common Nb and their N- or C-terminal FP fusions are expressed in mammalian cells regardless of the presence of cognate antigen, NIR-Fbs are allosterically unstable and degrade fast but become highly stabilized and, consequently, brightly fluoresce when bound to their cognate antigen. This unique feature of NIR-FbGFP allows background-free visualization of GFP-tagged proteins7, unlike N- or C-terminal fusions of Nbs to FPs, known as chromobodies. While, similarly to NIR-Fbs, chromobodies are advantageous in the cases where a direct fusion of a protein of interest with FPs alters behavior10, the latter ones suffer from the high background due to an excess of unbound chromobodies in the cell11. However, we found that NIR-FbGFP cannot bind biosensors based on GFP-derived FPs, likely because their binding epitope is hindered by analyte-binding parts12,13.
FPs, mainly GFP-derived FPs, serve as the major scaffold to design the majority of available intensiometric fluorescent biosensors that sense various ions14,15, analytes16,17, neurotransmitters18,19, receptors18, G-proteins20, and enzymes21. Among them, genetically encoded calcium biosensors are a remarkable example of GFP-based probes for studying brain functioning in vivo14,22. The most popular series of calcium biosensors is GCaMP23 and its improved versions14,24,25,26.
The ability to chronically monitor calcium dynamics in specific cell types is the main advantage of calcium biosensors. To this, several Thy1 transgenic mice with a high level of expression were generated25,27, which were successfully applied for recording and mapping neuronal activity in various brain regions with high spatiotemporal resolution28,29. However, transgenic mice express GCaMP with significant differences in the level and patterns of expression between lines due to stochasticity in the transgene cassette integration site. It was shown that Thy1-GCaMP2.2, Thy1-GCaMP3, Thy1-GCaMP6s, and Thy1-GCaMP6f transgenic mice express calcium biosensors in different cell fractions across different cortical regions25,27. Additionally, GCaMP transgenic mice exhibit mosaic expression with uneven transgene distribution across brain regions in one line25,27.
On the other hand, the basal calcium ions level may differ in various neuronal populations, and calcium biosensors differ in their affinity to calcium ions30. All this complicates an interpretation of data obtained during the imaging of multiple brain areas and requires the use of several transgenic lines for comparison. In addition, in vivo imaging may suffer from animal movements, photobleaching, and autofluorescence, especially when FPs with blue-green spectra are used. Therefore, the second fluorescent dye, which recolors cell populations heterogeneously expressing GFP and GFP-based probes and thus acting as a reference, could solve the problem of discrepancies in the patterns and levels of the probe expression and imaging conditions.
In this work, we develop a series of NIR-Fbs, named NIR-FbLAG16 and NIR-FbLAG30, which bind not only GFP-derived FPs but also GFP-based biosensors for calcium ions, pyruvate, and glucose. The binding of NIR-FbLAGs to biosensors has no pronounced effect on their dynamics and ability to recognize respective analytes in live cells. We show that NIR-FbLAGs effectively recognize biosensors and degrade in their absence, allowing background-free imaging in live cells and deep in the mouse brain in vivo. We also expand the spectral palette of Nbs by designing an additional NIR-Fb with emission at 718 nm, compared to 670 nm of the current NIR-Fbs, and apply it to multicolor NIR imaging of different antigens. We then design two modular NIR-FbGFP-based systems, which activate gene transcription only in the presence of GFP. Next, we validate NIR-FbGFP and both NIR-FbLAGs in the mouse brain of several transgenic mouse lines. Finally, we simultaneously monitor calcium activity and a stable reference signal of NIR-FbLAGs, bound to calcium biosensor, in GCaMP6f-expressing neurons in vivo.
Results
Design and screening of NIR Nbs for GFP-based biosensors
To determine whether previously developed NIR-FbGFP can bind GFP-based biosensors, we coexpressed it with GCaMP6s and GFP-derived blue EBFP2 targeted to the nucleus, and observed precise colocalization of NIR-FbGFP with EBFP2 but no NIR fluorescence in the cytoplasm where GCaMP6s was expressed (Supplementary Fig. 1). This indicated that NIR-FbGFP labels various GFP-derived FPs, such as EBFP2, but not GFP-based biosensors. Nevertheless, this NIR-FbGFP feature can be useful for NIR-targeting of different transgenic mouse and cell lines expressing proteins of interest fused with GFP-derived FPs, making them compatible with the co-expressed GFP-based biosensors.
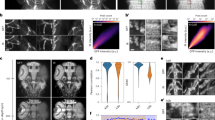
Indeed, according to the crystal structure of GBP1 Nb12,13,31 (PDP ID: 3OGO), which was used to design NIR-FbGFP7, it binds GFP in proximity to the chromophore hydroxyl12 and, therefore, does not bind GFP-based biosensors since analyte-binding part of the sensor interferes with the Nb: GFP interaction interface (Fig. 1a, b).
a Crystal structure of a Nb (PDB ID: 3OGO) with internally fused miRFP670nano3 (PDB ID: 7LSC) (NIR-Fb) bound to GFP-based biosensor GCaMP6m (PDB ID: 3WLD). Complementarity-determining regions (CDRs) are highlighted in violet. The position of miRFP670nano3 insertion to the anti-GFP Nb is indicated with a red arrow. b Crystal structure of GBP1 (PDB ID: 3OGO) Nbs to GFP. c Fluorescence intensity of cells transfected with eight different NIR-Fbs to GFP co-expressed with mEGFP (+) or mTagBFP2 (−). d Crystal structures of LAG16 (PDB ID: 6LR7) and LAG30 (PDB ID: 7SAI) Nbs to GFP. In a, b and d black arrows indicate chromophore. Red arrows indicate the position where miRFP670nano3 was inserted. e Far-red fluorescence intensity of cells transfected with NIR-FbLAG16, NIR-FbLAG30 or NIR-FbGFP and coexpressed with GCaMP6s biosensor in its apo or saturated states or with GCaMP6s with truncated M13 peptide. f Green fluorescence intensity of cells transfected with NIR-FbLAG16, NIR-FbLAG30 or NIR-FbGFP and co-expressed with GCaMP6s in its apo or saturated states or with GCaMP6s with truncated M13 peptide. In c, e and f fluorescence intensity was analyzed by flow cytometry using a 640 nm excitation laser and a 660/20 nm emission filter for NIR-Fbs, a 488 nm excitation laser and a 525/50 nm emission filter for mEGFP, a 405 nm excitation laser and a 450/50 nm emission filter for mTagBFP2. c, e, f Data are presented as mean values ± s.d. for n = 3 transfection experiments. Source data are provided as a Source Data file.
Therefore, to engineer a NIR Nb for GFP-based biosensors (Fig. 1a), we first analyzed crystal structures of several high-affinity anti-GFP Nbs. For further analysis, we have chosen two Nbs, called LAG16 and LAG30, with Kds values in the sub-nanomolar range (0.7 nM for LAG16 and 0.5 nM for LAG3032) and two Nbs for thermostable GFP (TGFP), called Sb44 and Sb92, with slightly lower Kds (4.4 and 3.3 nM, respectively33). We also used GBP2, GBP6, and GBP7 anti-GFP Nbs with unknown crystal structures but which recognize different epitopes compared to GBP113.
We inserted miRFP670nano3 into these seven Nbs between the residues, corresponding to NbGFP residues Ser65 and Val66 with Gly4Ser linkers (Fig. 1a and Supplementary Fig. 2). The internally tagged Nbs were co-expressed in HeLa cells with its cognate antigen mEGFP or with non-GFP-derived blue fluorescent protein mTagBFP234, as a negative control. As expected, no NIR fluorescence was detected in mTagBFP2 co-expressed cells, whereas the highest fluorescence level was observed for LAG16 and LAG30 Nbs (Fig. 1c and Supplementary Fig. 3). We named these constructs NIR-FbLAG16 and NIR-FbLAG30, respectively. Compared with parental NIR-FbGFP, the fluorescence intensity of NIR-FbLAG16 and NIR-FbLAG30 was 2.7- and 1.7-fold lower, respectively (Fig. 1c and Supplementary Fig. 4). LAG16 and LAG30 have 90% sequence identity (Supplementary Fig. 5) and bind GFP at closely located epitopes on the opposite to the chromophore hydroxyl side of the protein β-barrel35,36 (Fig. 1d). It makes NIR-FbLAG16 and NIR-FbLAG30 perfect candidates for binding GFP-based biosensors. A low NIR fluorescence was also observed for GBP6 with inserted miRFP670nano3 (Fig. 1c), though the affinity of GBP6 to GFP is much lower than that of GBP1, LAG16 and LAG3013,32. No fluorescence was observed for Sb44 and Sb92 with inserted miRFP670nano3 (Fig. 1c), likely because their affinity to GFP is ~10-fold lower compared to LAG16 and LAG3032,33.
Characterization of NIR-FbLAG constructs
To validate that inserted miRFP670nano3 did not change its spectra, we expressed NIR-FbLAG30 in bacteria and found that it preserved the parental spectral properties32, exhibiting fluorescence excitation and emission maxima at 645 nm and 670 nm, respectively (Supplementary Fig. 6a, b). When mEGFP was added to the Ni-NTA agarose preloaded with NIR-FbLAG30, the bright green and NIR fluorescence were observed only in the elution fraction (Supplementary Fig. 6c, d), confirming the ability to produce functional recombinant NIR-FbLAGs in bacteria.
To assess whether an antigen concentration affects the protein level of NIR-FbLAGs, we cotransfected cells with the fixed amount of plasmids encoding either NIR-FbLAG16 or NIR-FbLAG30 and decreasing amounts of a plasmid encoding EGFP antigen. Although we used the same amount of the NIR-FbGFP plasmid for cell transfection, the fluorescence intensity of NIR-FbLAGs varied and depended on the EGFP level (Supplementary Fig. 7), confirming that NIR-FbLAGs exhibit the antigen-dependence in a dose-dependent manner.
It has been shown that in the absence of EGFP antigen, NIR-FbGFP degrades by a ubiquitin-proteasome system7. To validate this mechanism for the NIR-FbLAGs nanobodies, we treated cells expressing NIR nanobodies in the absence of a cognate antigen with a proteasomal inhibitor bortezomib. While nontreated cells exhibited no NIR fluorescence, incubation of cells with 10 µM bortezomib for 20 h increased the fluorescence of NIR-FbLAGs by 75% (Supplementary Fig. 8), confirming that unbound NIR-FbLAGs degrade by the ubiquitin-proteasome system.
To assess if engineered NIR-FbLAG16 and NIR-FbLAG30 could bind GFP-based biosensors, we cotransfected them with GCaMP6s14 into HeLa cells. Cells were incubated in calcium-saturated (4 mM CaCl2) or calcium-depleted (2 mM EDTA) buffers to determine to which GCaMP6s conformation NIR-FbLAGs would predominantly bind. To eliminate conformation changes in calcium-binding parts upon binding calcium ions, we constructed a truncated GCaMP6s version with deleted M13 peptide, which did not respond to elevated calcium37. After 1 h incubation, no fluorescence was observed for NIR-FbGFP (Fig. 1e), indicating it predictably does not bind GCaMP6s because its epitope on GFP is structurally hidden by calcium-binding parts (Fig. 1a, b)38,39. Both NIR-FbLAGs effectively bound the GCaMP6s version without M13 (Fig. 1e). However, NIR-FbLAGs exhibited ~2.3–2.8-fold lower fluorescence in the calcium-saturated conditions than in calcium-free conditions, suggesting lower binding affinity of NIR-FbLAGs to calcium-bound GCaMP6s during a prolonged time scale. At the same time, the green fluorescence of GCaMP6s was ~2.2-fold higher in calcium-saturated conditions (Fig. 1f), which is consistent with its mechanism of action14.
To estimate whether NIR-FbLAGs binding affects the fast association and dissociation calcium kinetics of GCaMP6s, we performed stopped-flow experiments with purified GCaMP6s only or with GCaMP6s in the presence of NIR-FbLAG30. The fluorescence changes during rapid mixing of GCaMP6s, either with or without NIR-FbLAG30 with a series of Ca2+ buffers, provided observed relaxation rate constants (kobs). We analyzed the dependence of kobs from the free calcium ions concentration in the range of 0–350 nM (Supplementary Fig. 9a–c) and found that kobs were not affected by the NIR-FbLAG30 binding up to 250 nM of free calcium (Supplementary Fig. 9c). At 300 nM, GCaMP6s in the presence of NIR-FbLAG30 has a 1.3-fold slower calcium binding kinetics (Supplementary Fig. 9c). At the same time, we found that dissociation rate constants (koff) were rather similar for free and NIR-FbLAG30–bound GCaMP6s: 1.26 s−1 and 1.36 s−1, respectively (Supplementary Fig. 9d). We then assessed the effect of NIR-FbLAG30 binding to the GCaMP6s on the affinity of the indicator for calcium ions. According to the calcium titration experiment, GCaMP6s demonstrated comparable Kd in the presence and absence of NIR-FbLAG30: 148 ± 14 nM and 144 ± 4 nM, respectively (Supplementary Fig. 9e), which is consistent with the kinetics data. Because GCaMP6s is ≥90% saturated by bound calcium at 300 nM, differences in binding kinetics at calcium concentration above 300 nM should not contribute much to the fluorescence changes of NIR-FbLAG30-bound GCaMP6s. Overall, these data indicate that NIR-FbLAG30 does not interfere with the calcium binding and release and does not block GCaMP6s in “on” or “off” fluorescent states during fast changes of calcium concentration, which might be particularly valuable for in vivo brain imaging.
Performance of NIR-FbLAGs co-expressed with GCaMP6s
To study NIR-FbLAGs’ performance in live mammalian cells, they were co-expressed with mEGFP, GCaMP6s, or mTagBFP2 negative control and imaged with an epifluorescence microscope. NIR fluorescence was observed in the cells co-expressing mEGFP or GCaMP6s but not mTagBFP2 (Fig. 2a), suggesting that NIR-FbLAGs are stabilized only in the presence of its cognate antigen. The intracellular patterns of expression for NIR-FbLAGs coincided with those of mEGFP and GCaMP6s expression, confirming efficient NIR-FbLAG16 and NIR-FbLAG30 binding to their cognate antigen in live cells (Fig. 2a).
a Fluorescence images of HeLa cells coexpressing NIR-FbLAG16 or NIR-FbLAG30 anti-GFP Nb together with mTagBFP2 (negative control), mEGFP (positive control), and GCaMP6s. b Left, contrast of GCaMP6s only (n = 14) and GCaMP6s coexpressed with either NIR-FbLAG16 (n = 11) or NIR-FbLAG30 (n = 11) after the addition of 5 μM ionomycin. Right, the contrast of NIR-FbLAG16 and NIR-FbLAG30 for the data presented in the left graph. Data are presented as mean values ± s.e.m. for n = 3 transfection experiments. c Change in fluorescence intensity of the cells co-expressing GCaMP6s (green) and NIR-FbLAG16 (red) in response to 5 μM ionomycin. d Change in fluorescence intensity of the cells co-expressing GCaMP6s (green) and NIR-FbLAG30 (red) in response to 5 μM ionomycin. In c and d, data are shown for 3 cells, and the arrow indicates the time point when 5 μM ionomycin was added. For imaging of mTagBFP2, mEGFP or GCaMP6s and NIR-FbLAGs, 390/40 nm excitation and 460/40 nm emission, 480/40 nm excitation and 530/40 nm emission, and 605/40 nm excitation and 640LP nm emission filters were used, respectively. Scale bar, 40 μm. Source data are provided as a Source Data file.
To test whether NIR-FbLAG16 or NIR-FbLAG30 fluorescence will change upon calcium increase and whether NIR-FbLAGs interaction with GCaMP6s will change its ability to bind calcium ions, we added 5 μM ionomycin ionophore to the HeLa cell cultures co-expressing NIR-FbLAGs and GCaMP6s and time-lapsed imaged them. We observed that the maximal contrast of GCaMP6s was not substantially changed in cells co-expressing NIR-FbLAGs (Fig. 2b). Fluorescence intensity for both NIR-FbLAG16 and NIR-FbLAG30 did not change after ionomycin addition too (Fig. 2b–d), suggesting that NIR-FbLAGs do not influence GCaMP6s performance, and calcium binding does not affect NIR-FbLAGs interaction with GCaMP6s, reflected by the unchanged NIR fluorescence after calcium increase (Fig. 2c, d).
Targeting various types of GFP-biosensors with NIR-FbLAGs
To study whether NIR-FbLAGs can bind other GFP-based biosensors, we have chosen biosensors that detect chemical analytes other than calcium, such as Green Pegassos16, PyronicSF40, and iGlucoSnFR17,41. Green Pegassos and PyronicSF are pyruvate biosensors based on pyruvate dehydrogenase transcriptional regulator (PdhR) from E.coli inserted into GFP or linked to a circularly permuted version of GFP (cpGFP), respectively16,40. iGlucoSnFR is a glucose biosensor based on the insertion of cpGFP into a glucose-binding protein from Thermus thermophilus17.
We co-expressed Green Pegassos, PyronicSF, and iGlucoSnFR with either NIR-FbLAG16 or NIR-FbLAG30 and analyzed their fluorescence in live cells (Fig. 3a, b). We observed bright fluorescence of NIR-FbLAGs in cells co-expressing GFP-based biosensors or mEGFP but not mTagBFP2 negative control, suggesting their efficient antigen-dependent stabilization. Moreover, NIR-FbLAGs fully colocalized with the biosensors and exhibited no background fluorescence (Fig. 3a, b), confirming the NIR-FbLAGs ability to bind various GFP-based biosensors.
a Fluorescence images of HeLa cells co-expressing NIR-FbLAG16 anti-GFP Nb together with mTagBFP2 (negative control), mEGFP (positive control), pyruvate sensors Green Pegassos and PyronicSF and glucose sensor iGlucoSnFR. b Fluorescence images of HeLa cells co-expressing NIR-FbLAG30 anti-GFP Nb together with mTagBFP2 (negative control), mEGFP (positive control), pyruvate sensors Green Pegassos and PyronicSF and glucose sensor iGlucoSnFR. c Left, contrast of Green Pegassos only (n = 10) and Green Pegassos coexpressed with either NIR-FbLAG16 (n = 12) or NIR-FbLAG30 (n = 14) after the addition of 1 mM pyruvate. Right, the contrast of NIR-FbLAG16 and NIR-FbLAG30 for the data presented in the left graph. d Left, contrast of PyronicSF only (n = 12) and PyronicSF co-expressed with either NIR-FbLAG16 (n = 13) or NIR-FbLAG30 (n = 12) after addition of 10 mM pyruvate. Right, the contrast of NIR-FbLAG16 and NIR-FbLAG30 for the data presented in the left graph. e Left, contrast of iGlucoSnFR only (n = 16) and iGlucoSnFR co-expressed with either NIR-FbLAG16 (n = 14) or NIR-FbLAG30 (n = 14) after addition of 50 mM glucose. Right, the contrast of NIR-FbLAG16 and NIR-FbLAG30 for the data presented in the left graph. For imaging of mTagBFP2, mEGFP or Green Pegassos, PyronicSF and iGlucoSnFR, and NIR-FbLAGs, 390/40 nm excitation and 460/40 nm emission, 480/40 nm excitation and 530/40 nm emission, and 605/40 nm excitation and 640LP nm emission filters were used, respectively. Scale bar, 40 μm. c–e Data are presented as mean values ± s.e.m. for n = 3 transfection experiments. Source data are provided as a Source Data file.
To test if NIR-FbLAGs affect the ability of the biosensors to detect their analytes, we treated the co-expressing cells with 1 mM or 10 mM pyruvate, or 50 mM glucose, for Green Pegassos, PyronicSF, or iGlucoSnFR, respectively, and time-lapsed imaged them (Fig. 3c–e and Supplementary Figs. 10–12). As Green Pegassos and PyronicSF have different affinities for pyruvate (70 μM for Green Pegassos and 480 μM for PyroniSF)16,40, we used different pyruvate concentrations for these biosensors. No difference in the contrasts of biosensors’ fluorescence was observed without or with co-expressed NIR-FbLAG16 or NIR-FbLAG30 (Fig. 3c–e), suggesting that binding of NIR-FbLAGs may have a limited effect on the ability of GFP-based biosensors to detect their analytes.
Adding the second color to the NIR-Fb family
NIR-FbGFP, NIR-FbLAG16, and NIR-FbLAG30 have the inserted miRFP670nano3 (ex. 645 nm, em. 670 nm). For simultaneous labeling of several antigens, it would be beneficial to have distinct Nb colors.
Recently, cyanobacteriochrome-based NIR FP miRFP718nano (ex. 690 nm, em. 718 nm), spectrally resolvable from miRFP670nano3, was reported8. Since miRFP670nano3 and miRFP718nano have the same predecessor6, we have hypothesized that we could swap miRFP670nano3 for miRFP718nano in NIR-Fbs, including NIR-FbGFP (Fig. 4a and Supplementary Fig. 13), thus targeting two different antigens simultaneously. This red-shifted construct was named NIR-Fb(718). In cells co-expressing mEGFP, NIR-Fb(718) exhibited evenly distributed fluorescence whose pattern coincided with that of mEGFP expression (Fig. 4b), confirming that NIR-Fb(718) was dependent on the presence of the antigen.
a Design of NIR Nb for GFP (NbGFP) (PDB ID 3OGO) with indicated position for insertion of miRFP718nano (PDB ID 7LSD). CDRs of NbGFP are highlighted with violet and the position of miRFP718nano insertion is indicated in red. b Co-expression of NIR NbGFP containing inserted miRFP718nano (NIR-Fb(718)) with cognate mEGFP antigen in the cytoplasm (upper panels) or in the nucleus (bottom panels) of HeLa cells. c Upper panels, fluorescent images of HeLa cells co-expressing near-infrared Nb for p24 HIV protein with inserted miRFP670nano3 fluorescent protein (NIR-Fb59H10) and cognate antigen, NES-p24-mTagBFP2. Middle panels, fluorescent images of HeLa cells co-expressing NIR Nb for GFP with inserted miRFPnano718 (NIR-Fb(718)) and EGFP-H2B. Bottom panels, fluorescent images of HeLa cells co-expressing NIR-Fb59H10 and NIR-Fb(718) and two respective antigens, NES-p24-mTagBFP2 and EGFP-H2B. Patterns of co-expression are indicated with yellow and white arrows, respectively. For imaging of mTagBFP2 and mEGFP, 390/40 nm excitation and 460/40 nm emission and 480/40 nm excitation and 530/40 nm emission filters we used, respectively. For imaging of NIR-Fb59H10 and NIR-Fb(718), 605/40 excitation and 667/30 emission, and 682/12 excitation and 721/42 emission filters were used, respectively. Scale bar, 40 μm. For more details, see “Statistics and reproducibility” section of the Methods.
For targeting two antigens in a cell, we used a previously reported NIR-Fb59H107, which recognizes the p24 antigen of HIV (human immunodeficiency virus) and has internally fused miRFP670nano3. We fused p24 with mTagBFP2 and NES (nuclear exclusion signal) to direct it to cytosol and mEGFP with histone 2B (H2B) to localize it in the nucleus (Fig. 4c). When NIR-Fb(718) and NIR-Fb59H10 were co-expressed with these antigens, the NIR-Fb59H10 colocalized with p24-mTagBFP2 in the cytosol and NIR-Fb(718) colocalized with mEGFP in the nucleus (Fig. 4c), demonstrating the multicolor NIR imaging in the same cell.
Multicolor targeting GFP probes with NIR-Fbs
The developed NIR-Fbs of two distinct colors specific to GFP or GFP-based biosensors should be useful in designing various molecular tools. We have analyzed various GFP mutants and found that a pH-stable tandem dimer GFP (pH-tdGFP)42 could serve as the second antigen for the NIR-Fb(718) - NIR-FbLAG pair. Analyzing the dimerization interface of pH-tdGFP using EGFP crystal structure43, we hypothesized that only NIR-FbLAG30 would bind pH-tdGFP, whereas the epitope for NIR-Fb(718) would be buried in the dimerization interface and, thus, NIR-Fb(718) should be destabilized in the pH-tdGFP presence.
To check the possibility of multicolor imaging of two NIR-Fbs to GFP-like antigens, we transfected HeLa cells with NIR-FbLAG30 and NIR-Fb(718) and two antigens, pH-tdGFP with an NES signal and blue EBFP2 with a nuclear localization signal (NLS) in various combinations (Fig. 5). When either NIR-Fb(718) or NIR-FbLAG30 was co-expressed with NLS-EBFP2, both NIR-Fbs predictably exhibited NIR fluorescence in the nucleus (Fig. 5a, b). In the presence of NES-pH-tdGFP antigen, there was no NIR fluorescence when NIR-Fb(718) was cotransfected (Fig. 5c), whereas NIR-FbLAG30 exhibited bright NIR fluorescence in the cytosol (Fig. 5d), confirming our initial hypothesis. When two NIR-Fbs were cotransfected with both NES-pH-tdGFP and NLS-EBFP2 antigens, we observed cytosolic and nuclear localization of NIR-FbLAG30 and only nuclear localization of NIR-Fb(718) (Fig. 5e). These results demonstrated the ability of simultaneous tracking several GFP-based probes in the cell by combining different NIR Nbs.
Fluorescence images of HeLa cells co-expressing EBFP2-nuc with either (a) NIR-Fb for GFP with inserted miRFP718nano fluorescent protein (NIR-Fb(718)) or (b) with NIR-Fb for GFP with inserted miRFP670nano3 (NIR-FbLAG30). Fluorescence images of HeLa cells co-expressing pH-tdGFP with either (c) NIR-Fb(718) or (d) with NIR-FbLAG30. e Fluorescence images of cells co-expressing EBFP2-nuc, pH-tdGFP, NIR-FbLAG30, and NIR-Fb(718). Yellow arrows indicate patterns of co-expression for EBFP2-nuc with NIR-FbLAG30 and NIR-Fb(718) in the nucleus, white arrows indicate patterns of co-expression for pH-tdGFP with NIR-FbLAG30 in the cytosol. For imaging of EBFP2 and pH-tdGFP, 390/40 nm excitation and 460/40 nm emission and 480/40 nm excitation and 530/40 nm emission filters we used, respectively. For imaging of NIR-FbLAG30 and NIR-Fb(718), 605/40 excitation and 667/30 emission and 682/12 excitation and 721/42 emission filters were used, respectively. Scale bar, 40 μm. For more details, see “Statistics and reproducibility” section of the Methods.
NIR-Fb-based systems for transcription control
A unique feature of NIR-Fbs is their stabilization upon antigen binding and degradation in the absence of a cognate antigen, allowing the development of systems to control gene expression in an antigen-dependent manner. Since LAG16 (or LAG30) and GBP1 Nbs bind different GFP epitopes, it was shown that they can co-occupy GFP, forming complexes35,36. We decided to utilize this property and use superfolder GFP (sfGFP) as a dimerizer for two NIR-Fbs. To this, we first constructed a sfGFP version split at position 15744, in which each split fragment was fused to mTagBFP2 (Supplementary Fig. 14a). HeLa cells transfected with one sfGFP split fragment exhibited no green fluorescence, whereas two complementing sfGFP fragments fused to mTagBFP2 restored its green fluorescence (Supplementary Figs. 14a and 15a, b), confirming a weak dimerization ability of mTagBFP2. Analyzing the mEGFP crystal structure and published data12,35,36, we hypothesized that NIR-FbLAG30 would bind the N-split fragment (sfGFP(1-157)) (Supplementary Fig. 14b, c) while NIR-Fb(718) would bind only restored sfGFP, allowing two NIR-Fbs to bind the same sfGFP molecule simultaneously. When we co-expressed N- or C-end split fragments of sfGFP with NIR-FbLAG30 or NIR-Fb(718), respectively, NIR-fluorescence was observed only in the cells transfected with N-end split sfGFP and NIR-FbLAG30 (Supplementary Figs. 14d and 15c, d). When both split fragments were co-expressed, NIR fluorescence was observed for both NIR-FbLAG30 and NIR-Fb(718) (Supplementary Figs. 14e and 15c, d). This confirms the possibility of using GFP as a dimerizer for two NIR-Fbs binding distinct epitopes. Notably, since NIR-Fb(718) binds only restored sfGFP, it can be used as a NIR reporter of the split GFP complementation in the protein-protein interaction assays.
To study whether NIR-Fb could be used for gene regulation, we have chosen the GAL4-UAS system45. In the first approach, we fused the GAL4 DNA-binding domain to each of the split sfGFP fragments and NIR-Fb(718) to the VP16 activation domain. As a reporter, we used a pG12-mTagBFP2 with 12xUAS (upstream activating sequence; termed G12 here) repeats upstream of the reporter gene (Fig. 6a). In this design, binding of UAS sites by two GAL4 domains restores sfGFP molecule, then NIR-Fb(718) binds its sfGFP epitope, resulting in the formation of GAL4-VP16 complex and, consequently, activation of a mTagBFP2 reporter transcription (Fig. 6a). We observed blue reporter fluorescence only when cells were co-transfected with sfGFP(1-157)-GAL4, sfGFP(158-238)-GAL4, VP16-NIR-Fb(718), and pG12-mTagBFP2. The dynamic range of the system is ~78.8 ± 7.1 (Fig. 6c and Supplementary Fig. 16a). In the absence of one of the split sfGFP fragments, NIR-Fb(718) was destabilized, and no mTagBFP2 reporter fluorescence was observed (Fig. 6c).
a Scheme of NIR-Fb(718)-based system for activation of mTagBFP2 reporter transcription. Two fragments of split-sfGFP are fused to GAL4 DNA-binding domains, while NIR-Fb for GFP with inserted miRFP718nano (NIR-Fb(718)) is fused to VP16 transcriptional activation domain. In the presence of the fusions of GAL4 to the split sfGFP fragments and a VP16-NIR-Fb(718) fusion, sfGFP reconstitutes and can bind NIR-Fb(718). In turn, transcriptional activation domain VP16 forms a complex with GAL4 DNA-binding domain which binds to the UAS regulatory sequence and activates transcription of the mTagBFP2 reporter gene. b Scheme of NIR-FbLAG30- and NIR-Fb(718)-based system for activation of mTagBFP2 reporter transcription. Here GAL4 DNA-binding domain is fused to the NIR-FbLAG30. In the presence of mEGFP, both GAL4-NIR-FbLAG30 and VP16-NIR-Fb(718) bind different epitopes of mEGFP molecule and activate mTagBFP2 reporter transcription. c Fluorescence intensity of mTagBFP2 in HeLa cells transfected with a plasmid from the scheme (a). As a control, GAL4-VP16 plasmid with pG12-mTagBFP2 reporter plasmid or pG12-mTagBFP2 reporter plasmid only were used. d Fluorescence intensity of mTagBFP2 in HeLa cells transfected with plasmids from the scheme (b). Controls are the same as in c. Fluorescence intensity was analyzed by flow cytometry using a 640 nm excitation laser and a 660/20 nm emission filter for NIR-Fbs, a 488 nm excitation laser and a 525/50 nm emission filter for mEGFP, a 405 nm excitation laser and a 450/50 nm emission filter for mTagBFP2. Data are presented as mean values ± s.d. for n = 3 transfection experiments. Source data are provided as a Source Data file.
In the second approach, we fused GAL4 to the NIR-FbLAG30 and VP16 to NIR-Fb(718). In this system, mEGFP molecule acts as a dimerizer for two NIR-Fbs, stabilizing their fusions, resulting in the formation of GAL4-VP16 complex and, consequent, activation of mTagBFP2 reporter transcription with a dynamic range of ~100.0 ± 0.5 (Fig. 6b, d and Supplementary Fig. 16b). We also tested if this NIR-Fb-based system for transcription control could be triggered by Discosoma-derived FPs, such as mCherry46 and mOrange46. As expected, none of them induced gene transcription (Fig. 6d and Supplementary Fig. 16b). Both NIR-Fbs-based gene transcription systems can be further used for manipulation of GFP-expressing cell populations in numerous GFP transgenic organisms generated to date.
Targeting GFP-expressing cells with NIR-FbGFP in vivo
To demonstrate multiplex imaging of GFP-expressing cells targeted in vivo with NIR-FbGFP, we constructed a CAG promoter carrying AAV9 expression vector. The vector was injected into the somatosensory cortex of Thy1-GFP-M and Aldh1l1-GFP mice with fluorescently labeled neurons and astrocytes, respectively. 3–5 weeks after the injections, animals were prepared for in vivo two-photon imaging to determine how accurately NIR-FbGFP targets GFP-expressing neurons and astrocytes in the brain of transgenic mice. We found that NIR-FbGFP brightly fluoresced both in the transduced neurons and astrocytes (Fig. 7a, b and Supplementary Movies 1–4). NIR-FbGFP/GFP-positive neurons were visualized through the entire depth of the cortex (approximately 800 μm; Fig. 7c); and NIR-FbGFP/GFP-positive astrocytes were routinely visualized at the depth of 450 μm (Fig. 7d and Supplementary Movies 1–4).
a Left, example images from a xy fluorescence image stack showing GFP-expressing neurons and their processes (green) in the somatosensory cortex of an anesthetized 9.5-weeks-old Thy1-GFP-M mouse at the indicated depths (z) from the pial surface. Center, the same neurons show miRFP670nano3 expression three weeks after stereotactic AAV9-CAG-NIR-FbGFP vector delivery. Right, overlay of the simultaneously acquired two-photon images. Scale bar, 100 μm. b Left, example images from a xy fluorescence image stack showing GFP-expressing astrocytes and their processes (green) in the somatosensory cortex of an anesthetized 16-weeks-old Aldh1l1-GFP mouse at the indicated depths (z) from the pial surface. Center, the same astrocytes show miRFP670nano3 expression 4.5 weeks after stereotactic AAV9-CAG-NIR-FbGFP vector delivery. Right, overlay of the simultaneously acquired two-photon images. Scale bar, 100 μm. c Example maximum-intensity side projection (xz) from the image stack in panel a showing the GFP (left) and miRFP670nano3 (center) expression pattern across depth. An overlay image is shown on the right. Scale bar, 100 μm. d Example maximum-intensity side projection (xz) from the image stack in panel b showing the GFP (left) and miRFP670nano3 (center) expression pattern across depth. An overlay image is shown on the right. Scale bar, 100 μm. e Population analysis showing the percent overlap between miRFP670nano3 and GFP-positive cell bodies (NIR + GFP+: 96.8% ± 1.1%; NIR + GFP−: 3.2% ± 1.1%; n = 6 tissue sections from two mice). f Population analysis showing the percent overlap between miRFP670nano3 and GFP-positive cell bodies (NIR + GFP+: 94.4% ± 2.6%; NIR + GFP−: 5.6% ± 2.6%; n = 6 tissue sections from two mice). The entire xy fluorescence image stacks and xz side projections are shown in Supplementary Movies 1–4. e, f Data are presented as mean values ± s.d. Source data are provided as a Source Data file.
After two-photon imaging, animals were perfused and brain tissue was sectioned to 40 μm slices. Slices analysis demonstrated very good (>94%) correspondence between NIR-FbGFP- and GFP-expressing cell bodies both for neurons and astrocytes near the injection sites (Fig. 7e, f), indicating that NIR-FbGFP targeted various cell types expressing GFP in the mouse brain with precise accuracy.
Targeting GCaMP6-expressing neurons with NIR-FbLAGs in vivo
To evaluate the utility of the developed NIR-FbLAGs for targeting neurons expressing GFP-based genetically encoded biosensors, we constructed CAG promoter-carrying AAV9 vectors with either NIR-FbLAG16 or NIR-FbLAG30. The AAVs were then injected into the somatosensory cortex of Thy1-GCaMP6f mice with neurons expressing GCaMP6f. 3–4.5 weeks after the cortical injection, we performed two-photon imaging in awake mice on a spherical treadmill. Fluorescence of NIR-FbLAGs was detectable up to ~600 μm cortical depth and was co-localized with GCaMP6f expression (Supplementary Movies 5–8). NIR+/GCaMP6f+ neurons exhibited spontaneous and running-evoked calcium spiking with NIR fluorescence showing few signs of photobleaching during the extended (>10 min continuous) recordings (Fig. 8a, b, e, f and Supplementary Movies 9, 10). Vigorous mouse running led to motion-induced NIR+ and GCaMP6f+ fluorescence fluctuations (Fig. 8b, c, f, g). The stability of NIR fluorescence allowed us to use it as a ratiometric reference for the GCaMP6f signal. Calculating the ratio (R) of GCaMP6f and NIR-FbLAG fluorescence resulted in activity traces with noticeably reduced motion artifacts (Fig. 8a, d, e h). As shown in Fig. 8d h, normalizing the GCaMP6f trace to the NIR-FbLAG16 (or NIR-FbLAG30) trace from the same cell corrects the fluorescence baseline fluctuations, which often occur during animal locomotor activity (Fig. 8d, h). This, in turn, substantially improves the neuronal calcium spike estimation, for example, by helping to identify false positives and false negatives.
a Example two-photon fluorescence image from a dual-color time-lapse recording showing GCaMP6f- and miRFP670nano3-expressing neurons in the somatosensory cortex of a behaving Thy1-GCaMP6f mouse. Imaging was performed ~4.5 weeks after stereotactic AAV9-CAG-NIR-FbLAG16 injection. Recording depth (z) from the pial surface and seven somatic regions of interest (ROIs) are indicated. b Color-separated image for the NIR-FbLAG16 channel. c Color-separated image for the GCaMP6f channel. In a–c, right, fluorescence transients in the indicated ROIs are shown as ΔR/R (blue) and ΔF/F (red and gray) for the combined and individual channels, respectively. The simultaneously recorded mouse’s locomotor activity on a spherical treadmill is shown above the fluorescence traces. Scale bars, 50 μm (left), 100 mm/s, and 200% (right). d Zoom-ins of the two periods indicated in a–c show how the ΔR/R calculation corrects for fluorescence baseline variations caused by mouse locomotor activity and improves calcium spike estimation by identifying potential false positives and false negatives (arrowheads). e Example two-photon fluorescence image from a dual-color time-lapse recording for a Thy1-GCaMP6f mouse injected with AAV9-CAG-NIR-FbLAG30. f Color-separated image for the NIR-FbLAG30 channel. g Color-separated image for the GCaMP6f channel. In e–g, right, fluorescent transients in the indicated ROIs are shown as in a–c. The simultaneously recorded mouse’s locomotor activity on the spherical treadmill is shown atop. h Zoom-ins of the two periods indicated in e–g highlight the advantages of ratiometric imaging as in d. The corresponding time-lapse recordings are shown in Supplementary Movies 9, 10. xy fluorescence image stacks and xz side projections, including the recording sites, are shown in Supplementary Movies 5–8.
To more quantitatively assess NIR-FbLAGs targeting of GCaMP6f-expressing neurons, we prepared coronal slices from AAV9-CAG-NIR-FbLAG16 and AAV9-CAG-NIR-FbLAG30 injected Thy1-GCaMP6f mice. Near the injection sites, NIR fluorescence was seen in cell bodies and processes of GCaMP6f+ neurons (Fig. 9a, b) with ~99% overlap (Fig. 9c, d). Therefore, our CAG promoter-carrying AAV9 vectors with NIR-FbLAG16 and NIR-FbLAG30 enabled the targeting of GCaMP6f-expressing neurons with high specificity and penetrance (Supplementary Movies 5–10), permitting ratiometric functional imaging for improved spike estimation in behaving animals.
a Example confocal fluorescence images showing GCaMP6f- and miRFP670nano3-expressing neurons (center) in the cortex of an AAV9-CAG-NIR-FbLAG16-injected Thy1-GCaMP6f mouse. The uninfected control hemisphere of the same mouse is shown on the left. Right, zoom-ins of the indicated regions. Scale bars, 500 μm (center, left) and 25 μm (right). b Same as in panel a but for an AAV9-CAG-NIR-FbLAG30-injected Thy1-GCaMP6f mouse. c Population analysis showing the percent overlap between NIR-FbLAG16 and GCaMP6f-positive cell bodies (NIR+/GCaMP6f+: 99.4% ± 0.6%; NIR+/GCaMP6f−: 0.6% ± 0.6%; n = 9 tissue sections from three mice). d Population analysis showing the percent overlap between NIR-FbLAG30 and GCaMP6f-positive cell bodies (NIR+/GCaMP6f+: 99.5% ± 0.4%; NIR+/GCaMP6f-: 0.5% ± 0.4%; n = 9 tissue sections from three mice). c, d Data are presented as mean values ± s.d. Source data are provided as a Source Data file.
Discussion
By using the bright small cyanobacteriochrome-derived miRFP670nano3 as an internal tag and structural and experimental screening of anti-GFP Nbs, we developed a series of NIR-FbLAG constructs that recognize GFP-based biosensors (Figs. 1–3). NIR-FbLAGs preserve spectral properties of parental miRFP670nano3 and can be produced recombinantly in bacteria (Supplementary Fig. 6). Stabilization of NIR-FbLAGs by antigen binding in mammalian cells enables visualization of cell populations heterogeneously expressing GFP or GFP-based probes in a background-free manner.
We showed that NIR-FbLAGs bind a broad range of GFP-based biosensors, irrespective of the analyte they recognize, while having limited effect on the functionality of biosensors (Figs. 2b–d and 3c–e). At the same time, the bound NIR-FbLAGs provide stable longitudinal fluorescence. Although NIR-FbLAGs exhibit about twice higher affinity to the calcium-free conformation of GCaMP6s according to the flow cytometry data, we observed no change in NIR fluorescence after the increase in calcium level, suggesting the NIR-FbLAGs dissociation from calcium-bound GCaMP6s is quite slow at least for minutes after an increase in calcium concentration (Fig. 2c, d). This discrepancy may be due to the prolonged, 40–60 min, incubation of cellular cultures in calcium-saturated conditions before detection by flow cytometer, which may fasten dissociation of the NIR nanobody from GCaMP6s. At the same time, the fast calcium association and dissociation kinetics, as well as affinity to calcium ions of GCaMP6s, were not affected by NIR-FbLAG30 binding (Supplementary Fig. 9). This makes NIR-FbLAGs beneficial for ratiometric deep-brain mapping of fast neuronal activity in vivo by simply comparing green and NIR fluorescence intensities.
We expanded the spectral range of NIR-Fbs by developing the NIR-Fb(718) having ~50 nm red-shifted fluorescence as compared to the original NIR-Fbs7 (Fig. 4a, b) and demonstrated efficient multicolor targeting of two different proteins in live cells (Fig. 4c). Also, using the distinct-colors NIR-Fbs for GFP, we visualized two GFP-derived proteins in the cells, altogether demonstrating the wide possibilities of the NIR-Fbs with shifted emission for multiplexed imaging (Fig. 5).
Recently, adaptable turn-on maturation tools (ATOMs) based on circularly permuted monobodies (Mbs) or Nbs, inserted into GFP-like FPs, were developed and tested in cells, but not in vivo47. Similarly to NIR-Fbs, the fluorescence of ATOMs increases upon binding a cognate antigen. However, ATOMs do not degrade in the absence of antigen and, therefore, though non-fluorescent, accumulate in the cells. Also, depending on Mb stability, ATOM can be always in the “on” or “off” mode, thus, not reflecting a real increase in the antigen concentration. NIR-Fbs, on the other side, simply degrade in the absence of a cognate antigen.
The antigen-stabilization of NIR-Fbs can be used to control cellular processes in an antigen-dependent manner. Coupling our engineered NIR-Fbs with GFP-labeling, we developed two systems, in which gene transcription starts upon GFP expression (Fig. 6a, b). Unlike the previously reported approach48, the NIR-Fbs-fused fragments in our systems are rapidly degraded if unbound to GFP, allowing very tight control over gene transcription (Fig. 6c, d). These systems can be further applied to numerous available GFP-expressing transgenic animals for selective manipulations of GFP-labeled cells. Also, using NIR-FbLAGs as a destabilizing fusion partner it will be possible to target protein degradation in specific cell types expressing either GFP or GFP-based biosensors, unlike GFP only as with original NIR-FbGFP7.
NIR-FbLAGs offer advantages in in vivo imaging of transgenic animals expressing GFP or GFP-based biosensors (Figs. 7–9 and Supplementary Movies 1–10), addressing several challenges. First, transgenic mice express GFP-based probes heterogeneously in different cell populations due to random transgene integration into the genome27,28,49,50. Additionally, discrepancies in GFP-based biosensor expression levels, illumination conditions, and variations in analyte concentrations complicate data interpretation. NIR-FbLAGs could serve as orthogonal reference probes, enabling the normalization of functional fluorescence changes, including in situations where correction of image motion is infeasible or suboptimal (e.g., variable within-frame distortions during low frame-rate image acquisitions, or low signal-to-noise recordings from deep tissue regions; Fig. 8e–h and Supplementary Movie 10). Second, small bright NIR-Fbs could be ideal optical guide stars for deep-tissue light focusing in vivo, especially in scattering tissues such as the brain. Third, targeting NIR-FbLAGs to various intracellular compartments could provide precise localization for GFP-derived probes.
These and other advances of the engineered NIR-FbLAGs, NIR-Fb(718), and NIR-Fbs-based gene transcription systems highlight their great potential to enhance the accuracy and reliability of in vivo studies, marking a valuable step forward in functional imaging and control of cellular processes.
Methods
Ethical statement
All live animal procedures were performed following the National Institutes of Health (NIH) guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC) of the Salk Institute under the protocol number 13-00022.
Recombinant protein production in bacteria
The gene encoding the NIR-FbLAG30 protein was amplified by polymerase chain reaction (PCR) and inserted into the pET22 vector. All oligonucleotide PCR primers were purchased from Thermo Fisher Scientific. NIR Nb was expressed in Rosetta(DE3)pLysS bacterial cells, cotransformed with pWA23h plasmid, encoding heme oxygenase. Bacterial cells were grown to an optical density of 0.5–0.7 at 600 nm in LB/ampicillin/kanamycin/chloramphenicol medium supplemented with 0.02% rhamnose and, then, to induce NIR-FbLAG30 expression, 0.5 mM IPTG was added. Bacteria were cultured for 2–3 h at 37 °C and then for 36 h at 18 °C. Protein purification was performed with Ni-NTA agarose (Qiagen). Proteins were washed with wash buffer containing 20 mM PBS, 0.1 M NaCl, 1 mM β-mercaptoethanol and 20 mM imidazole and eluted with elution buffer containing 20 mM PBS, 0.1 M NaCl, 1 mM β-mercaptoethanol and 250 mM imidazole. For purification of NIR-FbLAG30 complex with mEGFP, the same protocol was used, except for Rosetta(DE3)pLysS cells were cotransformed with pET22-NIR-FbLAG30, pWA23h, and pWA23-mEGFP.
GCaMP6s calcium biosensor in the pBAD/HisB vector was transformed into an LMG194 bacterial host (Invitrogen/Thermo Fisher Scientific). The bacterial cells were grown to an optical density of 0.5–0.7 at 600 nm in LB/ampicillin medium, and then 0.02% arabinose was added to induce GCaMP6s expression. After 48 h culture at 30 °C, bacteria were centrifuged at 4650 × g for 10 min, and cell pellets were resuspended in PBS at pH 7.4 with 300 mM NaCl and lysed by French press. The recombinant protein was purified using Ni-NTA resin (Qiagen), followed by dialysis for 16–18 h against buffer solutions (50 mM MOPS, 100 mM KCl, pH 7.2 with either 1 mM EDTA or 1 μM CaCl2).
Fluorescence imaging of purified NIR-FbLAG30 fractions was performed on the IVIS Spectrum instrument (PerkinElmer). For mEGFP imaging, 465/20 nm excitation and 520/30 nm emission filters were used. For NIR-FbLAG30 imaging, 640/20 nm excitation and 720/30 nm emission filters were used.
Fluorescence and absorbance spectra of NIR-FbLAG30 were recorded with a FluoroMax-3 fluorimeter (Jobin Yvon) and a Hitachi U-3010 spectrophotometer, respectively. All measurements were performed in PBS (pH 7.4).
To determine the Kds of GCaMP6s and GCaMP6s with NIR-FbLAG30 for calcium ions, a series of buffers (100 mM KCl, 50 mM MOPS, pH 7.2), supplemented with either 10 mM EGTA or 10 mM CaEGTA, containing different amount of Ca2+ were prepared following the instructions provided in the Calcium Calibration Buffer Kit (Invitrogen). Purified proteins were then diluted to 5 μg/mL in the series of buffers with different concentrations of free Ca2+, and their fluorescence (excitation 480 nm; emission 490–650 nm) was measured with a FluoroMax-3 fluorimeter (Jobin Yvon). Binding data was fit with a Hill equation: y = Fmax × [Ca2+]^n/(Kd^n + [Ca2+]^n). The Kds were calculated as the concentration of Ca2+ when the fluorescence change for GCaMP6s or GCaMP6s in the presence of NIR-FbLAG30 was half its maximum value.
Stopped-flow spectroscopy of Ca2+-binding kinetics
A SX20 stopped-flow spectrofluorometer (Applied Photophysics) equipped with a 150 W xenon lamp and a photomultiplier was used for kinetic fluorescence measurements. The excitation wavelength was set to 490 nm, and a 495 nm cut-off filter was used. All measurements were done at 22 °C. Five replicates were averaged for analysis. Kinetics records were fitted to exponential using Origin 2017 SR1 software. To measure association kinetics, the GCaMP6s indicator (0.5 μM) or GCaMP6s with NIR-FbLAG30 (in molar concentration ratio of 1:2 for GCaMP6s: NIR-FbLAG30, incubated at 4 °C overnight before kinetic measurements) in 50 mM MOPS, 100 mM KCl, pH 7.2 and 1 mM EGTA were rapidly mixed (1:1) with a series of Ca2+ buffers. The change in the fluorescence signal during rapid mixing provided the relaxation rate constants (kobs) for the Ca2+ association reaction at various Ca2+ concentrations (from 0 nM to 350 nM).
To measure dissociation kinetics, protein solutions (0.5 μM; molar concentration ratio 1:2 for GCaMP6s: NIR-FbLAG30) in 50 mM MOPS, 100 mM KCl, pH 7.2 and 1 μM CaCl2 were rapidly mixed (1:1) with 50 mM MOPS, 100 mM KCl, pH 7.2 and 10 mM EGTA. Exponential fitting of the fluorescence signal changes over time provided the dissociation rate constants (koff).
Construction of mammalian plasmids
Anti-GFP Nbs internally fused with miRFP670nano3 or miRFP718nano were generated by overlap PCR, and the resulting constructs were inserted into the pcDNA plasmid at the KpnI/EcoRI sites. All oligonucleotide primers for PCR amplification were purchased from Thermo Fisher Scientific (Supplementary Table 1). LAG30 Nb to GFP was amplified from a pET21-pelB-LAG-30 plasmid (Addgene no.172754). LAG16 Nb to GFP was amplified from a pET21-pelB-LAG-16 plasmid (Addgene no.172746). GBP2, GBP6, and GBP7 Nbs to GFP were amplified from pCAG-Gal4DBD-GBP2 (Addgene no.49439), pCAG-p65AD-GBP6 (Addgene no.49440), and pCAG-GBP7-p65AD (Addgene no.49441) plasmids, respectively. Sb44 and Sb92 Nbs to TGFP were amplified from pSb-init-Sb44 (Addgene no.159421) and pSb-init-Sb92 (Addgene no.159424) plasmids, respectively.
GCaMP6s biosensor and its truncated version were amplified from a pAAV.CaMKII.GCaMP6s.WPRE.SV40 (Addgene no.107790) plasmid and inserted into the pcDNA plasmid at the HindIII/EcoRI sites. Glucose biosensor iGlucoSnFR was amplified from a pAAV.hSynap.(cyto)iGlucoSnFR-mRuby2 (Addgene no.164510) and inserted into the pcDNA plasmid at the KpnI/EcoRI sites.
To generate the NES-p24-mTagBFP2 plasmid, the mTagBFP2 gene was swapped with mEGFP in the NES-p24-mEGFP plasmid by the NEBuilder HiFi DNA Assembly. To generate NES-p24-mEGFP plasmid, a fusion of HIV p24 with mEGFP was amplified and inserted into the pcDNA plasmid with an NES signal (Addgene no.164052) at the BamHI/XhoI sites. To construct an NES-pH-tdGFP plasmid, pH-tdGFP was amplified from a pH-tdGFP plasmid (Addgene no.74322) and inserted into the pcDNA plasmid with an NES signal (Addgene no.164052) at the BamHI/XhoI sites.
To generate mTagBFP2-sfGFP fusions, mTagBFP2 was fused to the N-end part of the sfGFP (nucleotides 1–157) via 8 a.a. long linker (Gly2SerGlySerGly2Ser) by overlap PCR and C-end part of the sfGFP (nucleotides 158-238) was fused to mTagBFP2 via 8 a.a. long linker (Gly2SerGlySerGly2Ser) by overlap PCR. Amplified fusions were then inserted into the pcDNA plasmids by the KpnI/NotI sites.
To generate GAL4 fusions with sfGFP and NIR-Fb fusions with GAL4 and VP16, a 15-residue linker (Gly3SerGly4SerGly3SerGly2) was inserted into a pcDNA plasmid by the BamHI/NotI sites. For the construction of sfGFP(1-157) or sfGFP(158-238) fused to GAL4, GAL4 was amplified from a pQP-T2A plasmid (Addgene no.102583) and inserted into a pcDNA3.1 plasmid, containing the 15 aa-long linker at the KpnI/BamHI sites, and sfGFP(1–157) or sfGFP(158-238) was inserted at the NotI/XbaI sites. To generate NIR-FbLAG30 fused to GAL4, GAL4 was amplified from a pQP-T2A plasmid (Addgene no.102583) and inserted into a pcDNA3.1 plasmid containing the 15 a.a. long linker at the KpnI/BamHI sites, and NIR-FbLAG30 was inserted at the NotI/XbaI sites. To engineer NIR-Fb(718) fused to VP16, VP16 was amplified from a pQP-T2A plasmid (Addgene no.102583) and inserted into a pcDNA3.1 plasmid containing the 15 a.a. long linker at the KpnI/BamHI sites, and NIR-Fb(718) was inserted at the NotI/XbaI sites. A nuclear localization signal (NLS) was added to VP16-NIR-Fb(718) fusion by PCR amplification. For the construction of a pG12-mTagBFP2 reporter plasmid, mTagBFP2 was PCR-amplified and inserted into pG12-SEAP plasmid (Addgene no.170271) by BamHI/NotI sites.
Mammalian cell culture and transfection
HeLa (CCL-2) cells were obtained from the ATCC. Cells were cultured in a DMEM medium (Corning) supplemented with 10% FBS (Gembio), 0.5% penicillin-streptomycin (HyClone) and 2 mM glutamine at 37 °C. For live-cell fluorescence microscopy, cells were plated in 35 mm glass-bottom Petri dishes (Greiner Bio-One). Transient transfections were performed using polyethylenimine51 or Effectene reagent (Qiagen).
Green Pegassos (Addgene no.163114) and PyronicSF (Addgene no. 124812) plasmids were used for transfection in GFP-based biosensors targeting experiments.
For multi-color imaging of two different NIR-Fbs, HeLa cells were cotransfected with plasmids encoding NIR-Fb59H10, NIR-Fb(718), and cognate antigens, NES-p24-mTagBFP2 and H2B-EGFP, in a 1:5:1:1 ratio. For multi-color imaging of two NIR-Fbs for GFP, HeLa cells were cotransfected with plasmids encoding NIR-FbLAG30, NIR-Fb(718), NES-pH-tdGFP and EBFP2-nuc, in a 1:5:10:1 ratio. For imaging and flow cytometry of NIR-Fb-based systems for transcription activation, HeLa cells were cotransfected with plasmids encoding GAL4-sfGFP(1-157), GAL4-sfGFP(158-238), VP16-NIR-Fb(718), and reporter plasmid pUAS-mTagBFP2 (12xUAS) in the 1:1:5:10 ratio or with plasmids encoding VP16-NIR-Fb(718), GAL4-NIR-FbLAG30, pUAS-mTagBFP2 (12xUAS) and mEGFP in the 1:1:1:10 ratio. As controls, cells transfected with GAL-VP16 (Addgene no.71728) and a reporter plasmid pUAS-mTagBFP2 or with pUAS-mTagBFP2 reporter plasmid only were used.
To study protein stability in mammalian cells, HeLa cells were incubated for 20 h with 10 µM bortezomib (Millipore).
No biliverdin was exogenously supplied in all mammalian cell experiments.
Epifluorescence live-cell microscopy
Live mammalian cells were imaged using an Olympus IX81 inverted epifluorescence microscope equipped with a Lambda LS Xenon light source (Sutter). An ORCA-Flash4.0 V3 camera (Hamamatsu) was used for image acquisition. Cells were imaged using a 60 × 1.35 NA oil immersion objective lens (UPlanSApo, Olympus) and 390/40 nm, 480/40 nm, 570/30 nm, 605/40 nm, or 682/12 nm excitation filters and 460/40 nm, 530/40 nm, 615/40 nm, 667/30 nm, or 721/42 nm emission filters, respectively. Light power densities measured at the rear aperture of the objective lens were 0.76 mW cm−2 for 390/40 nm, 3.73 mW cm−2 for 480/40 nm, 2.1 mW cm−2 for 570/30 nm, 3.41 mW cm−2 for 605/40 nm, and 0.67 mW cm−2 for 682/12 nm excitation filters, which gives an estimate (FN is 26.5 for the objective lens) for the light power densities at the front focal objective plane of 0.47 W cm−2, 2.32 W cm−2, 1.3 W cm−2, 2.12 W cm−2, and 0.42 W cm−2, respectively. During imaging, HeLa cells were incubated in a DMEM medium and kept at 37 °C. The microscope was operated with a SlideBook v.6.0.8 software (Intelligent Imaging Innovations). For imaging performance of NIR-Fbs coexpressed with GCaMP6s biosensor, HeLa cells were washed twice with pre-warmed 20 mM HEPES-buffered Hanks’ Balanced Salt Solution (HHBSS) (Corning) before imaging and incubated in HHBSS supplemented with 4 mM CaCl2. After the first frame was imaged, 5 μM ionomycin (Cayman Chemical) was added. Cells were maintained at 35 °C using the environmental chamber.
For imaging pyruvate biosensors, Green Pegassos and PyronicSF, HeLa cells were incubated in HHBSS 30 min before imaging. After the first frame was imaged, a 1 mM or 10 mM solution of sodium pyruvate (Gibco) was added, respectively. For imaging glucose biosensor, iGlucoSnFr, HeLa cells were incubated in a DMEM medium without glucose and pyruvate (Gibco). After the first frame was imaged, a 50 mM glucose solution was added.
For imaging of NIR-FbLAG16, NIR-FbLAG30, and NIR-Fb59H10, 605/40 nm excitation and 667/30 nm or 640LP emission filters (Chroma) were used. For imaging NIR-Fb(718), 682/12 nm excitation and 721/42 nm emission filters (Chroma) were used. For imaging mEGFP, GCaMP6s, Green Pegassos, PyronicSF, iGlucoSnFr and pH-tdGFP, 480/40 nm excitation and 530/40 nm emission filters (Chroma) were used. For imaging mTagBFP2 and EBFP2, 390/40 nm excitation and 460/40 nm emission filters (Chroma) were used. The data were analyzed using SlideBook v. 6.0.8 (Intelligent Imaging Innovations), GraphPad Prism v.5.04, and Origin 2017 SR1 software.
Flow cytometry
Flow cytometry analysis was performed using an LSR II (BD Biosciences) flow cytometer. Before analysis, live cells were washed with cold PBS, trypsinized for 5 min at 37 °C and diluted in cold PBS containing 4% FBS and 2 mM EDTA to a density of 500,000 cells per ml.
For analysis of cells co-expressing GCaMP6s in their Ca2+-saturated or Ca2+-depleted states and NIR-FbLAGs, samples were washed with cold HHBSS and trypsinized in 0.1% Soybean Trypsin Inhibitor (Thermo Fisher) for 5 min at 37 °C. Cells then were centrifuged at 4 °C, and pellets were washed with cold HHBSS, diluted in cold HHBSS containing 4 mM CaCl2 or 2 mM EDTA to a density of 500,000 cells per ml and incubated for 40-60 min. At least 30,000 cells per sample were recorded. Data were collected using BD FACSDiva v.8.0.1 (BD Biosciences) software. The fluorescence intensity of cells expressing Nbs to GFP with internally fused miRFP670nano3 was analyzed using the 640 nm excitation laser and 660/20 nm emission filter. The fluorescence intensity of cells expressing Nb to GFP with internally fused miRFP718nano was analyzed using the 640 nm excitation laser and 780/60 nm emission filter. The fluorescence intensity of mEGFP was analyzed using a 488 nm laser for excitation, and its fluorescence was detected with a 525/50 nm emission filter. The fluorescence intensity of mTagBFP2 was analyzed using a 405 nm excitation laser and a 450/50 nm emission filter. The fluorescence intensity of mCherry was analyzed using a 561 nm excitation laser and a 610/20 nm emission filter. The data were analyzed using FlowJo v.7.6.2 software.
Animal subjects
Live animal procedures were performed following the NIH guidelines and approved by the IACUC of the Salk Institute. We used two male Thy1-GFP-M (Jackson Laboratories; RRID: IMSR_JAX:007788), two male Aldh1l1-EGFP (GENSAT; RRID: MMRRC_011015-UCD), and eight female Thy1-GCaMP6f mice (Jackson Laboratories; RRID: IMSR_JAX:025393) with a typical age of ~10–17 weeks at the time of imaging (~3–4.5 weeks after stereotactic injection; two injections per hemisphere). All mice were on a C57BL/6J background. Mice were group-housed, provided with bedding and nesting material, and maintained on a 12-h light-dark cycle in a temperature (22 ± 1 °C) and humidity controlled (45–65%) environment.
Virus production and stereotactic injection
The NIR-FbGFP, NIR-FbLAG16, and NIR-FbLAG30 genes were PCR amplified and subcloned into an AAV transfer vector downstream of the CAG promoter. These vectors were cotransfected into HEK293-AAV cells (Vector Biolabs) along with a pAdeno-helper vector and a pRC-AAV9 rep-cap plasmid. Recombinant AAV9 production was then carried out either in-house or by the Viral Vector Core at the Salk Institute. The recombinant AAV9-CAG-NIR-FbGFP, AAV9-CAG-NIR-FbLAG16, and AAV9-CAG-NIR-FbLAG30 viruses had a titer of 2.4E + 13 GC ml−1, 2.4E + 14 GC ml−1, and 2.6E + 14 GC ml−1, respectively.
0.4 μl of diluted AAV (1:10 and 1:50 for NIR-FbGFP and NIR-FbLAGs, respectively) was injected into the somatosensory cortex (coordinates: AP −0.5–(−1.85) mm; ML 1.4–1.65 mm; DV 0.3–0.95 mm). Surgical procedures closely followed previously established protocols7,52. Briefly, thin-wall glass pipettes were pulled on a Sutter Flaming/Brown micropipette puller (model P-97). Pipette tips were cut at an acute angle under 10× magnification using a sterile technique. Tip diameters were typically 15–20 μm. Pipettes that did not result in sharp bevels nor had larger tip diameters were discarded. Millimeter tick marks were made on each pulled needle to measure the virus volume injected into the brain.
Mice were anesthetized with isoflurane (4–5% for induction; 1–1.5% for maintenance) and positioned in a computer-assisted stereotactic system with digital coordinate readout and atlas targeting (Leica Angle Two). Body temperature was maintained at 36–37 °C with a direct current (DC) temperature controller and ophthalmic ointment was used to prevent eyes from drying. A small amount of depilator cream (Nair) was used to remove hair over the dorsal areas of the injection site. The skin was cleaned and sterilized with a two-stage scrub of betadine and 70% ethanol.
A midline incision was made beginning just posterior to the eyes and ending just past the lambda suture. The scalp was pulled open and the periosteum-cleaned using a scalpel and forceps to expose the desired hemisphere for calibrating the digital atlas and coordinate marking. Once reference points (bregma and lambda) were positioned using the pipet needle and entered into the program, the desired target was set on the digital atlas. The injection pipette was carefully moved to the target site (using AP and ML coordinates). Next, the craniotomy site was marked and an electrical micro-drill with a fluted bit (0.5 mm tip diameter) was used to thin a 0.5–1 mm diameter part of the bone over the target injection site. Once the bone was thin enough to flex gently, a 30 G needle with an attached syringe was used to carefully cut and lift a small (0.3–0.4 mm) segment of bone.
For injection, a drop of AAV was carefully pipetted onto parafilm (2–3 μl) to fill the pulled injection needle with the desired volume. Once loaded with sufficient volume, the injection needle was slowly lowered into the brain until the target depth was reached. Manual pressure was applied using a 30-ml syringe connected by shrink tubing and 0.4 μl of the virus was slowly injected over 5–10 min. Once the virus was injected, the syringe’s pressure valve was locked. The position was maintained for approximately 10 min to allow the virus to spread and avoid backflow upon needle retraction. Each animal received two injections (~1 mm apart) in one hemisphere. Following the injections, head clamps were removed, muscles were approximated, and the skin was sutured along the incision. Mice were given subcutaneous Buprenorphine SR (0.5 mg per kg) and allowed to recover before placement in their cage.
Animal preparation for in vivo two-photon imaging
Surgical procedures closely followed established protocols7,52,53. Briefly, mice were anesthetized with isoflurane (4–5% for induction; 1–1.5% for maintenance) on a custom surgical bed (Thorlabs). Body temperature was maintained at 36–37 °C with a DC temperature control system and ophthalmic ointment was used to prevent eyes from drying. Depilator cream (Nair) was used to remove hair above the imaging site. The skin was thoroughly cleansed and disinfected with a two-stage scrub of betadine and 70% ethanol.
A scalp portion was surgically removed to expose frontal, parietal, and interparietal skull segments. Scalp edges were attached to the lateral sides of the skull using a tissue-compatible adhesive (3 M Vetbond). A custom-machined metal plate was affixed to the skull with dental cement (Coltene Whaledent, cat. no. H00335), allowing the head to be stabilized with a custom holder. Animals intended for awake imaging were allowed to recover for at least three days and then habituated to head fixation on a spherical treadmill (typically three sessions over three consecutive days). On the day of imaging, an approximately 3 mm diameter craniotomy was made over the AAV injection sites. A ~1.5% agarose solution and coverslip were applied to the exposed tissue. The coverslip was affixed to the skull with dental cement to control tissue motion. Mice imaged under awake conditions were allowed to recover from isoflurane anesthesia for at least 1–1.5 h before recordings commenced.
In vivo two-photon microscopy
Live animal imaging was performed 3–4.5 weeks after AAV injection, as previously described7,52,53. Briefly, a Sutter Movable Objective Microscope equipped with a pulsed femtosecond Ti:Sapphire laser (Chameleon Ultra II, Coherent) and two fluorescence detection channels were used for imaging (green emission filter: ET-525/70M (Chroma); near-infrared emission filter: ET645/75M (Chroma) supplemented with a FF01-720/SP (Semrock); dichroic beamsplitter: 565DCXR (Chroma); photomultiplier tubes: H7422-40 GaAsP (Hamamatsu)). The laser excitation wavelength was set between 880 and 920 nm. The average laser power was <10–15 mW at the tissue surface and adjusted with depth as needed to compensate for signal loss due to scattering and absorption. At the excitation intensities and durations used in this study, no signs of phototoxicity, such as a gradual increase in baseline fluorescence, lasting changes in activity rate, or blebbing of labeled cells, were apparent in our recordings. An Olympus 20× 1.0 NA water immersion objective was used for light delivery and collection. Z-stacks included up to 900 images, acquired at 1 μm axial step size, using a two- to four-frame average, 512 ×512-pixel resolution, and 1.0–2.5× zoom (corresponding to 701–282 μm fields of view). Time-lapse recordings used a 256 ×256-pixel resolution, 1.0× zoom (corresponding to a 348 μm field of view), and 8.14 Hz frame rate. Awake recordings were performed in head-fixed mice on a spherical treadmill equipped with an optical encoder (E7PD-720-118; US Digital). Analog and imaging data were acquired synchronously, allowing the animal’s running speed to be related to cellular fluorescence changes (Fig. 8 and Supplemental Movies 9, 10).
Brain tissue fixation, slicing and immunostaining
Around 3–4.5 weeks after AAV injection, mice were euthanized in their home cage following American Veterinary Medical Association (AVMA) guidelines. Transcardial perfusion was performed using 10% sucrose followed by 4% paraformaldehyde (PFA). Brain tissue was carefully extracted and incubated in 4% PFA overnight at 4 °C. The tissue was then washed on a shaker with 1× PBS three times (15 min per wash). A Leica VT1000S vibratome was used to prepare 40-μm-thick coronal tissue sections. Immunostaining was performed on floating sections using standard techniques. Primary and secondary antibodies included GFP (1:250 dilution; Thermo Fisher Scientific; RRID: AB_2534023) and Alexa Fluor 488 goat anti-chicken (1:100 dilution; Thermo Fisher Scientific; RRID: AB_2534096), respectively.
Confocal microscopy
Confocal imaging of brain sections was performed on a Zeiss LSM 710 with ZEN Black software (v2011) and an Olympus 20 × 0.8 NA air-matched objective. Two-channel, 4–6 × 4–6 tiled z-stacks (~15 images at 1 μm axial step size) were acquired using 488 nm and 633 nm laser lines. Each z-stack image had a 1024 × 1024-pixel resolution.
Image and analog data processing
Data were processed, analyzed, and plotted in Fiji (v2.0.0c; SciJava), Imaris (v9.2; Oxford Instruments), Igor Pro (v9.04; WaveMetrics), and Prism software (v8.4.3; GraphPad). xz projections were created from xy fluorescence image stacks using the ‘Reslice’ and ‘3D Project’ functions in Fiji. Motion artifacts in time-lapse recordings were reduced using the ‘Image Stabilizer’ and ‘TurboReg’ plugins. Analog data from the spherical treadmill encoder were processed using Igor Pro. Supplemental Movies 1–10 were created using Igor Pro. Immunostained tissue was quantified by generating digital representations of labeled cells with Imaris’ spot creation wizard. To quantify the overlap of NIR+ and GFP+ or GCaMP6f+ cell bodies, cells were classified as “Spots.” Spots were created on background-subtracted images. A 10 μm “XY Diameter” was used as the spot size filter. The ‘Quality’ filter was set to include the top 2% and 6% of spots for GFP+ and GCaMP6f+ cells, respectively, accounting for differences in labeling intensity and uniformity.
Statistics and reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses for all experiments, except for histology. For histology, slices with weak or uneven/irregular staining were excluded from data analysis. The experiments were not randomized. The investigators were not blinded to allocation during the experiments and outcome assessment. No sample-size estimation was performed to ensure adequate power to detect a pre-specified effect size. For flow cytometry, at least 30,000 cells per sample were recorded. For animal experiments, the sample size included 2–6 mice for each experiment. Sex as a biological variable was not considered in the research design and analyses, as the study’s primary goal was to demonstrate the approach’s technical capabilities.
Standard n > 3 independent experiments were performed for all presented nanobodies, cognate antigens, and genetically encoded sensors, unless noted in the figure legend. All attempts at replication were successful. For all experiments involving live animals, the results were replicated successfully within and across animals. For histology experiments, 2-3 biological replicates were used with 3 or 6 slices per animal quantified. Fluorescence imaging of NIR-Fb(718) (Fig. 4b), multicolor imaging of two different NIR-Fbs (Fig. 4c) and multicolor imaging of two NIR-Fbs to GFP (Fig. 5a–e) show typical data of n = 3 independent experiments. Fluorescence imaging of NIR-Fbs binding to split-sfGFP (Supplementary Fig. 14a, d, e) and fluorescence imaging of NIR-FbGFP-based systems for gene transcription activation (Supplementary Fig. 16a, b) show typical data of n = 2 independent experiments. For other experiments n is provided in the Figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the article, its Supplementary Information and Source Data file. The major plasmids constructed in this study, their maps and sequences are deposited at Addgene (#220739 - #220750). Protein structures used in this work include PDB IDs 3OGO, 7LSC, 3WLD, 6LR7, 7SAI, 7LSD. Source data are provided with this paper.
References
Weissleder, R. & Ntziachristos, V. Shedding light onto live molecular targets. Nat. Med. 9, 123–128 (2003).
Shcherbakova, D. M., Stepanenko, O. V., Turoverov, K. K. & Verkhusha, V. V. Near-Infrared Fluorescent Proteins: Multiplexing and Optogenetics across Scales. Trends Biotechnol. 36, 1230–1243 (2018).
Wang, K. et al. Direct wavefront sensing for high-resolution in vivo imaging in scattering tissue. Nat. Commun. 6, 7276 (2015).
Yang, J. et al. Focusing light inside live tissue using reversibly switchable bacterial phytochrome as a genetically encoded photochromic guide star. Sci. Adv. 5, eaay1211 (2019).
Li, L. et al. Small near-infrared photochromic protein for photoacoustic multi-contrast imaging and detection of protein interactions in vivo. Nat. Commun. 9, 2734 (2018).
Oliinyk, O. S., Shemetov, A. A., Pletnev, S., Shcherbakova, D. M. & Verkhusha, V. V. Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing. Nat. Commun. 10, 279 (2019).
Oliinyk, O. S. et al. Single-domain near-infrared protein provides a scaffold for antigen-dependent fluorescent nanobodies. Nat. Methods 19, 740–750 (2022).
Oliinyk, O. S. et al. Deep-tissue SWIR imaging using rationally designed small red-shifted near-infrared fluorescent protein. Nat. Methods 20, 70–74 (2023).
Oliinyk, O. S., Pletnev, S., Baloban, M. & Verkhusha, V. V. Development of bright red-shifted miRFP704nano using structural analysis of miRFPnano proteins. Protein Sci. Publ. Protein Soc. 32, e4709 (2023).
Snapp, E. Design and use of fluorescent fusion proteins in cell biology. Current protocols in cell biology Chapter 21, 21 24 21-21 24 13 (2005).
Traenkle, B. & Rothbauer, U. Under the Microscope: Single-Domain Antibodies for Live-Cell Imaging and Super-Resolution Microscopy. Front. Immunol. 8, 1030 (2017).
Kubala, M. H., Kovtun, O., Alexandrov, K. & Collins, B. M. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. Publ. Protein Soc. 19, 2389–2401 (2010).
Kirchhofer, A. et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–138 (2010).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Shen, Y. et al. Genetically encoded fluorescent indicators for imaging intracellular potassium ion concentration. Commun. Biol. 2, 18 (2019).
Harada, K. et al. Green fluorescent protein-based lactate and pyruvate indicators suitable for biochemical assays and live cell imaging. Sci. Rep. 10, 19562 (2020).
Keller, J. P. et al. In vivo glucose imaging in multiple model organisms with an engineered single-wavelength sensor. Cell Rep. 35, 109284 (2021).
Patriarchi, T. et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360 (2018).
Marvin, J. S. et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10, 162–170 (2013).
Kim, J. et al. Intensiometric biosensors visualize the activity of multiple small GTPases in vivo. Nat. Commun. 10, 211 (2019).
Mehta, S. et al. Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat. Cell Biol. 20, 1215–1225 (2018).
Akerboom, J. et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 6, 2 (2013).
Nakai, J., Ohkura, M. & Imoto, K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137–141 (2001).
Tian, L. et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 6, 875–881 (2009).
Akerboom, J. et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 32, 13819–13840 (2012).
Zhang, Y. et al. Fast and sensitive GCaMP calcium indicators for imaging neural populations. Nature 615, 884–891 (2023).
Dana, H. et al. Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PloS One 9, e108697 (2014).
Diez-Garcia, J. et al. Activation of cerebellar parallel fibers monitored in transgenic mice expressing a fluorescent Ca2+ indicator protein. Eur. J. Neurosci. 22, 627–635 (2005).
Ma, Y. et al. Resting-state hemodynamics are spatiotemporally coupled to synchronized and symmetric neural activity in excitatory neurons. Proc. Natl Acad. Sci. USA 113, E8463–E8471 (2016).
Grienberger, C. & Konnerth, A. Imaging calcium in neurons. Neuron 73, 862–885 (2012).
Caussinus, E., Kanca, O. & Affolter, M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19, 117–121 (2011).
Fridy, P. C. et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260 (2014).
Cai, H. et al. An improved fluorescent tag and its nanobodies for membrane protein expression, stability assay, and purification. Commun. Biol. 3, 753 (2020).
Subach, O. M., Cranfill, P. J., Davidson, M. W. & Verkhusha, V. V. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PloS One 6, e28674 (2011).
Zhang, Z., Wang, Y., Ding, Y. & Hattori, M. Structure-based engineering of anti-GFP nanobody tandems as ultra-high-affinity reagents for purification. Sci. Rep. 10, 6239 (2020).
Cong, A. T. Q., Witter, T. L. & Schellenberg, M. J. High-efficiency recombinant protein purification using mCherry and YFP nanobody affinity matrices. Protein Sci. Publ. Protein Soc. 31, e4383 (2022).
Barykina, N. V. et al. FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity. Int. J. Mol. Sci. 21 (2020).
Ding, J., Luo, A. F., Hu, L., Wang, D. & Shao, F. Structural basis of the ultrasensitive calcium indicator GCaMP6. Sci. China Life Sci. 57, 269–274 (2014).
Akerboom, J. et al. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J. Biol. Chem. 284, 6455–6464 (2009).
Arce-Molina, R. et al. A highly responsive pyruvate sensor reveals pathway-regulatory role of the mitochondrial pyruvate carrier MPC. eLife 9, e53917 (2020).
Diaz-Garcia, C. M. et al. Quantitative in vivo imaging of neuronal glucose concentrations with a genetically encoded fluorescence lifetime sensor. J. Neurosci. Res. 97, 946–960 (2019).
Roberts, T. M. et al. Identification and Characterisation of a pH-stable GFP. Sci. Rep. 6, 28166 (2016).
Tsien, R. Y. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998).
Chernov, K. G., Neuvonen, M., Brock, I., Ikonen, E. & Verkhusha, V. V. Introducing inducible fluorescent split cholesterol oxidase to mammalian cells. J. Biol. Chem. 292, 8811–8822 (2017).
Sadowski, I., Ma, J., Triezenberg, S. & Ptashne, M. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335, 563–564 (1988).
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004).
Sekhon, H. et al. Adaptable, turn-on maturation (ATOM) fluorescent biosensors for multiplexed detection in cells. Nat. Methods 20, 1920–1929 (2023).
Tang, J. C. et al. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell 154, 928–939 (2013).
Arenkiel, B. R. et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron 54, 205–218 (2007).
Madisen, L. et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015).
Longo, P. A., Kavran, J. M., Kim, M. S. & Leahy, D. J. Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529, 227–240 (2013).
Tufail, Y. et al. Phosphatidylserine Exposure Controls Viral Innate Immune Responses by Microglia. Neuron 93, 574–586.e578 (2017).
Nimmerjahn, A., Kirchhoff, F. & Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005).
Acknowledgements
We thank Sviatlana Smolskaya (Albert Einstein College of Medicine) for help with in vitro protein characterization, Fedor Subach (National Research Center “Kurchatov Institute”) for pBAD/HisB-GCaMP6s vector, Daniel Groom (Albert Einstein College of Medicine) for help with stopped-flow spectroscopy, Vladlena Kharchenko (Albert Einstein College of Medicine) for assistance with use of French press, the Flow Cytometry Core (Albert Einstein College of Medicine) and the Viral Vector Core (Salk Institute for Biological Studies) for virus production. This work was supported by grants GM122567 (V.V.V.), NS123719 (A.N.), and NS112959 (A.N.) from the US National Institutes of Health, 220011 from the Jane and Aatos Erkko Foundation (V.V.V.), the Magnus Ehrnrooth Foundation (V.V.V.), and the Sol Goldman Charitable Trust (A.N.).
Author information
Authors and Affiliations
Contributions
N.V.B. together with O.S.O. developed the NIR-Fbs, N.V.B. characterized NIR-Fbs in cells. E.C. performed the AAV injections and mouse surgeries. A.N. planned and conducted mouse surgeries and two-photon in vivo imaging. V.V.V. planned and supervised the whole project, and together with N.V.B. and A.N. designed the experiments, analyzed the data, and wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Barykina, N.V., Carey, E.M., Oliinyk, O.S. et al. Destabilized near-infrared fluorescent nanobodies enable background-free targeting of GFP-based biosensors for imaging and manipulation. Nat Commun 15, 7788 (2024). https://doi.org/10.1038/s41467-024-51857-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51857-x