Abstract
The human gut virome, which is mainly composed of bacteriophages, also includes viruses infecting archaea, yet their role remains poorly understood due to lack of isolates. Here, we characterize a temperate archaeal virus (MSTV1) infecting Methanobrevibacter smithii, the dominant methanogenic archaeon of the human gut. The MSTV1 genome is integrated in the host chromosome as a provirus which is sporadically induced, resulting in virion release. Using cryo-electron tomography, we capture several intracellular virion assembly intermediates and confirm that only a small fraction of the host population actively produces virions in vitro. Similar low frequency of induction is observed in a mouse colonization model, using mice harboring a stable consortium of 12 bacterial species (OMM12). Transcriptomic analysis suggests a regulatory lysogeny-lysis switch involving an interplay between viral proteins to maintain virus-host equilibrium, ensuring host survival and viral persistence. Thus, our study sheds light on archaeal virus-host interactions and highlights similarities with bacteriophages in establishing stable coexistence with their hosts in the gut.
Similar content being viewed by others
Introduction
The human gastrointestinal tract (GIT) harbors a diverse population of viruses which plays a key role in shaping microbial populations and maintaining GIT homeostasis1,2,3,4. The composition of the human GIT virome is remarkably stable over time, highly individual-specific, and is modulated by several factors, including age, diet, lifestyle, and drug consumption5,6,7,8,9,10,11,12. Various microscopy analyses have shown that the human GIT virome encompasses diverse viral morphotypes, with counts ranging from ∼109 to 1010 virus-like particles (VLPs) per gram of intestinal contents13,14,15. More recently, metagenomic analyses provided complementary insights into the diversity and uniqueness of the GIT virome, showing that no more than 57% of viral contigs can be taxonomically assigned or linked to putative hosts, underscoring our limited understanding of the viral component within the human GIT microbiome7,16,17,18. Among the classified contigs, nearly 97% correspond to bacteriophages (viruses that infect bacteria), indicating their dominance in this environment5,7.
It has been estimated that in most environments phages outnumber their hosts with a virus-to-microbe ratio (VMR) ranging from 13:1 to 2:119,20. However, the human GIT presents a contrasting scenario, with a much lower VMR estimated at 0.1:18,21. The low VMR, combined with the remarkable stability of the GIT virome over time and the predominance of lysogens in the microbiome of healthy adults, has prompted researchers to suggest that the adult GIT virome exhibits a “piggyback the winner” dynamic22,23,24. Under this model, the high-density bacterial populations, such as those found in the GIT, promote lysogeny over lytic phage replication. Temperate phages could take advantage of the proliferation of their hosts by replicating together and may also contribute to host survival through mechanisms such as superinfection exclusion or the introduction of new genes coding for antibiotic resistance, virulence factors, or novel metabolic functions21,22,23,24,25,26.
Although much of the research on viruses in the human GIT has focused on bacteriophages, recent metagenomics studies have shown that viruses infecting archaea represent an integral part of the human GIT virome27,28,29. An analysis of microbial genomes from the human GIT collected across 24 countries has revealed that archaea account for 1.2% of the total sequencing reads29, with methane-producing archaea, known as methanogens, constituting the dominant part of the human GIT archaeome30,31. Among them, Methanobrevibacter smithii stands out as the major component of the archaeal population in the GIT, accounting for over 90% of its composition31,32,33,34. Consistently, multiple M. smithii strains from the human GIT samples have been isolated and successfully cultivated under laboratory conditions35,36,37,38. M. smithii is a strict anaerobe that obtains energy by reducing carbon dioxide (CO2) into methane (CH4) using molecular hydrogen (H2) as electron donor37. Unlike most archaea, members of the genus Methanobrevibacter contain a cell wall which is analogous to the peptidoglycan of bacteria. Based on CRISPR spacer matching, several metagenomics studies have suggested that the human virome includes viruses capable of infecting Methanobrevibacter species27,28,29. However, no archaeal virus infecting human GIT-associated archaea has been isolated so far, precluding studies on virus-host interactions and limiting our understanding on the impact of viruses on the GIT archaea.
Methanobrevibacter smithii strain PS (ATCC 35061), isolated from a sewage digester, carries a provirus39, hereinafter referred to as Methanobrevibacter smithii tailed virus 1 (MSTV1, see below), which based on metagenomic analysis represents one of the most abundant archaeal viruses in the human GIT27. Here, we demonstrate that MSTV1 is an active virus and characterize its relationship with the M. smithii host both in vitro and in vivo. Using cryo-electron tomography, we captured several virion assembly intermediates inside the infected cells and confirmed that only a small fraction of M. smithii cells produces the virions, characterized by an isometric capsid and a long non-contractile tail. We show that MSTV1, similar to GIT phages, coexists with its host in a stable equilibrium, with average virus-to-host ratio maintained at ~0.1 both in vitro and in vivo. Leveraging the available transcriptomic data, we propose a regulatory mechanism underlying the switch between the temperate and lytic cycles which involves an interplay between viral proteins, likely acting as the repressor and activator of the lytic cycle, respectively. Collectively, our results indicate that viruses infecting hosts from the two prokaryotic domains, Bacteria and Archaea, in the human GIT have converged on similar strategies ensuring stable coexistence with their hosts.
Results
MSTV1-like viruses are globally distributed and form a family-level group
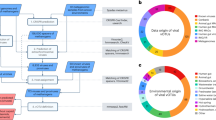
MSTV1 was identified as a provirus integrated in the genome of M. smithii strain PS39. Searches in a catalog of 1167 genomes from the human GIT archaeome database29 revealed that MSTV1 is present in 20% of all available M. smithii genomes (n = 465) and is globally distributed (Fig. 1a). Given that M. smithii is the most prevalent archaeal species in the human GIT31,32,33, MSTV1 might be one of the main human-associated archaeal viruses. Notably, MSTV1 could not be detected in any of the genomes of Candidatus Methanobrevibacter intestini (n = 108), a recently proposed Methanobrevibacter species inhabiting the GIT29, suggesting that the host range of MSTV1 is restricted to M. smithii strains. Many M. smithii strains contain CRISPR spacers targeting MSTV1 and hence are likely to be resistant to this virus (Fig. S1).
a Geographical distribution of MSTV1. The map was created using the packages “maps” and “ggplot2” of R studio v.4.3.2106. b Proteomic tree of head-tailed viruses infecting methanogenic and hyperhalophilic archaea42. Families of viruses associated with methanogenic archaea are highlighted with colored background. Branch lengths are log-scaled and the branch length for family-level demarcation is around 0.05. MSTV1 is indicated in bold within the newly proposed virus family ‘Usuviridae’. c Alignment of MSTV1-like viruses originating from human and animal GIT. ORFs are depicted by arrows that indicate the direction of transcription. Functional annotations are shown above the corresponding ORFs. Homologous genes are depicted with the same colors and are connected by shading in grayscale, with intensity reflecting the amino acid sequence identity. Cdc6: AAA+ ATPase Orc1/Cdc6; HEPN: higher eukaryotes and prokaryotes nucleotide-binding domain-containing protein; int: integrase; MCP: major capsid protein; mCP: minor capsid protein; MTase: methyltransferase; NTN-hydrolase: N‐terminal nucleophile hydrolase; Prot: serine protease; RHH: ribbon-helix-helix protein; TAC: tail assembly chaperone; TerL: terminase large subunit; TerS: terminase small subunit; TMP: tail tape measure protein; TTP: tail tube protein; wHTH: winged helix-turn-helix domain; ZF: zinc finger domain-containing protein. d Schematic representation of a programmed −1 translational frameshift in the MCP sequence of MSTV1-like viruses, resulting in the translation of an Ig-like domain. The slippery sequence is boxed. e Structural models of the dominant and the Ig-like domain-containing frameshifted MSTV1 MCPs.
To gain further insights into the diversity and distribution of MSTV1-like viruses, we explored the recently assembled database of 282 high-quality genomes of (pro)viruses associated with methanogenic archaea discovered in silico27. We identified nine complete and nearly complete MSTV1-like virus genomes sharing <95% average nucleotide identity (ANI) and thus representing different virus species (Supplementary Data 1). Four of the viruses were found in the human GIT metagenomes, and the other five were from GIT metagenomes of diverse animals, including cows, goats, gorillas and pigs. The predicted hosts of the nine MSTV1-like viruses belong to the order Methanobacteriales, the main components of the animal and human GIT archaeome29,40 (Supplementary Data 1). Notably, whereas some Methanobacteriales strains carrying MSTV1 or related proviruses lack CRISPR-Cas defense (e.g., hosts of vir406 and vir075), others possess type I-B (e.g., host of MSTV1) or III-A (e.g., host of vir362) CRISPR-Cas systems. The MSTV1-like proviruses also display different integration sites within the M. smithii strains. For instance, MSTV1 is integrated into the 3′-distal region of a gene encoding the CzcD family heavy metal cation efflux system protein39, whereas other proviruses target different intergenic regions.
Whole-proteome-based phylogenomic analysis places MSTV1-like viruses in a distinct clade, outside of the recently established families of tailed viruses associated with methanogenic archaea27 (Fig. 1b). Therefore, we propose to classify MSTV1 and related viruses into a new virus family, tentatively named ‘Usuviridae’ (after ‘usus’, intestine in Indonesian). The genomes of usuviruses can be divided into two regions, a conserved part including the structural genes and a hypervariable region including an array of small genes likely implicated in various aspects of virus-host interactions (Fig. 1c). The structural module of MSTV1-like viruses includes genes for the hallmark proteins conserved in bacterial and archaeal viruses of the class Caudoviricetes27,39,41,42, namely, the HK97-like major capsid protein (MCP), the portal protein, the large subunit of the terminase (TerL) and several tail components (Fig. 1c, Supplementary Data 2, 3). The hypervariable region of MSTV1 encodes several proteins related to defense and counterdefense. In particular, ORF2-ORF4 appear to be co-transcribed and likely encode a toxin-antitoxin system, with ORF4 encoding a putative HEPN (Higher Eukaryotes and Prokaryotes Nucleotide-binding) domain toxin. In addition, ORF5 and ORF8 encode VapB-like and MazE-like antitoxins, which could counteract the host toxin-antitoxin systems and/or mediate superinfection immunity as has been demonstrated in the case of a MazE-like antitoxin encoded by the haloarchaeal virus SNJ143. In other usuviruses, this region also contains DNA methyltransferases, which could protect the virus against the host restriction-modification systems, and many short genes of unknown function. The hypervariability of the locus including defense and counterdefense genes points to a highly dynamic arms race in the human GIT environment.
Bacteriophages in the GIT encode a repertoire of proteins containing immunoglobulin (Ig)-like domains that bind to glycans and have been shown to play a role in attaching to the mucosal layer of the GIT epithelial cells44,45,46,47. A recent study has revealed that viruses of Methanobacteriales from the GIT encode significantly more Ig-like domains than viruses of environmental Methanobacteriales, suggesting a possible role of Ig-like domains in the adaptation of viruses to the GIT conditions27. Consistently, the capsids of MSTV1-like viruses are also likely to be decorated with Ig-like domains. In the majority of usuviruses, the Ig-like domain is translated through a programmed -1 frameshifting in the MCP gene sequence, with the frameshift site located in the slippery A AAG GAA sequence (Fig. 1d, Fig. S2). As a result of frameshifting, a fraction of synthesized MCPs is predicted to contain a C-terminal Ig-like domain (Fig. 1e). Notably, one of the ten MSTV1-like viruses (vir272) apparently employs a different mechanism – the MCP is directly fused to an Ig-like domain (Fig. 1c, Fig. S2).
The “Usuviridae” clade splits into human- and animal-associated subclades (Fig. 1b, Supplementary Data 1). Viruses from the two subclades display distinct neck-tail modules, with the corresponding proteins showing low or no sequence similarity (Fig. 1c). Similarly, animal GIT-associated viruses vir406, vir075 and vir128 encode a distinct TerL variant (Fig. 1c). Phylogenetic analysis of TerL from tailed viruses infecting environmental and host-associated methanogens revealed that TerL proteins of the three viruses fall into a different clade than the other MSTV1-like viruses (Fig. S3), indicating non-orthologous gene replacement and recombination between different groups of viruses of methanogens, likely facilitated by shared environment and host range. Finally, the human and animal GIT-associated viruses differ in their endolysins implicated in digestion of the archaeal cell wall during virion egress at the end of the infection cycle27,48,49. Whereas MSTV1 and other human-associated usuviruses encode PeiW-like endolysins (Peptidase_C71 family; Fig. 1c, Supplementary Data 2, 3), usuviruses recovered from the animal GIT encode PeiR-like endolysins (Peptidase_C39 family; Fig. 1c, Supplementary Data 3)50. These differences between human and animal GIT-associated usuviruses likely reflect distinct adaptations to the respective hosts and ecological contexts. Accordingly, we propose classifying the human- and animal-associated usuviruses into distinct genera, ‘Manusuvirus’ and ‘Hewusuvirus’, respectively (Fig. 1b).
MSTV1 is an active virus which exists in a stable equilibrium with its host
To gain insights into the activity and impact of MSTV1-like viruses in the human GIT, we focused on MSTV1 and its host M. smithii PS. We first verified whether MSTV1 is an active virus by performing polymerase chain reaction (PCR) analyses with primers targeting the excised form of the provirus. The results confirmed that the excised form of the MSTV1 genome is actively replicated in the host and is released into the medium (Fig. 2a). Analysis of the cell-free culture supernatants by transmission electron microscopy (TEM) confirmed that MSTV1 is a viable virus with a siphovirus-like morphology, featuring an icosahedral head of ~65 nm (n = 51) in diameter and a long, flexible, non-contractile tail of ~270 nm in length (Fig. 2b). The viral genome was extracted from the purified virus-like particles (VLPs) and sequenced using the Illumina platform. Assembly of the sequencing reads yielded a circular 38,824 bp-long contig, which exhibited 100% identity to the provirus integrated in the M. smithii PS chromosome. Given that in all members of the Caudoviricetes the genomes are packaged as linear dsDNA molecules, the circular assembly is likely a result of a terminal redundancy and circular permutation due to head-full packaging mechanism51.
a Detection of MSTV1 in cell and supernatant fractions of M. smithii PS cultures. The agarose gel electrophoresis displays the PCR amplified products: Lane 1, provirus integrated within the host chromosome (amplification across the chimeric left (attL) attachment site, 180 bp); lane 2, the excised form of the MSTV1 genome (216 bp). b Transmission electron micrographs of MSTV1 virions. The top image displays an MSTV1 virion attached to the M. smithii surface. Scale bar, 200 nm. The bottom image shows a representative MSTV1 virion observed in the supernatants of M. smithii cultures (the experiment was repeated more than 5 times). Scale bar, 100 nm. c M. smithii PS growth and virus production over a period of 120 h. 16S rRNA gene copies/mL and free MSTV1 genome copies/mL from cell and supernatant fractions, respectively, were assessed by qPCR. The virions/cell ratio was calculated per each time point. Error bars represent standard deviation from three independent measurements. Data are presented as mean values. d M. smithii PS growth and virus production over a period of 21 days. Cultures were supplemented with H2 and CO2 at the time points marked with gray triangles. Error bars represent standard deviation from three independent measurements. Data are presented as mean values. Source data for Fig. 2c, d are provided as a Source Data file. e Visualization of transcriptomic data related to MSTV1 recovered from public repositories. Samples correspond to cultures of the provirus-containing strain M. smithii PS grown in presence of H2 under high and low concentrations of formate (2.8 and 44.1 mM)61. The read counts were aligned to MSTV1 genes, normalized and visualized. f Schematic diagram of the hypothetical transition from MSTV1 lysogenic to lytic cycle. In the left panel, the provirus is integrated into the host chromosome in a linear form. In this state, we hypothesize that the wHTH DNA-binding protein encoded by MSTV1 ORF6 represses the expression of the integrase (ORF1), thereby maintaining the virus in a lysogenic state with no production of virions. In an induced state (right panel), our hypothesis suggests that an unknown signal triggers the expression of Orc1/Cdc6, encoded by MSTV1 ORF7, which, directly or indirectly, antagonizes the action of the repressor. Consequently, the integrase gene is expressed, initiating the excision of the virus genome, and leading to the production of MSTV1 viral particles. The illustration was created with Corel Draw Graphics Suite 2021.
The detection of MSTV1 virions in the supernatants of M. smithii cultures suggests that the virus is actively induced. Various approaches to establish a plaque assay were attempted, but none were successful. Therefore, the number of viral genome copies released from the M. smithii cultures was monitored over time through quantitative PCR (qPCR). The results confirmed that MSTV1 virions are constantly produced and released into the medium without addition of exogenous inducers, resembling the spontaneous release reported for phages infecting GIT bacteria22,52,53,54, although induction under unknown conditions specific to the virus-host system cannot be ruled out. After 24 h of incubation, corresponding to the onset of the exponential phase, the concentration of MSTV1 and M. smithii cells reached 7.7 × 106 genome copies/mL and 9.9 × 107 16S rRNA gene copies/mL, respectively (corresponding to a virus-to-cell ratio of ~0.1) (Fig. 2c). Approximately 72 h later, when cells reached the stationary phase, the virus-to-cell ratio remained ~0.1, indicating that MSTV1 is released at a constant rate regardless of the growth phase of the host (Fig. 2c). As the population progressed into the death phase, a decline in cell number was observed, while the virus count remained stable, suggesting that cell death is likely a result of nutrient and energy source depletion rather than caused by virus-mediated lysis. Consistently, the provirus-free M. smithii DSM 2375 strain also entered the death phase after prolonged incubation (Fig. S4), indicating that cell death is not associated with virus infection.
Stable relationships have been described for phage-bacteria systems in the mammalian GIT8,9,11,22,55. For instance, CrAss-like phages, the most abundant phage family in the GIT, are reported to stably persist in the human GIT viromes for up to 4 years5,56,57. Likewise, the isolated ΦcrAss001 and ΦcrAss002 phages persist in cultures in vitro for extended periods without affecting the growth of their Bacteroidetes hosts58,59,60. To investigate whether MSTV1 establishes a long-term relationship with its host, we monitored the production of the virus in M. smithii cultures for 21 days. M. smithii cultures were regularly supplied with H2 and CO2 to maintain the cells metabolically active (Fig. 2d). Our results show that the populations of both extracellular viruses and cells remained stable throughout the experiment, with counts of around 107 genome copies/mL and 108 16S rRNA gene copies/mL, respectively (Fig. 2d).
The relatively constant virus-to-host ratio observed in our experiments suggests that MSTV1 exists in a stable equilibrium with its host, wherein the virus is sporadically produced without dramatically impacting the growth of the host population. As in the case of bacteriophages, this strategy could ensure the survival of most of the host population, while also promoting maintenance of the provirus in an active state.
Transcriptomics provides clues on the lysogeny-lysis switch
To gain insights into the molecular mechanism underlying the repression of the lytic cycle and the long-term coexistence of MSTV1 with its host, we leveraged the 14 transcriptomes available for M. smithii PS strain61. The number of reads aligning to the MSTV1 genes from each of the transcriptomes was normalized according to the total number of reads mapping to the provirus region (Fig. 2e, Supplementary Data 4). The genes encoding viral structural and lysis-related proteins exhibit low expression level with average transcript abundances per gene being lower than 0.5 reads per kilobase per million (RPKM) (Fig. 2e, Supplementary Data 4). However, four loci (L1-L4) in the MSTV1 genome were consistently expressed in most of the samples.
Loci L3 and L4 correspond to ORF26 and ORF35, which encode a zinc finger protein and a protein without recognizable functional domains, respectively. Whereas ORF26 is moderately expressed (RPKM > 5), ORF35 is one of the most highly expressed genes (average transcript abundance of 10.5 RPKM). The functions of these two genes are hard to predict with confidence, but their consistent expression suggests their involvement in silencing of the host defense systems or in superinfection exclusion.
Locus 1 includes ORFs 2-4 which encode a putative toxin-antitoxin system and a stand-alone VapB-like antitoxin (ORF5). ORF2 and ORF4 were highly expressed (average RPM > 8), whereas the expression of ORF5 was considerably lower (2.7 RPM). We hypothesize that the toxin-antitoxin system centered around the HEPN ribonuclease toxin functions as an addiction module and ensures stable maintenance of the provirus, so that loss of the provirus would result in post-segregational killing of the provirus-free cells. Consistently, when the exponentially growing M. smithii PS culture was plated on solid medium and 73 isolated colonies were analyzed by PCR for the presence of the provirus, we found that the provirus was not lost from any of the isolated colonies, further supporting stable association between MSTV1 and its host.
Finally, locus 2 includes ORF6 (the most highly expressed ORF, with an average RPM of 11.8) and ORF7 (average RPM of 4.7), which encode a winged helix-turn-helix (wHTH) DNA binding protein and a homolog of the Orc1/Cdc6 AAA+ ATPase, respectively (Fig. 2e). This gene cassette resembles the regulatory circuit controlling the switch between the temperate and replicative states of the haloarchaeal pleomorphic SNJ2 virus62. In this system, the temperate life cycle is maintained by the SNJ2 encoded wHTH DNA binding protein Orf4, which represses the expression of the viral integrase gene. Upon mitomycin C-induced DNA damage, the viral Orc1/Cdc6 protein is activated, triggering the expression of a third protein, Orf7, which in turn relieves the repression of the integrase gene by Orf4, leading to viral genome excision and active replication62. Similarly, the expression of the putative MSTV1 wHTH DNA binding protein (ORF6) and Orc1/Cdc6 (ORF7) appears to be antagonistic: when ORF6 is highly expressed, the expression of ORF7 is low, and vice versa (Fig. 2e, Supplementary Data 4). We hypothesize that ORFs 6 and 7 function as the repressor and activator of the MSTV1 induction, respectively. In 13 out of 14 analyzed transcriptomes, ORF6 and ORF7 exhibit medium-to-high and medium-to-low expressions, respectively, consistent with the repression of the lytic cycle in the majority of the cells in the population (Fig. 2e, f). Notably, the pattern of transcription is different in the transcriptome SRR073618, where ORF6 is not expressed (0 RPKM), while ORF7 is highly expressed (10.9 RPM) (Fig. 2e, f). Consistent with our hypothesis, high expression of ORF7 coincides with higher transcription levels of the viral integrase (4.9 RPKM), strongly suggesting that the viral Orc1/Cdc6 is an activator of the integrase transcription and, therefore, the lytic lifecycle. It remains unclear whether MSTV1 ORF7-encoded Orc1/Cdc6 directly removes the ORF6-encoded repressor, or if a third unidentified protein plays a role in activating the lytic cycle, as described for SNJ2. Furthermore, the signal that induces the switch between the temperate and lytic cycles is also unknown. Notably, other MSTV1-like viruses do not encode homologs of ORFs 6 and 7, suggesting the existence of different regulatory mechanisms in closely related viruses. However, we identified homologs of MSTV1 Orc1/Cdc6 in the genomes of two unclassified viruses recovered from human metagenomes (Fig. S5)63. This finding suggests that the proposed mechanism for the regulation of the lysogeny-lysis switch is not unique to MSTV1 and M. smithii PS but is also used by other viruses in the intestinal environment.
Natural compounds in the GIT stimulate virus release
To elucidate the factors that promote the lytic cycle of MSTV1, the effects of 34 chemical, physical and biological agents were tested (Supplementary Data 5). The released viral genome copies were assessed at 0-, 4-, 8-, and 16- hours post-induction (hpi). We first considered a range of chemical and physical inducers. The chemical inducers included different concentrations of mitomycin C and hydrogen peroxide (H2O2) as well as known inhibitors of methanogenesis, such as 2-bromoethanesulfonate (BES) and lauric acid64,65,66. As physical stressors we considered variations in growth temperature, H2 deprivation, nutrient limitation, and oxygen exposure in liquid cultures. In contrast to SNJ2 and many bacteriophages62,67, the DNA-damaging agent mitomycin C showed no significant effect on MSTV1 production (Fig. 3, Supplementary Data 5), whereas reduced temperature (30 °C) had modest but significant impact on virus production, with ~1-fold increase at 16 hpi when compared to the non-induced cultures.
Fold change was determined by quantifying the number of free virus genome copies at 4, 8 and 16 h post induction (hpi) relative to the number of free virus genome copies at time zero. Error bars denote the standard deviation from three independent measurements. Data are presented as mean values. Stars indicate the significance levels based on the two-tailed t-test: *p < 0.05 and **p < 0.01. The significant p-values are the following: incubation at 30 °C at 16 hpi: 0.0450; colon content of germ-free mice (GFM) at 4 and 8 hpi: 0.0484 and 0.0470, respectively; mixture of bile acids 0.2% at 4, 8 and 16 hpi: 0.0229, 0.0256 and 0.0255, respectively. Source data are provided as a Source Data file.
We next considered whether metabolites, toxins, and/or other molecules secreted by bacteria and archaea inhabiting the human GIT could trigger provirus induction. For this, supernatants from species belonging to the families Bacteroidaceae, Lachnospiraceae, Veillonellaceae, Enterobacteriaceae, Lactobacillaceae, and Methanobacteriaceae were added to M. smithii PS cultures. In addition, to mimic the environment of the mammalian GIT, we tested the effects of filtered extracts of colon and caecum contents from germ-free mice (GFM) and specific-pathogen free (SPF) mice (Supplementary Data 5). Although the supernatants from the human GIT microorganisms did not show any significant effect on MSTV1 induction, the colon extract from GFM exhibited a 1-fold increase at 4 and 8 hpi. The observed increase in the number of viral genome copies in the supernatants after only 4 hpi suggests that the MSTV1 replication cycle lasts less than 4 h, similar to what has been described for tailed phages in the GIT, but considerably shorter than for most characterized tailed archaeal viruses59,68,69,70,71.
To further understand the role of compounds naturally occurring in the mammalian GIT in virus induction, the two primary bile acids synthesized in the human liver, cholic acid (CA) and chenodeoxycholic acid (CDCA), along with the two predominant secondary bile acids, deoxycholic acid (DCA) and lithocholic acid (LCA), were used as potential inducers72. A commercial mixture of bile acids was also tested. While no evident impact on virus production was observed when individual bile acids were assessed, a ~ 2-fold increase throughout the experiment was observed when a mixture of bile acids (0.2% wt/vol) was used (Fig. 3, Supplementary Data 5).
The significant induction of MSTV1 release observed when M. smithii cultures were treated with either the GFM colon extracts or the mixture of bile acids suggests that the virus has evolved the ability to sense GIT-specific changes in the environment, although the exact mechanism remains to be elucidated.
In vivo experiments recapitulate the virus-host dynamics observed in vitro
To assess how the above in vitro results relate to the conditions in vivo, the production of MSTV1 was monitored in a murine model. To this end, the provirus-containing M. smithii PS strain was introduced into the isobiotic Oligo-Mouse-Microbiota (OMM12) mice, which harbor a stable consortium of 12 bacterial species representing the five most prevalent and abundant phyla in the murine GIT and containing 11 active prophages73,74. Mice were orally gavaged with M. smithii on days 0 and 10, receiving doses of 108 cells and 109 cells, respectively, and GIT colonization with M. smithii and MSTV1 production were monitored in stool samples using qPCR (Fig. 4a).
a Schematic diagram of the experimental design. Four mice were inoculated by oral gavage with M. smithii PS cells at days 0 (108 cells) and 10 (109 cells). Fresh stool samples were collected at the indicated time points. b qPCR quantification of the number of 16S rRNA gene copies of M. smithii in the stool samples of each OMM12 mouse (n = 4). c qPCR quantification of MSTV1 genome copies in the stool samples of each OMM12 mouse (n = 4). Yellow dots in (b, c) correspond to individual values and vertical bars represent the average per time point. The arrows in (b, c) indicates the second gavage performed at day 10. Source data for Fig. 4b, c are provided as a Source Data file.
After the initial gavage, M. smithii was consistently detected in mice fecal samples, with the highest average count of 2.5 × 107 16S rRNA gene copies/g stool observed 6 h post-gavage (Fig. 4b). Over the following days, the count of M. smithii declined, stabilizing on day 4 at an average count of 3.5 × 103 gene copies/g stool (Fig. 4b). A second gavage with a higher cell dose administered on day 10 (Fig. 4a) resulted in similar colonization dynamics, with rapid decrease in the M. smithii titer, followed by stabilization on days 14 and 17, with 3.1 × 103 and of 4.9 × 102 16S gene copies/g stool, respectively (Fig. 4b). Previous attempts to colonize the murine GIT with M. smithii resulted in higher M. smithii titers (up to 107–108 cells/g of fecal or caecum/colon content)75,76. However, in both reported cases, the mice lacked the natural microbiota (either germ free mice were used, or mice were pretreated with antibiotics). Thus, the observed decrease in M. smithii counts might be attributed to competitive interactions between the stable bacterial consortium of the OMM12 murine model and M. smithii. We note that for the aim of our experiment, the suppression of the OMM12 bacterial consortium was not desirable because antibiotics could potentially hamper normal induction dynamics of MSTV1 and affect the propagation of the host within a healthy mammalian GIT. Regardless, the achieved level of colonization was adequate for our experiments.
The copy number of the excised form of the MSTV1 genome detected in the mice fecal samples closely followed the pattern observed for the host (Fig. 4c). Six hours after the first gavage, an average of 9.0 × 105 MSTV1 genome copies/g stool were detected in the fecal samples. As in the case of M. smithii, the MSTV1 titer stabilized on day 4 of the experiment at 1.3 × 102 genome copies/g stool. Following the second gavage, viral counts on days 14 and 17 reached similar levels to those detected 4 days after the initial gavage, with counts of 1.8 × 102 and 1.7 × 101 genome copies/g stool, respectively. No notable peaks in viral genome copies, which would signify virus induction, were observed during the analyzed time points, suggesting that, akin to the in vitro conditions, MSTV1 and its host exist in a stable relationship, which is not significantly perturbed by the conditions of the murine GIT.
Cryo-ET provides the first experimental insights into in situ assembly of tailed archaeal viruses
We hypothesized that the low virus-to-host ratio consistently observed in vitro and in vivo results from the lysis of only a small fraction of the M. smithii PS population. To test this hypothesis, we analyzed exponentially growing cells using cryo-electron tomography (cryo-ET), which allows observing subcellular structures including virus particles directly inside intact cells, preserved in a close to native state. Cryo-tomography tilt series of 53 M. smithii cells were collected on a Titan Krios cryo-electron microscope and reconstructed in 3D to generate the final tomographic volumes (Fig. 5a, b). The imaged cells exhibited the characteristic ovococcus morphology of M. smithii, with some cells apparently undergoing cell division (Fig. S6). Among the 53 visualized cells, only one cell showed evidence of the MSTV1 virion assembly (Fig. S6), confirming our initial hypothesis.
a Cryo-ET raw slices of a M. smithii PS cell containing MSTV1 virions. Red triangles show full capsids, blue triangles represent empty capsids and pink triangles depict arrays of virion tails. Scale bar, 200 nm. Out of 53 visualized M. smithii cells, only one showed evidence of MSTV1 virion assembly. b A segmented and surface-rendering display of the reconstructed tomogram shown in panel 5a. The reconstruction displays the following cellular and viral components: membrane (light green), cell wall (dark green), ribosomes (yellow), empty capsids (blue), full capsids (red), and viral tails (purple). Scale bar, 200 nm. c Schematic representation of the head-tailed virus assembly, as known from bacteriophages. The process initiates with the assembly of an empty procapsid through the binding of multiple copies of the capsid/scaffold complex to the portal protein. Subsequently, the scaffolding protein undergoes proteolytic cleavage by the viral maturation/prohead protease, leading to the expansion of the procapsid. Concomitantly with or prior to the assembly of the expanded procapsid, DNA is translocated into the capsid with the assistance of the packaging complex, resulting in the formation of a mature capsid. The tail is independently assembled and then attached to the mature capsid, thereby producing a mature virion. Schematic modified from ref. 41 using Corel Draw Graphics Suite 2021. d Closer view of the various stages of capsid assembly visualized in the tomogram shown in panel 5b; I, empty procapsid; II, expanded procapsid, III, mature capsid, and IV, empty extracellular virion. e Surface area of the capsids at the four stages identified in 5d. n corresponds to the number of capsids counted at the different stages. The box plot displays the median (middle line), the 25th and 75th percentiles (box), minimum and maximum values (whiskers), and individual data points (dots). Source data are provided as a Source Data file.
Assembly of tailed bacteriophages has been extensively studied for decades77, and proceeds through several sequential maturation stages, whereby capsids and tails are assembled separately and attached to each other following genome packaging (Fig. 5c). The assembly of the MCP is promoted by the scaffolding protein, yielding empty capsids with a rounded appearance (stage I). Subsequently, the scaffolding protein is proteolyzed by the phage-encoded protease and the capsids assume more stable, angular appearance (stage II), which is followed by genome packaging powered by the terminase complex, yielding DNA-containing capsids (stage III). The attachment of the separately preassembled tails completes the assembly of mature virions (stage IV)77. However, no tailed archaeal virus has been studied in this respect, and our current understanding of virion assembly hinges nearly exclusively on comparative genomics41.
Reconstruction and analysis of the tomogram of M. smithii actively producing virions provided valuable insights into the in situ assembly of the MSTV1 virions. We detected multiple viral capsids (n = 33) and assembled tails within the cell. The observed capsids could be categorized into four distinct maturation stages (Fig. 5d, e), resembling those described for tailed bacteriophages (Fig. 5c). Empty capsids displayed two distinct shapes, smaller rounded ones (2/33, 6%) and larger angular ones (3/33; 9%), indicative of two different stages (I and II) of capsid maturation, respectively (Fig. 5d). Comparison of the calculated surface areas confirms that empty procapsids are significantly smaller than the expanded empty capsids (Fig. 5e). It remains unclear whether capsid expansion and DNA packaging occur sequentially or simultaneously78. However, the identification of empty expanded capsids suggests that, in the case of MSTV1, capsid expansion and gain of angular appearance precede DNA packaging. A similar assembly model has been suggested for Syn5, a tailed bacteriophage infecting cyanobacteria79. Most of the observed capsids (n = 27, 82%) exhibited angular appearance and contained a clearly visible internal density attributable to viral DNA, but the tails were not yet attached (Fig. 5a). The prevalence of DNA-containing capsids suggests an advanced stage (III) in the virion assembly process. The tomographic reconstruction also allowed identification of a DNA-free extracellular MSTV1 virion on the M. smithii cell surface, likely representing a mature capsid (stage IV) after injecting its DNA into the host (Fig. 5d, e). Stacks of partially assembled tails were observed in proximity to the capsids (Fig. 5b). Unfortunately, due to limitations in resolution, it was not possible to determine their length or whether they are connected to the DNA-containing capsids.
Collectively, these data confirm the similarity between the virion assembly mechanisms used by tailed bacterial and archaeal viruses, as previously predicted41. Moreover, the presence of assembling MSTV1 virions in only one of the imaged cells supports the hypothesis that, at any given time, only a small fraction of the M. smithii population is undergoing the lytic cycle and produces the virus.
Discussion
Compared to other archaea, viruses of methanogenic species have not been extensively studied, and even less information is available on viruses infecting archaea in the GIT. Here, we characterized MSTV1, a virus infecting Methanobrevibacter smithii, the dominant archaeon in the human GIT. Our results show that MSTV1 and related viruses are globally distributed, being present in 20% of available M. smithii genomes, and constitute the newly proposed virus family ‘Usuviridae’. MSTV1 is induced under both in vitro and in vivo conditions, without significantly impacting the host population growth dynamics. Such mode of propagation mirrors the known behavior of bacteriophages in the GIT. For instance, a recent analysis of deep-sequencing data from a healthy individual who was sampled over 2.4 years revealed that active prophages were spontaneously induced and constantly present at low levels in the GIT microbiota as extracellular phage particles22. Further studies have shown that spontaneous prophage induction is common in murine and human GIT bacteria54,80. It is increasingly recognized that bacteriophages in the GIT commonly depart from the traditional ‘kill-the-winner’ model, whereby the virus kills the dominant species in the population, and instead favor the ‘piggy-back-the-winner’ dynamics22,81,82,83. Under this model, high-density microbial populations, such as those found in the GIT, promote lysogeny over lytic phage replication. Our results suggest that viruses infecting GIT bacteria and archaea have converged on a similar strategy of ‘keeping a low profile’, by maintaining a stable equilibrium under which virus replication does not exert prohibitive burden on host fitness.
In this context, the tight control of the switch between lysogeny and lysis is therefore of prime importance for both the virus and the cell. Hence, viruses have evolved to sense diverse environmental and cellular cues to ensure optimal life choices under given conditions. We tested a broad panel of conditions to induce the lytic replication of MSTV1; however, most of these conditions had no significant impact on virus production. In this respect, MSTV1 again appears to largely mirror GIT bacteriophages. Although production of some GIT prophages appears to be stimulated by certain signals84, a recent large-scale study showed that the induction of 125 active prophages originating from human GIT bacteria using well-known induction agents resulted only in marginal increase in virus replication compared to standard growth conditions54. Our results suggest that spontaneous prophage induction may be common not only in bacteria but also characteristic of archaea in the GIT. Despite not being completely understood, spontaneous induction is believed to be caused by different factors such as stochastic gene expression, sporadic DNA damage, or stalled replication forks85. For example, it has been shown that excision of some mobile genetic elements, such as pathogenicity islands and integrative and conjugative elements, is driven by stochastic noise that modulates their activation frequency85,86. Although we cannot rule out the existence of an unknown specific signal triggering MSTV1 induction, it is also possible that stochastic gene expression of transcription regulators involved in the life cycle switch leads to activation of the lytic cycle. Analysis of multiple M. smithii PS transcriptomes allowed to pinpoint the likely molecular players responsible for stable maintenance of the MSTV1 provirus through a toxin-antitoxin addiction module, as well as regulation of the lysogeny-lysis switch through a pair of virus-encoded wHTH and Orc1/Cdc6 proteins.
The absence of a genetic system for Methanobrevibacter species currently prevents experimental verification of the postulated model. In situ cryo-ET allowed to partly overcome these limitations and obtain important insights into the assembly of the MSTV1 virions. Our data strongly suggests that virion assembly of tailed archaeal viruses follows the same blueprint described for tailed bacteriophages, with capsid undergoing several stages of maturation, followed by genome packaging and tail attachment. Finally, cryo-ET provided explanation for the low virus-to-cell ratio of 0.1, as only a small fraction of the population (1 cell out of 53 in our experiment) actively replicates and releases the virus progeny. Even if the 33 observed capsids within one cell yielded infectious virions, upon their release the virus-to-cell ratio would still be below unity. More research is needed to further unravel the intricate circuit regulating the lysogeny-lysis decision of MSTV1 and other archaeal and bacterial viruses in the GIT environment.
Methods
Identification of MSTV1-like homologs
MSTV1-like related viruses were recovered from a recent published database containing 282 high-quality (pro)viral sequences associated with methanogenic archaea27 using vConTACT gene-sharing network87. Using an average nucleotide identity (ANI) of 95% as a threshold, nine complete or nearly complete viral operational taxonomic units (vOTUs) were identified (Supplementary Data 1). ORFs were predicted Prokka v.1.14.588. Searches for distant homologs were performed using HHpred against PFAM (Database of Protein Families), PDB (Protein Data Bank) and CDD (Conserved Domains Database) databases89. Genomes of the MSTV1-like viruses were compared using Clinker v.0.0.2390. The whole-proteome-based phylogenomic analysis of head-tailed viruses related to Methanobacteriales, including MSTV1-like viruses was generated by the ViPTree web server 3.091. To determine the distribution of MSTV1 in M. smithii sequences, the virus sequence was searched in the archaeome database29. Spacer sequences were retrieved from27. Spacers were mapped on the MSTV1 genome using blastn (word size 8, e-value 0,001, identity >0.9).
Terminase phylogeny
The maximum-likelihood phylogenetic tree of the terminase large-subunit of viruses related to Methanobacteriales, including MSTV1-like viruses, was inferred using IQTree (best model Q.pfam+F + I + R4)92 based on alignment generated by mafft93. The tree was visualized using iTOL94.
Identification of Ig-like domains and structural models
The programmed -1 frameshift in the MSTV1 MCP sequence was identified in UGENE95. The structural model of the MCP of MSTV1 was downloaded from the AlphaFold database96. The Ig-like domain was modeled using AlphaFold296 through ColabFold v1.5.597 “alphafold2_multimer_v3” model with six recycles. The AlphaFold model of the MSTV1 MCP without the Ig-domain was used as a template for modeling. The models were visualized using ChimeraX v1.7.198.
M. smithii growth conditions
Methanobrevibacter smithii PS (ATCC 35061/DSM 861) cultures were grown at 37 °C in serum bottles and Hungate tubes under strict anaerobic conditions in modified DSM 119 Methanobacterium medium that contained 0.5 g/L KH2PO4, 0.4 g/L MgSO4 x 7H2O, 0.4 g/L NaCl, 0.4 g/L NH4Cl, 0.05 g/L CaCl2 x 2H2O, 2 mg/L FeSO4 x 7H2O, 1 mL trace element solution SL-10 (from DSM 320 medium), 1 g/L yeast extract, 1 g/L Na-acetate, 2 g/L Na-formate, 0.5 g/L tryptone, 0.5 mL/L Na-resazurin solution 0.1% w/v, 4 g/L NaHCO3, 0.5 g/L L-Cysteine-HCl, 0.5 g/L Na2S x 9H2O, and 10 mL vitamin solution (from DSM 141 medium). The medium was prepared as described previously99. The pH was adjusted to 7 with HCl and transferred into serum bottles and Hungate tubes. The gas phase utilized for the growth of the strain comprised 80% H2 and 20% CO2 at 2.0 bar, with shaking at 140 rpm. Cultures were grown for approximately 7–14 days and periodically gassed with H2 and CO2 maintaining the ratio 80:20.
Detection of MSTV1 by PCR
Polymerase chain reactions (PCRs) with primers targeting the integrated (F: 5′- TTGATGATGTTAATAATGGTGATGA-3′, R: 5′-AGGATTTCTTCATTGGTTCTCA-3′; expected size: 180 bp) and excised (F: 5′-GGGTTTAATTTTGGGGGATA-3′, R: 5′- AGGATTTCTTCATTGGTTCTCATA-3′; expected size: 216 bp) forms of MSTV1 were performed on washed M. smithii cells and cell-free supernatants of M. smithii cultures. Cultures were grown as described above. After incubation, the supernatants and cells were separated by low-speed centrifugation (Eppendorf F-35-6-30 rotor, 7745 × g, 20 min, 20 °C). Supernatants were recovered and pelleted cells were resuspended in PBS buffer and washed 3 times. PCRs were performed using DreamTaq Green DNA Polymerase with the following steps: 95 °C × 3 min followed by 35 cycles of 95 °C ×30 s, 57 °C ×30 s, and 75 °C × 1 min, and a final extension step at 72 °C × 10 min.
Host and virus quantification by qPCR
M. smithii 16S rRNA gene copies and viral genome copies were estimated by quantitative (q)PCR. Primers targeting the 16S rRNA gene (Mbs-955B F 5′- GCCAGGTTGATGACTTTGCTTG-3′, Mbs-1162 R: 5′-GCGTGTTGCCCAGAGGATTC-3′) and the excised form of MSTV1 (F: 5′-GGGTTTAATTTTGGGGGATA-3′, R: 5′- AGGATTTCTTCATTGGTTCTTCTCATA-3′) were used. One µL of the sample (supernatant or pelleted cells resuspended in PBS buffer), together with 0.5 µL of each primer (10 µM), were mixed with the qPCR kit to a final volume of 20 µL (Luna Universal qPCR Master Mix, New England Biolabs). qPCR was performed in a Bio-Rad CFX96 Touch Real-Time PCR Detection System with the following steps: 95 °C × 1 min followed by 40 cycles of 95 °C × 15 s, 57 °C × 30 s, and 68 °C × 20 s. A standard curve using 10-fold serial dilutions of the pGEM®-T Vector containing the M. smithii PS 16S rRNA or the excised form of the virus were prepared per each run. For the standards preparation, PCR products were cloned into a pGEM-T vector according to the manufacturer’s instructions (Promega, Charbonnières-les-Bains, France). A melting curve for each pair of primers was performed. All quantifications were performed in triplicate.
Host growth and virus production
1 mL of an exponentially growing culture of M. smithii PS was inoculated into 5 mL of the modified 119 Methanobacterium medium and incubated at 37 °C with agitation (140 rpm). Cultures were gassed with H2 and CO2 at a ratio of 80:20 at the day 0. Aliquots were collected at defined time points. The number of viruses and cells at each time point was estimated by qPCR as described above. The virus-to-host ratio was calculated with the results obtained by qPCR. For the long-term experiment, cultures were gassed with H2 and CO2 at a ratio of 80:20 at days 0, 4, 7, 15 and 18. Experiments were conducted in three biological replicates.
Concentration of virus particles
Cultures of M. smithii PS (45–50 mL) were grown as described above for approximately 14 days. After incubation, cells were removed by low-speed centrifugation (Eppendorf F-35-6-30 rotor, 7745 × g, 20 min, 20 °C). The cell-free supernatants were filtered through a 0.45 µm filter (Merck Millipore) and virus-like particles (VLPs) were concentrated by ultracentrifugation (80,000 × g, 2 h, 15 °C, Beckman 45 Ti rotor). After the run, the supernatant was removed and the pellet was resuspended in 250 μL of sodium-magnesium (SM) buffer (200 mM NaCl2, 10 mM MgSO4,50 mM Tris-HCl, pH 7.5).
Transmission electron microscopy (TEM)
5 µL of the concentrated viral particles were applied to carbon‐coated copper grids and negatively stained using 2% uranyl acetate (wt/vol). Samples were imaged with the transmission electron microscope FEI Spirit Tecnai Biotwin operate at 120 kV. The dimensions of the negatively stained capsids and tails of the virus particles were determined using ImageJ100.
Viral DNA extraction, sequencing and bioinformatic analyses
Before extraction, the concentrated virus preparation was treated with DNase I (Roche) to remove cellular DNA. Subsequently, the virus preparation was treated with SDS and Proteinase K with final concentrations of 0.5% wt/vol and 100 µg ml−1, respectively, for 30 min at 55 °C. Viral DNA was extracted from the concentrated virions with phenol/chloroform/isoamyl alcohol (25:24:1 vol/vol). Sequencing libraries were prepared and sequenced on Illumina MiSeq platform with 150-bp paired-end read lengths (Institut Pasteur, France). Raw sequence reads were processed with Trimmomatic v.0.3.6 and assembled with SPAdes v3.11.1 with default parameters. Open reading frames (ORF) were predicted by RAST v2.0 and Prokka v.1.14.588. The in silico-translated protein sequences were analyzed by BLASTP against the non-redundant protein database at the NCBI with an upper threshold E-value of 1e-03. Searches for distant homologs were performed using HHpred against PFAM, PDB and CDD databases89. The MSTV1 and vir075 genome sequences were deposited to GenBank under accession numbers PP537965 and BK068243, respectively.
Mining available transcriptomic data of the MSTV1-containing strain M. smithii PS
Transcriptomic data of M. smithii PS available in public repositories was analyzed61. A total of 14 transcriptomes of M. smithii that were grown in presence of hydrogen under low and high concentrations of formate (2.8 and 44.1 mM, respectively) were analyzed61. The total read counts aligning to MSTV1 genes from each of the transcriptomes were normalized based on the total number of reads mapping to the provirus region. The reads per kilobase per million (RPKM) were calculated using featureCounts function from R library Rsubread101 and normalized by gene length. The visualization was made in R using Gviz package102.
Induction assays
Thirty-four chemical, physical and biological conditions were employed as stressor agents to test the induction of MSTV1. The full list of conditions tested is found in Supplementary Data 5. All stressors were applied to cultures of M. smithii PS grown in Hungate tubes at an optical density (OD600) of 0.15-0.20. Non-induced cultures were used as a control. Aliquots were collected under all tested conditions at 0-, 4-, 8- and 16-hours post-induction (hpi). The amount of viral genome copies was estimated by qPCR, as described above. For all conditions and time points, the number of viral particles was normalized to the amount of viral genome copies at the time zero of the assay. The experiment was performed in triplicates.
Chemical inducers
Mitomycin C, a known phage inducer, was tested at final concentrations of 0.5, 1.5 and 10 µg/mL67. Similarly, hydrogen peroxide (H2O2) at concentrations of 2 and 10 mM was added to grown M. smithii cultures. A minimal medium was elaborated and tested by preparing the DSM 119 medium without the addition of yeast extract and tryptone. Known inhibitors of methanogenesis, such as 2-bromoethanesulfonate (BES) and lauric acid were also evaluated at final concentrations of 10 mM and 0.8 mg/mL, respectively64,65,66. The natural sweetener Stevia (SweetLeaf, USA), which has been found to induce several intestinal phages, was also tested at a concentration of 10% v/v103. The stock solutions of the agents tested were prepared dissolving the powder/liquid in water, DMSO or M. smithii medium under anaerobic conditions. Finally, the depletion of H2 and CO2 was also tested by replacing both gases in M. smithii cultures with N2.
Physical inducers
Regarding physical inducers, variation of the growth temperature, cold shock and air exposure were tested. Cultures were grown at 25, 30 and 42 °C under agitation (140 rpm). In the case of cold shock, cultures were placed on ice for 30 min. Subsequently, cultures were returned to 37 °C with agitation. Exposure to air was also tested by injecting 1 and 3 mL of air into M. smithii cultures using a syringe.
Biological inducers
The following bacteria were grown at 37 °C in anaerobic Hungate tubes until late exponential phase: Bacteroides thetaiotaomicron DSM2079 (medium M104), Anaerostipes caccae DSM 14662 (medium BHI4, 29950699), Megasphaera elsdenii DSM 20460 (medium M104), Lactobacillus rhamnosus GG ATCC 53103 (medium MRS), E. coli Nissle 1917 (medium Luria-Bertani LB) under anaerobic conditions. Archaeal strains M. smithii DSM 2375 and M. stadtmanae DSM 3091 were grown in the modified DSM 119 Methanobacterium medium for seven days at 140 rpm and under gasing with H2:CO2 at a 80:20% ratio. Medium for M. stadtmanae was further complemented with methanol at a final concentration of 0.5% v/v prior inoculation. Cultures were centrifuged in an anaerobic chamber for 10 min at 5000 × g and the supernatants were recovered, filtered at 0.2 µm and kept at 4 °C in Hungate tubes until use. Supernatants were added to M. smithii cultures at a final concentration of 5% v/v.
The following bile acids were added to M. smithii cultures at a final concentration of 0.1 mM: cholic acid, deoxycholic acid, chenodeoxycholic acid, and lithocholic acid. Additionally, a bile acid mixture (B8381, Sigma Aldrich) at final concentrations of 0.1% and 0.2% was tested. The solutions of single bile acids as well as the mixture were prepared by dissolving the respective powders in DMSO.
Colon and caecum luminal content of adults germ-free (GFM) and specific-pathogen free (SPF) C57BL/6 mice were also tested as potential inducers. Organs were extracted shortly after the death of animals and place on ice. Colon and caecum of GFM were flushed with 5 and 6 ml sterile PBS, respectively, under biosafety cabinet and place in an anaerobic chamber. Colon and caecum of SPF were placed in an anaerobic container before processing by flushing with 4 ml and 3 ml PBS respectively. Flushed materials were centrifuged for 15 min at 5000 × g to recover the supernatant. The supernatants were filter at 0.2 µm, flushed with N2 and kept at 4 °C until use.
Animals and ethics
A total of four OMM12 mice (two female and two males; seven to nine-week-old) were reared at Institut Pasteur (Paris, France). The animals were housed in the gnotobiotic facility in accordance with the Institut Pasteur guidelines and European recommendations. Food and drinking water were provided ad libitum. Protocols were approved by the committee on animal experimentation of the Institut Pasteur (Ref.#18.271) and the National Ethics Committee (APAFIS#26874-2020081309052574 v4).
The four OMM12 mice received a first oral dose via gavage of 200 µL of 108 M. smithii PS cells resuspended in sterile sodium bicarbonate solution (2.6% sodium bicarbonate in PBS buffer). M. smithii cells were harvested from seven-day cultures by low-speed centrifugation under anaerobic conditions. Fecal samples were collected 5 days and 6 h before gavage, then 6 h, 2, 3, 4 and 7 days after gavage. On day 10, the mice received a second oral gavage with 109 M. smithii cells resuspended in sterile sodium bicarbonate solution. Fecal samples were collected on days 11, 14 and 17 post-gavage. Fecal pellets were weighted, and total DNA was extracted using the QIAamp PowerFecal DNA Kit, with modifications previously described in40. Briefly, cells were lysed twice in the buffer provided by the PowerFecal Kit using a FastPrep Tissue Homogenizer (MP Biomedicals), with the “faecal sample” default setting. M. smithii cells as well as the viral particles were quantified using qPCR as described above.
Cryo-ET: sample preparation
A solution of bovine serum albumin–gold tracer (Aurion) containing 10-nm-diameter colloidal gold particles was added to a fresh culture of M. smithii in its exponential phase with a final ratio of 1:1. A small amount of the sample was applied to the front (3 µl) and the back (1 µl) of carbon-coated copper grids (Cu 200 mesh Quantifoil R2/2), previously glow discharged 2 mA and 1.5–1.8 × 10−1mbar for 1 min in a glow discharge system (ELMO, Corduan). The excess liquid was then removed by blotting with filter paper the back side of the grids for 8 s at 18°C and 98% humidity and then the sample was rapidly frozen in liquid ethane using a Leica EMGP system. The grids were stored in liquid nitrogen until image acquisition in the transmission electron microscope.
Cryo-ET: Tilt series acquisition
Tilt series were collected on a 300 kV Titan Krios G3 transmission electron microscope (Thermo Fisher Scientific) equipped with a X-FEG Tip, a Gatan K3 Direct Electron Detector and a Gatan BioQuantum LS Imaging Filter with slit width of 20 eV and a single-tilt axis holder. Tilt series were acquired with Tomography software v.5.6 (Thermo Fisher Scientific) using a dose-symmetric scheme104, with an angular range of ±60°, 2° angular increment, -8um defocus, pixel size of 3.4 Å (26000x). The total dose was set at 140 e/Ų at a dose rate of 41.6 e/pix/sec in vacuum, with C2 and Objectif Apertures of 100 um. The 2D projections were saved as separate stacks of frames and subsequently motion-corrected using the alignframes function in IMOD, in combination with home-made scripts. 3D tomographic reconstructions were calculated in IMOD by weighted back projection using the SIRT-like filter with 9 iterations105.
Cryo-ET: Segmentation and analysis of tomographic data
Tomograms were analyzed using the 3dmod interface of IMOD105. The membrane and pseudopeptidoglycan were manually modeled by tracing them every 30 slides and the subsequent use of the interpolator tool. Both closed and open contours were employed, depending on whether the full cell was displayed in the field of view. Ribosomes, capsids and tails were modeled by manual tracing in all the slices where they were present. All traces were merged through the “merge” tool. Final segmentation results were visualized as iso-surfaces. The surface area (A) of the visualized capsids was calculated using ImageJ100 by using the formula for a regular icosahedron: \({{\rm{A}}}=5\sqrt{3}{{{\rm{l}}}}^{2}\), where \({{\rm{l}}}\) corresponds to the edge length.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The genomic sequences of MSTV1 and vir075 were deposited to GenBank under the accession numbers PP537965 and BK068243, respectively. Source data are provided with this paper.
References
Cao, Z. et al. The gut virome: a new microbiome component in health and disease. EBioMedicine 81, 104113 (2022).
De Sordi, L., Lourenço, M. & Debarbieux, L. The battle within: interactions of bacteriophages and bacteria in the gastrointestinal tract. Cell Host Microbe 25, 210–218 (2019).
Popescu, M., Van Belleghem, J. D., Khosravi, A. & Bollyky, P. L. Bacteriophages and the immune system. Annu. Rev. Virol. 8, 415–435 (2021).
Draper, L. A. et al. Autochthonous faecal viral transfer (FVT) impacts the murine microbiome after antibiotic perturbation. BMC Biol. 18, 173 (2020).
Shkoporov, A. N. et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26, 527–541.e525 (2019).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Gregory, A. C. et al. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe 28, 724–740.e728 (2020).
Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010).
Garmaeva, S. et al. Stability of the human gut virome and effect of gluten-free diet. Cell Rep. 35, 109132 (2021).
Liang, G. & Bushman, F. D. The human virome: assembly, composition and host interactions. Nat. Rev. Microbiol. 19, 514–527 (2021).
Minot, S. et al. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. USA 110, 12450–12455 (2013).
Shalon, D. et al. Profiling the human intestinal environment under physiological conditions. Nature 617, 581–591 (2023).
Hoyles, L. et al. Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res. Microbiol. 165, 803–812 (2014).
Kim, M. S., Park, E. J., Roh, S. W. & Bae, J. W. Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 77, 8062–8070 (2011).
Lepage, P. et al. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 57, 424–425 (2008).
Aggarwala, V., Liang, G. & Bushman, F. D. Viral communities of the human gut: metagenomic analysis of composition and dynamics. Mob. DNA 8, 12 (2017).
Camarillo-Guerrero, L. F., Almeida, A., Rangel-Pineros, G., Finn, R. D. & Lawley, T. D. Massive expansion of human gut bacteriophage diversity. Cell 184, 1098–1109.e1099 (2021).
Nayfach, S. et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 6, 960–970 (2021).
Chibani-Chennoufi, S., Bruttin, A., Dillmann, M. L., & Brüssow, H. Phage-host interaction: an ecological perspective. J. Bacteriol. 186, 3677–3686 (2004).
López-García, P. et al. Metagenome-derived virus-microbe ratios across ecosystems. ISME J. 17, 1552–1563 (2023).
Shkoporov, A. N. & Hill, C. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25, 195–209 (2019).
Sutcliffe, S. G., Reyes, A. & Maurice, C. F. Bacteriophages playing nice: lysogenic bacteriophage replication stable in the human gut microbiota. iScience 26, 106007 (2023).
Silveira, C. B. & Rohwer, F. L. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes 2, 16010 (2016).
Silveira, C. B., Luque, A. & Rohwer, F. The landscape of lysogeny across microbial community density, diversity and energetics. Environ. Microbiol. 23, 4098–4111 (2021).
Kirsch, J. M. et al. Bacteriophage-bacteria interactions in the gut: from invertebrates to mammals. Annu. Rev. Virol. 8, 95–113 (2021).
Guerin, E. & Hill, C. Shining light on human gut bacteriophages. Front. Cell. Infect. Microbiol. 10, 481 (2020).
Medvedeva, S., Borrel, G., Krupovic, M. & Gribaldo, S. A compendium of viruses from methanogenic archaea reveals their diversity and adaptations to the gut environment. Nat. Microbiol. 8, 2170–2182 (2023).
Li, R., Wang, Y., Hu, H., Tan, Y. & Ma, Y. Metagenomic analysis reveals unexplored diversity of archaeal virome in the human gut. Nat. Commun. 13, 7978 (2022).
Chibani, C. M. et al. A catalogue of 1167 genomes from the human gut archaeome. Nat. Microbiol. 7, 48–61 (2022).
Borrel, G. et al. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. ISME J. 11, 2059–2074 (2017).
Dridi, B., Henry, M., El Khéchine, A., Raoult, D. & Drancourt, M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PloS One 4, e7063 (2009).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Mihajlovski, A., Doré, J., Levenez, F., Alric, M. & Brugère, J. F. Molecular evaluation of the human gut methanogenic archaeal microbiota reveals an age-associated increase of the diversity. Environ. Microbiol. Rep. 2, 272–280 (2010).
Dridi, B., Henry, M., Richet, H., Raoult, D. & Drancourt, M. Age-related prevalence of Methanomassiliicoccus luminyensis in the human gut microbiome. APMIS 120, 773–777 (2012).
Miller, T. L. & Wolin, M. J. Enumeration of Methanobrevibacter smithii in human feces. Arch. Microbiol. 131, 14–18 (1982).
Dermoumi, H. L. & Ansorg, R. A. Isolation and antimicrobial susceptibility testing of fecal strains of the archaeon Methanobrevibacter smithii. Chemotherapy 47, 177–183 (2001).
Miller, T. L., Wolin, M. J., Conway, de Macario, E. & Macario, A. J. Isolation of Methanobrevibacter smithii from human feces. Appl. Environ. Microbiol. 43, 227–232 (1982).
Khelaifia, S., Raoult, D. & Drancourt, M. A versatile medium for cultivating methanogenic archaea. PloS One 8, e61563 (2013).
Krupovic, M., Forterre, P. & Bamford, D. H. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J. Mol. Biol. 397, 144–160 (2010).
Thomas, C. M., Desmond-Le Quéméner, E., Gribaldo, S. & Borrel, G. Factors shaping the abundance and diversity of the gut archaeome across the animal kingdom. Nat. Commun. 13, 3358 (2022).
Baquero, D. P. et al. Structure and assembly of archaeal viruses. Adv. Virus Res. 108, 127–164 (2020).
Liu, Y. et al. Diversity, taxonomy, and evolution of archaeal viruses of the class Caudoviricetes. PLoS Biol. 19, e3001442 (2021).
Chen, B. et al. ORF4 of the temperate archaeal virus SNJ1 governs the lysis-lysogeny switch and superinfection immunity. J. Virol. 94, e00841-20 (2020).
Barr, J. J. et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. USA 110, 10771–10776 (2013).
Barr, J. J. et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc. Natl. Acad. Sci. USA 112, 13675–13680 (2015).
Chin, W. H. et al. Bacteriophages evolve enhanced persistence to a mucosal surface. Proc. Natl. Acad. Sci. USA 119, e2116197119 (2022).
Van Belleghem, J. D., Dąbrowska, K., Vaneechoutte, M., Barr, J. J. & Bollyky, P. L. Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses 11, 10 (2018).
Schofield, L. R., Beattie, A. K., Tootill, C. M., Dey, D. & Ronimus, R. S. Biochemical characterisation of phage pseudomurein endoisopeptidases PeiW and PeiP using synthetic peptides. Archaea 2015, 828693 (2015).
Visweswaran, G. R., Dijkstra, B. W. & Kok, J. Two major archaeal pseudomurein endoisopeptidases: PeiW and PeiP. Archaea 2010, 480492 (2010).
Altermann, E., Schofield, L. R., Ronimus, R. S., Beatty, A. K. & Reilly, K. Inhibition of rumen methanogens by a novel archaeal lytic enzyme displayed on tailored bionanoparticles. Front. Microbiol. 9, 2378 (2018).
Casjens, S. R. & Gilcrease, E. B. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol. Biol. 502, 91–111 (2009).
Mathieu, A. et al. Virulent coliphages in 1-year-old children fecal samples are fewer, but more infectious than temperate coliphages. Nat. Commun. 11, 378 (2020).
Mavrich, T. N. et al. Characterization and induction of prophages in human gut-associated Bifidobacterium hosts. Sci. Rep. 8, 12772 (2018).
Dahlman, S. et al. Temperate gut phages are prevalent, diverse, and predominantly inactive. bioRxiv: the preprint server for biology, 2023.2008.2017.553642, https://doi.org/10.1101/2023.08.17.553642 (2023).
Lourenço, M. et al. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28, 390–401.e395 (2020).
Smith, L., Goldobina, E., Govi, B. & Shkoporov, A. N. Bacteriophages of the order crassvirales: what do we currently know about this keystone component of the human gut virome? Biomolecules 13, 584 (2023).
Gulyaeva, A. et al. Discovery, diversity, and functional associations of crAss-like phages in human gut metagenomes from four Dutch cohorts. Cell Rep. 38, 110204 (2022).
Guerin, E. et al. Isolation and characterisation of ΦcrAss002, a crAss-like phage from the human gut that infects Bacteroides xylanisolvens. Microbiome 9, 89 (2021).
Shkoporov, A. N. et al. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 9, 4781 (2018).
Shkoporov, A. N. et al. Long-term persistence of crAss-like phage crAss001 is associated with phase variation in Bacteroides intestinalis. BMC Biol. 19, 163 (2021).
Hansen, E. E. et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl. Acad. Sci. USA 108, 4599–4606 (2011).
Chen, Z. et al. A virus-borne DNA damage signaling pathway controls the lysogeny-induction switch in a group of temperate pleolipoviruses. Nucleic Acids Res. 51, 3270–3287 (2023).
Tisza, M. J. & Buck, C. B. A catalog of tens of thousands of viruses from human metagenomes reveals hidden associations with chronic diseases. Proc. Natl. Acad. Sci. USA 118, e2023202118 (2021).
Zhou, X. et al. The transcriptome response of the ruminal methanogen Methanobrevibacter ruminantium strain M1 to the inhibitor lauric acid. BMC Res. Notes 11, 135 (2018).
Nollet, L., Demeyer, D. & Verstraete, W. Effect of 2-bromoethanesulfonic acid and Peptostreptococcus productus ATCC 35244 addition on stimulation of reductive acetogenesis in the ruminal ecosystem by selective inhibition of methanogenesis. Appl. Environ. Microbiol. 63, 194–200 (1997).
Zhou, X., Meile, L., Kreuzer, M. & Zeitz, J. O. The effect of saturated fatty acids on methanogenesis and cell viability of Methanobrevibacter ruminantium. Archaea 2013, 106916 (2013).
Levine, M. Effect of mitomycin C on interactions between temperate phages and bacteria. Virology 13, 493–499 (1961).
Dalmasso, M. et al. Three new escherichia coli phages from the human gut show promising potential for phage therapy. PloS One 11, e0156773 (2016).
Pietilä, M. K. et al. Insights into head-tailed viruses infecting extremely halophilic archaea. J. Virol. 87, 3248–3260 (2013).
Zhao, X. et al. Enrichment culture but not metagenomic sequencing identified a highly prevalent phage infecting lactiplantibacillus plantarum in human feces. Microbiol. Spectr. 11, e0434022 (2023).
Svirskaitė, J., Oksanen, H. M., Daugelavičius, R. & Bamford, D. H. Monitoring physiological changes in haloarchaeal cell during virus release. Viruses 8, 59 (2016).
Just, S. et al. The gut microbiota drives the impact of bile acids and fat source in diet on mouse metabolism. Microbiome 6, 134 (2018).
Lamy-Besnier, Q. et al. Chromosome folding and prophage activation reveal specific genomic architecture for intestinal bacteria. Microbiome 11, 111 (2023).
Brugiroux, S. et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2, 16215 (2016).
Ramezani, A. et al. Gut colonization with methanogenic archaea lowers plasma trimethylamine n-oxide concentrations in apolipoprotein e-/- mice. Sci. Rep. 8, 14752 (2018).
Samuel, B. S. & Gordon, J. I. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc. Natl. Acad. Sci. USA 103, 10011–10016 (2006).
Fokine, A. & Rossmann, M. G. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 4, e28281 (2014).
Aksyuk, A. A. & Rossmann, M. G. Bacteriophage assembly. Viruses 3, 172–203 (2011).
Dai, W. et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature 502, 707–710 (2013).
Kim, M. S. & Bae, J. W. Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J. 12, 1127–1141 (2018).
Shkoporov, A. N., Turkington, C. J. & Hill, C. Mutualistic interplay between bacteriophages and bacteria in the human gut. Nat. Rev. Microbiol. 20, 737–749 (2022).
De Sordi, L., Lourenço, M. & Debarbieux, L. "I will survive": a tale of bacteriophage-bacteria coevolution in the gut. Gut Microbes 10, 92–99 (2019).
Avellaneda-Franco, L., Dahlman, S. & Barr, J. J. The gut virome and the relevance of temperate phages in human health. Front. Cell. Infect. Microbiol. 13, 1241058 (2023).
Henrot, C. & Petit, M. A. Signals triggering prophage induction in the gut microbiota. Mol. Microbiol. 118, 494–502 (2022).
Nanda, A. M., Thormann, K. & Frunzke, J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 197, 410–419 (2015).
Miyazaki, R. et al. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet. 8, e1002818 (2012).
Bin Jang, H. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632–639 (2019).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Söding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005).
Gilchrist, C. L. M. & Chooi, Y. H. Clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 37, 2473–2475 (2021).
Nishimura, Y. et al. ViPTree: the viral proteomic tree server. Bioinformatics 33, 2379–2380 (2017).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Okonechnikov, K., Golosova, O. & Fursov, M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167 (2012).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. Publ. Protein Soc. 32, e4792 (2023).
Pende, N. et al. SepF is the FtsZ anchor in archaea, with features of an ancestral cell division system. Nat. Commun. 12, 3214 (2021).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinforma. 18, 529 (2017).
Liao, Y., Smyth, G. K. & Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019).
Hahne, F. & Ivanek, R. Visualizing genomic data using gviz and Bioconductor. Methods Mol. Biol. 1418, 335–351 (2016).
Boling, L. et al. Dietary prophage inducers and antimicrobials: toward landscaping the human gut microbiome. Gut Microbes 11, 721–734 (2020).
Hagen, W. J. H., Wan, W. & Briggs, J. A. G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197, 191–198 (2017).
Mastronarde, D. N. & Held, S. R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017).
Wickham, H. Ggplot2: Elegant Graphics for Data Analysis (Springer New York, 2009).
Acknowledgements
This work was supported by Agence Nationale de la Recherche (grant ANR-23-CE02-0022 to S.G. and M.K. and grant ANR-20-CE92-0048 to L.D.). C.M.G. was supported by an FRM Retour en France fellowship. N.P. was supported by a Pasteur-Roux Postdoctoral Fellowship from the Institut Pasteur (Paris) and the Austrian Science Fond (FWF) Elise Richter Fellowship (FWF project V 931-B). We acknowledge the cryo-ET expertise and assistance of the Institut Pasteur’s NanoImaging Core facility, created and supported by a PIA grant (EquipEx CACSICE: ANR-11-EQPX-0008). We also acknowledge E. Turc and L. Lemée from the Biomics Platform, C2RT, Institut Pasteur, Paris, France, supported by France Génomique (ANR−10-INBS-09) and IBISA. We are grateful for support for Ultrastructural BioImaging Core Facility equipment from the GIS-IBISA, the French Government Program Investissements d’Avenir France BioImaging (FBI, N° ANR−10-INSB-04-01) and the French government (Agence Nationale de la Recherche) Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (ANR-10-LABX-62-IBEID).
Author information
Authors and Affiliations
Contributions
M.K., S.G. and G.B. conceived the study; D.P.B. performed the experiments; S.M., D.P.B. and M.K. performed the bioinformatic analyses; D.P.B., M.K. and L.D. conceived the in vivo experiment; D.P.B., C.M. and T.P. performed the in vivo experiment; N.P., A.S. and S.T. prepared the samples and collected the cryo-EM data; D.P.B. reconstructed the tomogram. D.P.B., M.K., S.G. and G.B. wrote the manuscript. All authors revised and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Susanne Erdmann and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Baquero, D.P., Medvedeva, S., Martin-Gallausiaux, C. et al. Stable coexistence between an archaeal virus and the dominant methanogen of the human gut. Nat Commun 15, 7702 (2024). https://doi.org/10.1038/s41467-024-51946-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51946-x
This article is cited by
-
Methanobrevibacter smithii strain U29 whole genome sequence delineates M. smithii intermediate cell variants
BMC Research Notes (2025)
-
A systematic review on the dynamic interplay between the immune system and gut microbiome in inflammatory bowel disease patients
Future Journal of Pharmaceutical Sciences (2025)
-
Biogenesis of DNA-carrying extracellular vesicles by the dominant human gut methanogenic archaeon
Nature Communications (2025)
-
The nitrite bottleneck revisited: exploring the potential of phage-mediated microbial regulation for advancing novel nitrogen removal strategies
npj Clean Water (2025)